Automatic functional analysis of left ventricle in cardiac cine MRI
Introduction
To quantitatively analyze global and regional cardiac function of the left ventricle (LV) from cine magnetic resonance (MR) images, clinical parameters such as ejection fraction (EF), systolic and diastolic volumes, and myocardial mass are required (1,2). Calculation of these parameters depends upon accurate delineation of endocardial, epicardial, papillary and trabecular muscles’ contours. Manual delineation is time-consuming and has high interobserver variability. Moreover, in clinical practice the manual delineation is typically limited to the end diastolic (ED) and end systolic (ES) phases due to time constraints, which is insufficient to fully analyze wall motion or compute the peak ejection and filling rates. As a result, an accurate, rapid, fully automatic LV contour detection (segmentation) algorithm analyzing all phases of the cardiac cycle is highly desirable.
A number of methods have been proposed for (semi-) automatic LV segmentation including: probability atlas (3), dynamic programming (4), fuzzy clustering (5), deformable model (6,7), active appearance model (8), active shape model (9), variational and level sets (10,11), graph cuts (12), and image-driven approaches (13,14). For a complete review of recent literatures describing cardiac segmentation techniques see (15). Despite recent advances, accurate LV segmentation is still acknowledged as a difