Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI
Introduction
Prostate cancer (PCa) is one of the most prevalent malignant neoplasms in elderly males worldwide (1). Currently, with the rapid increase in the incidence rate in China (2), PCa has become a major health concern that affects many families. According to the 2017 European Association of Urology PCa Guidelines, patients with a Gleason score (GS) <7 PCa are recommended to undergo active surveillance and watchful waiting. In contrast, patients with a GS ≥7 PCa have an increased risk of progression and a shorter overall survival and should adopt timely treatment. Thus, accurate risk assessment is critical for selecting the optimal treatments for these patients (3,4).
Total prostate-specific antigen (tPSA), free PSA (fPSA), PSA density (PSAD) and the ratio of free-to-total PSA (f/t PSA) are clinically used indicators for PCa detection and grading (5-7). To date, there is still controversy regarding which indicators are more suitable for the diagnosis and grading of PCa, and no consensus has been reached (8,9). Furthermore, the clinical utility of these indicators has certain inadequacies, such as overdiagnosis and subsequent overtreatment (5). Therefore, there is an urgent need for a new method for early and accurate PCa risk stratification to achieve a good prognosis for patients.
In recent years, multi-parameter MRI (mp-MRI) has been increasingly applied in PCa localization, qualitative assessment and staging diagnosis (10,11). The proposal of PI-RADS is to better standardize the examination and interpretation of prostate MRI. The PI-RADS version 2.1 proposes a concept of biparametric MRI (bp-MRI) (including only T2WI and DWI) to simplifies prostate MRI sequences (12). Radiomics based on MRI as a trending and promising area of research, has been widely used to evaluate tumour heterogeneity and has achieved high diagnostic efficiency. Radiomics is defined as the high-throughput extraction of large numbers of medical imaging features that are then converted to mineable high-dimensional data, quantitative analysis of these data can provide an unprecedented opportunity to improve clinical decision-making (13-15). Currently, many previous studies have focused on radiomics feature analysis to assess and classify PCa lesions (16-18). More specifically, these studies analysed cancerous pixels in quantified regions of interest by using the feature extraction function, and combined numerous radiomics features with machine learning methods have shown high accuracy on PCa differentiation and aggressiveness (19-21). However, the research methods of previous articles used different standards and the results are inconsistent. Some studies have shown that radiomics plays an important role in GS score grading of PCa (22,23); some clinical indicators have certain implications for the invasiveness of PCa, but not many researches combine radiomics features with clinical factors together to conduct risk assessment. Therefore, we constructed three models based on radiomics signatures and clinical factors separately and together to predict clinically significant PCa and compared whether the combination of the two can help improve the diagnosis performance.
Methods
Patients
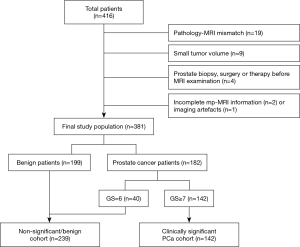
This retrospective study was approved by the Institutional Ethics Committee of our hospital, which waived the requirement for written informed consent. A total of 416 patients were included in our study between December 2014 and March 2017 in our hospital. The inclusion criteria were as follows: (I) patients with clinical symptoms indicative of PCa (frequent micturition, urgency of urination, pain while urinating or dysuria, etc.) or elevated PSA levels; (II) patients with a prostate 3.0T MRI examination before ultrasound-guided biopsy; and (III) biopsy with confirmed pathological results. The exclusion criteria were as follows: (I) pathological biopsy of lesions that were difficult to delineate on MRI (pathological results show that the lesions' location cannot be displayed on the magnetic resonance image) (n=19); (II) tumour volume that was too small (maximum diameter <5 mm) (n=9); (III) prostate biopsy, surgery, radiation therapy or endocrine therapy performed before MRI examination (n=4); or (IV) incomplete MRI data (n=2) or the presence of imaging artefacts preventing segmentation of cancerous lesions (n=1). Ultimately, the study population consisted of 381 patients including 182 PCa patients and 199 patients without any histological evidence of cancer. According to the final pathological diagnosis results, we defined clinically significant PCa as patients with a GS ≥7 PCa, whereas patients with a GS <7 (including non-clinically significant cancer and benign lesions) were classified as having no significant PCa. The details of patient selection are shown in Figure 1.
Pathology
All patients underwent transrectal ultrasound-guided (TRUS) systematic prostate biopsy. The area of systematic prostate biopsy was divided as follows: the prostate gland was divided into 3 sections (the basal region, the body and the apex) from top to bottom, and each part was divided into the left and right regions. The right and left regions of the basal and body regions were divided into the inner and outer regions, and the whole prostate gland was divided into 10 areas. Each area was punctured with 1 needle, and 2–4 additional needles were used to puncture the suspicious area.
Clinical data
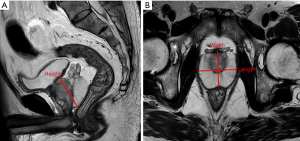
We collected clinical information including age, prostate volume (PV), serum PSA [including tPSA and free PSA (Fpsa)], f/t PSA, PSAD, and prostate biopsy pathology from the selected patients. The PV was calculated as the width × length × height ×0.52 on T2-weighted images (24), and detailed measurements are shown in Figure S1. The PSAD was calculated as total PSA/PV.
MRI acquisition and preprocessing
All patients underwent 3.0-T MRI scanner (Philips Ingenia, The Netherlands) with a 32-channel body phased array coil as the receiving coil. Scan sequences included sagittal T2-weighted imaging (T2WI), axial T2WI, T1-weighted imaging (T1WI), diffusion-weighted imaging (DWI) (b values of 0 and 1,000 sec/mm2) and dynamic contrast-enhanced MRI (DCE-MRI) The details regarding MRI acquisition are listed in Table S1. The details of the imaging sequence parameters, including sequence type, section, echo time (TR), repetition time (TE), slice thickness, slice gap, field of view (FOV) and matrix, are summarized in Table S2.

Full table
Full table
Bp-MRI sequences, including axial T2WI and ADC images, were chosen to extract radiomics features due to the availability and emphasis in PI-RADS v2.1. The obtained T2WI and ADC images (DICOM format) of each case were imported into A.K. [Artificial Intelligence Kit, V3.0.1, independently developed by GE Healthcare (China)] software. Prior to tumour segmentation, three preprocessing techniques were applied to standardized images in order to improve texture recognition. Firstly, the µ ± 3σ method was adopted for image intensity normalization and the intensity signals beyond 3σ were eliminated, which could enhance the difference between two classes; then, 64 grey levels was used for grey-level quantization. Finally, we used a voxel size of 1×1×1 mm3 for image resampling isotopically (25,26).
MRI lesion segmentation
For consistency between ROIs in both pre-processed T2WI and ADC images, all depicted ROIs were strictly delineated with the same criteria and visually validated by the same expert. The location and size of the lesions were determined as follows: (I) the detailed records of the prostate system punctures (injection site and depth) and pathological diagnostic results were used to determine the location and nature of the lesion; (II) the location described by pathology was matched to the corresponding lesion on the MRI images; and (III) combined with the PI-RADS V2 scoring diagnostic criteria (27), the range of lesions was determined. Peripheral zone (PZ) lesions were mainly based on the ADC map, supplemented by DWI and DCE-MRI sequences, and transitional zone (TZ) lesions were mainly based on T2WI, supplemented by DWI and the ADC map. The ROIs were manually delineated layer-by-layer along the lesion boundary, obtaining a three-dimensional data. There are several notable points in the process of sketching ROI: (I) the urethra, ejaculation tube, and seminal vesicle root structure were avoided; (II) to truly reflect tumour heterogeneity, ROI delineation included areas of necrosis, haemorrhage, cystic tissue and calcification; (III) for multifocal PCa, the ROI of lesions with the highest GS were selected and confirmed by biopsy pathology; if the GSs were the same, the ROI of the lesions with the maximum diameter was selected; and (IV) for patients with non-cancerous prostate regions confirmed by biopsy pathology, ROIs were also delineated using a volume of no less than 0.5 mL. An example of lesion segmentation in a PCa patient is shown in Figure 2.
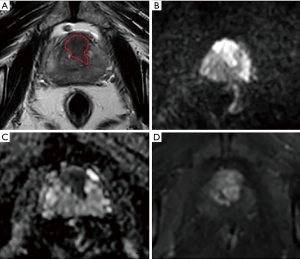
The intra-observer and inter-observer repeatability of lesion segmentation was based on the repeatability of feature extraction. To evaluate the intra- and inter-observer repeatability in feature extraction, 40 patients were randomly selected, and their radiomics features were extracted by two radiologists who were blinded to their clinical data. Radiologists A and B had 9 and 3 years of clinical experience in the diagnosis of prostate MRI, respectively, and the same 2 years of experience in feature extraction. Radiologist A repeated the feature extraction for the same 40 patients’ MR images over a 2-week period.
Radiomics feature extraction and feature selection
Feature extraction and feature selection were also performed using A.K. software. The kinds of computer-derived features included gradient-based histogram features, grey-level co-occurrence matrix (GLCM), run-length matrix (RLM), and grey-level size zone matrix (GLSZM), which were calculated based on the voxels in the delineated ROI. Although large numbers of features were extracted, not all features were helpful in predicting clinically significant PCa; therefore, we used two feature selection methods to determine the best discriminating feature set. First, mRMR was performed to eliminate the redundant and irrelevant features. Then, least absolute shrinkage and selection operator (LASSO) analysis was conducted to choose the optimized subset of features to construct the final model.
Model construction
Clinical factors and radiomic features were used to identify biomarkers of PCa. We randomly assigned 60% of the patients to the training set (n=229) and the remaining 40% to the test set (n=152). In terms of clinical factors, univariate and multivariate logistic analyses were used to establish the logistic regression model to find the associations between clinical features and significant PCa. For the radiomics model, a logistic regression model was trained using the selected features through feature selection methods to classify clinically significant PCa. A formula called the radiomics score (RAD-SCORE) was generated using a logistic regression analysis of selected features that were weighted by their coefficients. A multivariable logistic regression method was used to establish a combined clinical-radiomics model with a combined weight of radiomic features and clinical risk factors.
Statistical analysis
For clinical model establishment, univariate logistic regression was used to select clinical risk factors, then clinical features with P<0.05 were introduced into a multivariate logistic regression to build a clinical model. In logistic regression, a backward stepwise selection was applied using a likelihood ratio test with Akaike’s information criterion as the stopping rule. Meanwhile, the variance inflation factor (VIF) was used to evaluate collinearity; features with VIF >10 were excluded. Finally, a clinical model was established. The performance of each model was quantified by the area under the receiver operating characteristic (ROC) curve (AUC) with 95% confidence intervals (95% CIs). The DeLong test was used to determine whether significant differences existed in terms of the AUC values among these three models. The nomogram of the clinical-radiomics model was constructed to assist in using the abovementioned signature to improve decision making. Decision curve analysis was conducted to determine the clinical usefulness of the radiomics-based nomogram by quantifying the clinical net benefits at different threshold probabilities in the test set (28). Decision curves were also plotted for the clinical, radiomics and combined models. The abovementioned analyses were performed using R software (version 3.4.3, http://www.Rproject.org).
The entire workflow of this research is presented in Figure 3.
Results
Patient characteristics
For all 381 patients, 199 patients (52%) were negative (benign lesions), and the other 182 patients were positive (PCa). Biopsy showed a GS <7 PCa in 40 patients (11%) and a GS ≥7 PCa in 142 patients (37%). The GS distribution of all patients was as follows: 3+3=6 (40 patients); 3+4/4+3=7 (57 patients); 4+4=8 (46 patients); 4+5/5+4=9 (31 patients); and 5+5=10 (8 patients). The characteristics of all patients are listed in Table 1. The sample distribution of this study is listed in Table S3.
Full table

Full table
Clinical model
In terms of clinical factors, univariate logistic analysis showed that patient age, PV, tPSA, fPSA and PSAD were significant factors for predicting clinically significant PCa. The multivariate logistic analysis showed that age, tPSA and PSAD were significant (P<0.05), and these three clinical factors could be used as independent predictors. The results of the univariate and multivariate logistic regression analyses are shown in Table 2. Finally, the logistic regression classifier was established based on the selected clinical features. The AUC, accuracy rate, sensitivity and specificity of the training set were 0.76 (95% CI: 0.70–0.82), 0.78, 0.81, and 0.76, respectively, and the AUC, accuracy rate, sensitivity and specificity of the test set were 0.79 (95% CI: 0.70–0.88), 0.74, 0.76, and 0.74, respectively.
Full table
Radiomics model
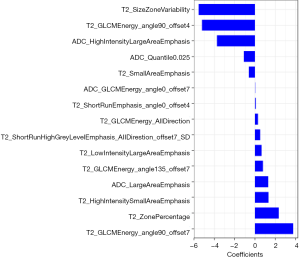
Intra- and inter-class correlation coefficients (ICCs) were used to evaluate the intra- and inter-observer agreement of feature extraction, with an ICC greater than 0.75 indicating good agreement. The intra-observer ICCs based on radiologist A’s two stages of feature extraction ranged from 0.83 to 0.96 and 0.82 to 0.97 for T2WI and ADC mapping, respectively, and the inter-observer ICCs based on the feature extraction of radiologist A (the first time) and radiologist B ranged from 0.76 to 0.93 and 0.78 to 0.90 for T2WI and ADC mapping, respectively. Radiologist A completed the process for the remaining images, and the radiomics-based models were built with the features extracted by radiologist A. Through feature selection, 396 features were reduced to 15 potential predictors with non-zero coefficients using the LASSO logistic regression classifier. The relative important features are shown in Figure S2. These features were incorporated into the RAD-SCORE, which indicated the sum of the weighted features as follows:
RAD-SCORE =-5.555×T2_SizeZoneVariability +1.33× T2_HighIntensitySmallAreaEmphasis +0.783× T2_GLCMEnergy_angle135_offset7−3.751×ADC_HighIntensityLargeAreaEmphasis +3.752×T2_GLCMEnergy_angle90_offset7+0.512×T2_ShortRunHighGreyLevelEmphasis_AllDirection_offset7_SD +0.658×T2_LowIntensityLargeAreaEmphasis +0.023×ADC_GLCMEnergy_angle0_offset7+2.333×T2_ZonePercentage +0.285×T2_GLCMEnergy_AllDirection_offset7+0.085×T2_ShortRunEmphasis_angle0_offset4-1.094×ADC_Quantile0.025-5.209×T2_GLCMEnergy_angle90_offset4-0.603×T2_SmallAreaEmphasis +1.314×ADC_LargeAreaEmphasis −3.991.
The radiomics signature showed high predictive efficiency with AUC values of 0.99 (95% CI: 0.98–1.00) in the training set and 0.98 (95% CI: 0.97–1.00) in the test set.
Clinical-radiomics combined model
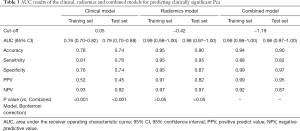
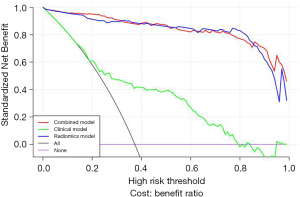
The clinical-radiomics nomogram incorporating age, tPSA, PSAD and RAD-SCORE is shown in Figure 4. The clinical-radiomics combined model also showed good predictive ability with high AUC values of 0.99 (95% CI: 0.98–1.00) in the training set and 0.98 (95% CI: 0.97–1.00) in the test set. The diagnostic efficacy of the clinical-radiomics model was the same as the radiomics model. The AUC, accuracy, sensitivity and specificity of the three models are listed in Table 3. A comparison of the ROC curves of these three models is shown in Figure 5.
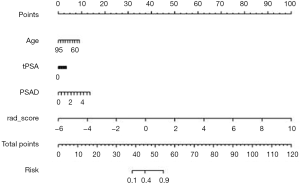
Full table
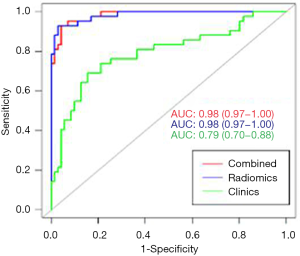
Decision curve
The decision curves for the clinical, radiomics and combined models are presented in Figure 6. The decision curves show that either the radiomics or combined model to predict clinically significant PCa adds more benefit than using the clinical model.
Discussion
This study extends the analysis of individual imaging texture features to an “omics”-based risk level estimation approach. By extracting a large number of quantitative imaging features and efficiently selecting these features, radiomics models based on logistic regression analysis were established. Then, the RAD-SCORE combined with clinical factors was used to construct a clinical-radiomics combined model. The radiomics model and the clinical-radiomics combined model showed higher performance than the clinical model in terms of clinical net benefit and AUC for predicting clinically significant PCa, both in the training set and test set. This conclusion shows that including the RAD-SCORE in the clinical model improved the diagnostic efficiency of clinical factors, which demonstrates the distinctive value of the radiomics signature for predicting clinically significant PCa.
In previous studies, researchers have attempted to identify better prostate-related clinical factors for the diagnosis of PCa and assessment of aggressiveness; however, to date, there is no clear consensus. Xiao et al. (29) combined PSA, digital rectal examination (DRE) texture, DRE nodules and B-ultrasound results in a nomogram and created a classifier to predict PCa patients with a GS ≥7; their model had an AUC of 0.831. Niu et al. (30) indicated that age, Prostate Imaging-Reporting and Data System (PI-RADS) version 2 score, and adjusted PSAD were independent predictors of high-grade PCa (HGPCa), with an AUC of 0.83. Fang et al. (31) utilized clinical factors (age, PSA, fPSA, PV, and TRUS) with or without MRI results to predict the presence of PCa and HGPCa. The AUC values with or without MRI for predicting PCa were 0.875 and 0.841, respectively, whereas those for predicting HGPCa were 0.872 and 0.850, respectively. Through univariate and multivariate logistic analyses, our research shows that clinical factors including age, tPSA and fPSA were significant factors for predicting clinically significant PCa, with AUC values of 0.911 for the training set and 0.842 for the test set, which were also different from the results of the above-mentioned studies. The possible reasons are first, the clinical factors selected by each study are different; in addition, this study grouped benign prostate cases into the non-clinically significant PCa group, which was different from the above study. Therefore, a more convenient and accurate method should be sought for the GS grading of preoperative PCa evaluation.
Different pathological grades of PCa show differences in internal cellular components, liquid contents, collagen levels, and fibromuscular stromal features. Non-significant PCa is well differentiated and has some remaining glandular structures that preserve some intercellular space. Clinically significant PCa has poor differentiation and is characterized by high cellularity and reduced extracellular space. These differences in histopathological features can be reflected by quantitative analysis and radiomics methods. Many studies have shown that extracted texture parameters can be used for PCa diagnosis and risk classification. Wibmer et al. (32) used T2WI and ADC maps and certified that the five parameters of the Haralick texture analysis (energy, entropy, correlation, homogeneity, and inertia) were beneficial for evaluating PCa and GS; in particular, ADC maps of energy and entropy were significantly different in GS ≤3+4 versus ≥4+3 cancers. Sidhu et al. (33) showed that the textural features of ADC kurtosis and T1 entropy can discriminate significant TZ PCa, and the combination of these parameters had an AUCof 0.86. Nketiah et al. (34) showed that combining T2WI textural features [angular second moment (ASM), contrast, correlation, and entropy] resulted in a higher classification performance (AUC: 0.82) than the use of MRI-based physiological parameters [ADC and DCE pharmacokinetic parameters (Ktrans and Ve)] combined (AUC: 0.75). However, the number of patients included in these studies was relatively small, ranging from a few dozen to more than one hundred patients, and the results may be less stable. In addition, these research methods only focused on extracting several texture features, and the information provided was relatively limited. Therefore, to increase the amount of information and enhance the stability of the results, the present study utilized a larger sample size (enrolled 381 cases) and expanded the types of extracted image texture features to establish a radiomics model based on bpMRI images to achieve an accurate preoperative diagnosis of clinically significant PCa.
At present, some studies have compared or combined radiomics methods with common methods to evaluate the diagnostic value of PCa. Wang et al. (21) showed that a radiomics model was more effective than the PI-RADS score in the diagnosis of PCa of either in the PZ or TZ, and the combined diagnostic efficiency of the two methods was significantly increased. Litjens et al. (35) also proved that the diagnostic efficiency of a computer-aided diagnostic model combined with the PI-RADS score in identifying PCa was better than that of either of the two models alone, and the AUC reached 0.88. Bonekamp et al. (36) showed that the diagnostic efficacy of quantitative methods (radiomics and the ADC value) was higher than that of qualitative methods (PI-RADS score) in differentiating benign and malignant prostate tissues, but there was no significant difference between radiomics and the ADC value. Chen et al. (19) indicated that radiomics models based on T2WI and ADC maps had high diagnostic efficacy and was superior to the PI-RADS v2 scores in distinguishing cancerous vs. noncancerous prostate tissue and high-grade vs. low-grade PCa. In the current study, radiomics methods were combined with clinical factors related to PCa diagnosis to construct a combined model for the diagnosis of clinically significant PCa. In the test set, the radiomics model (AUC value: 0.98) and the combined model (AUC value: 0.98) were significantly better than the clinical model (AUC value: 0.79) in the diagnostic efficacy of clinically significant PCa, and the difference was significant (P<0.05). At the same time, the two models based on radiomics (the radiomics model and the combined model) also had a greater clinical net benefit than that of clinical model. It is obvious that the addition of the RAD-SCORE into the clinical model improved the diagnostic efficacy and clinical net benefit of the clinical model in the diagnosis of clinically significant PCa; the model based on radiomics features has a clear application value in the diagnosis of clinically significant PCa.
In recent years, the nomogram figure forecast model has been widely used in clinical medicine, and there have been many studies related to this model published in high-impact clinical professional journals (37-39). The nomogram figure forecast model uses a risk score to represent multiple disease risk factors and predict patient outcomes, which is more clear, concise and easier to be understood. At the same time, it can be effectively applied to clinical work and is advantageous for doctor-patient communication, improving the doctor-patient relationship. The present study developed the nomogram of a clinical-radiomics combined model, which provided an intuitive and convenient method for doctors to diagnose clinically significant PCa and is expected to become a new means of auxiliary diagnosis in clinical work.
Despite its promising results, our study still has several limitations. First, it was a retrospective study performed in a single institution. Although we extracted partial data from this group of patients as a test set to validate the models, multicentre validation with a larger sample size is warranted to acquire better evidence for clinical application. Second, the pathological gold standard of the enrolled patients in this study was TRUS systematic prostate biopsy, instead of the large pathological sections of the specimens after radical prostatectomy were compared. When delineating the ROIs of lesions, it was difficult to completely match the MRI images with the pathological sections, which had a certain impact on the accuracy of ROI delineation. In addition, the puncture pathology may underestimate the accuracy of the PCa score and may not accurately reflect the true pathological status. Therefore, we will enrol PCa patients with large pathological sections after radical prostatectomy in further research. Third, our study did not separate the PZ of the prostate from the TZ because some patients had highly malignant disease in both zones. Therefore, further research is needed to increase the size of the studied population and to separate the PZ and TZ of the prostate differently.
Conclusions
In this study, a radiomics model was developed to predict the presence of clinically significant PCa with high diagnostic efficacy and clinical net benefit. In addition, the radiomics signature added value to PCa-related clinical risk factors for estimating the aggressiveness of PCa. The incorporation of radiomics features and clinical factors into the nomogram can provide a quantitative and intuitive approach for radiologists that could more confidently predict clinically significant PCa. Although the radiomics model established in this study is quantitative and relatively objective and has achieved good results, it still needs to be verified by comparison with subjective methods, such as the PI-RADS score, which is our future research direction.
Acknowledgments
Funding: This work was supported by the Suzhou Science and Technology Development Plan [grant number SS201534]; Advantageous Clinical Discipline Group of the Second Affiliated Hospital of Soochow University [grant number XKQ2015009]; and Suzhou industry technological innovation [grant number SYSD2017018]; and Youth Pre-Research Fund of the Second Affiliated Hospital of Soochow University [grant number SDFEYQN1814].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This retrospective study was approved by the Institutional Ethics Committee of our hospital, which waived the requirement for written informed consent.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M, Fossati N, Gross T, Henry AM, Joniau S, Lam TB, Mason MD, Matveev VB, Moldovan PC, van den Bergh RCN, Van den Broeck T, van der Poel HG, van der Kwast TH, Rouviere O, Schoots IG, Wiegel T, Cornford P. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol 2017;71:618-29. [Crossref] [PubMed]
- Cornford P, Bellmunt J, Bolla M, Briers E, De Santis M, Gross T, Henry AM, Joniau S, Lam TB, Mason MD, van der Poel HG, van der Kwast TH, Rouviere O, Wiegel T, Mottet N. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. Eur Urol 2017;71:630-42. [Crossref] [PubMed]
- Salami SS, Vira MA, Turkbey B, Fakhoury M, Yaskiv O, Villani R, Ben-Levi E, Rastinehad AR. Multiparametric magnetic resonance imaging outperforms the Prostate Cancer Prevention Trial risk calculator in predicting clinically significant prostate cancer. Cancer 2014;120:2876-82. [Crossref] [PubMed]
- Chen Y, Fan Y, Yang Y, Jin J, Zhou L, He Z, Zhao Z, He Q, Wang X, Yu W, Wu S. Are prostate biopsies necessary for all patients 75years and older? J Geriatr Oncol 2018;9:124-9. [Crossref] [PubMed]
- Braun K, Sjoberg DD, Vickers AJ, Lilja H, Bjartell AS. A Four-kallikrein Panel Predicts High-grade Cancer on Biopsy: Independent Validation in a Community Cohort. Eur Urol 2016;69:505-11. [Crossref] [PubMed]
- Wang R, Wang J, Gao G, Hu J, Jiang Y, Zhao Z, Zhang X, Zhang YD, Wang X. Prebiopsy mp-MRI Can Help to Improve the Predictive Performance in Prostate Cancer: A Prospective Study in 1,478 Consecutive Patients. Clin Cancer Res 2017;23:3692-9. [Crossref] [PubMed]
- MacAskill F, Lee SM. Diagnostic value of MRI-based PSA density in predicting transperineal sector-guided prostate biopsy outcomes. Int Urol Nephrol 2017;49:1335-42. [Crossref] [PubMed]
- Ueno Y, Tamada T, Bist V, Reinhold C, Miyake H, Tanaka U, Kitajima K, Sugimura K, Takahashi S. Multiparametric magnetic resonance imaging: Current role in prostate cancer management. Int J Urol 2016;23:550-7. [Crossref] [PubMed]
- Aydm H, Kizilgoz V, Tekin BO. Overview of current multiparametric magnetic resonance imaging approach in the diagnosis and staging of prostate cancer. Kaohsiung J Med Sci 2015;31:167-78. [Crossref] [PubMed]
- Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, Tempany CM, Choyke PL, Cornud F, Margolis DJ, Thoeny HC, Verma S, Barentsz J, Weinreb JC. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur Urol 2019;76:340-51. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Rozenberg R, Thornhill RE, Flood TA, Hakim SW, Lim C, Schieda N. Whole-Tumor Quantitative Apparent Diffusion Coefficient Histogram and Texture Analysis to Predict Gleason Score Upgrading in Intermediate-Risk 3 + 4 = 7 Prostate Cancer. AJR Am J Roentgenol 2016;206:775-82. [Crossref] [PubMed]
- Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ, Bostrom PJ, Bohm M, Haynes AM, Brenner P, Delprado W, Thompson J, Pulbrock M, Taimen P, Villani R, Stricker P, Rastinehad AR, Jambor I, Madabhushi A. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. J Magn Reson Imaging 2017;46:184-93. [Crossref] [PubMed]
- Algohary A, Viswanath S, Shiradkar R, Ghose S, Pahwa S, Moses D. Radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: Preliminary findings. J Magn Reson Imaging 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Chen T, Li M, Gu Y, Zhang Y, Yang S, Wei C, Wu J, Li X, Zhao W, Shen J. Prostate Cancer Differentiation and Aggressiveness: Assessment With a Radiomic-Based Model vs. PI-RADS v2. J Magn Reson Imaging 2019;49:875-84. [Crossref] [PubMed]
- Fehr D, Veeraraghavan H. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A 2015;112:E6265-73. [Crossref] [PubMed]
- Wang J, Wu CJ, Bao ML, Zhang J, Wang XN, Zhang YD. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol 2017;27:4082-90. [Crossref] [PubMed]
- Chaddad A, Kucharczyk MJ. Multimodal Radiomic Features for the Predicting Gleason Score of Prostate Cancer. Cancers (Basel) 2018. [Crossref] [PubMed]
- Chaddad A, Niazi T, Probst S, Bladou F, Anidjar M, Bahoric B. Predicting Gleason Score of Prostate Cancer Patients Using Radiomic Analysis. Front Oncol 2018;8:630. [Crossref] [PubMed]
- Giubilei G, Ponchietti R, Biscioni S, Fanfani A, Ciatto S. F DIL, Gavazzi A, Mondaini N. Accuracy of prostate volume measurements using transrectal multiplanar three-dimensional sonography. Int J Urol 2005;12:936-8. [Crossref] [PubMed]
- Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 2004;22:81-91. [Crossref] [PubMed]
- Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 2003;50:92-8. [Crossref] [PubMed]
- Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, Thoeny HC, Verma S. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol 2016;69:16-40. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. [Crossref] [PubMed]
- Li X, Pan Y, Huang Y, Wang J, Zhang C, Wu J, Cheng G, Qin C, Hua L, Wang Z. Developing a model for forecasting Gleason score >/=7 in potential prostate cancer patients to reduce unnecessary prostate biopsies. Int Urol Nephrol 2016;48:535-40. [Crossref] [PubMed]
- Niu XK, He WF, Zhang Y, Das SK, Li J, Xiong Y, Wang YH. Developing a new PI-RADS v2-based nomogram for forecasting high-grade prostate cancer. Clin Radiol 2017;72:458-64. [Crossref] [PubMed]
- Fang D, Zhao C, Ren D, Yu W, Wang R, Wang H, Li X, Yin W, Yu X, Yang K, Liu P, Shan G, Li S, He Q, Wang X, Xin Z, Zhou L. Could Magnetic Resonance Imaging Help to Identify the Presence of Prostate Cancer Before Initial Biopsy? The Development of Nomogram Predicting the Outcomes of Prostate Biopsy in the Chinese Population. Ann Surg Oncol 2016;23:4284-92. [Crossref] [PubMed]
- Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, Zheng J, Goldman D, Moskowitz C, Fine SW, Reuter VE, Eastham J, Sala E, Vargas HA. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 2015;25:2840-50. [Crossref] [PubMed]
- Sidhu HS, Benigno S, Ganeshan B, Dikaios N, Johnston EW, Allen C, Kirkham A, Groves AM, Ahmed HU, Emberton M, Taylor SA, Halligan S, Punwani S. Textural analysis of multiparametric MRI detects transition zone prostate cancer. Eur Radiol 2017;27:2348-58. [Crossref] [PubMed]
- Nketiah G, Elschot M, Kim E, Teruel JR, Scheenen TW, Bathen TF, Selnaes KM. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: preliminary results. Eur Radiol 2017;27:3050-9. [Crossref] [PubMed]
- Litjens GJ, Barentsz JO, Karssemeijer N, Huisman HJ. Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur Radiol 2015;25:3187-99. [Crossref] [PubMed]
- Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Gotz M, Kickingereder P, Yaqubi K, Hitthaler B, Gahlert N, Kuder TA, Deister F, Freitag M, Hohenfellner M, Hadaschik BA, Schlemmer HP, Maier-Hein KH. Radiomic Machine Learning for Characterization of Prostate Lesions with MRI: Comparison to ADC Values. Radiology 2018;289:128-37. [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, Wang Z, Zhu Z, Deng Q, Xiong X, Shao W, Shi X, He J. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]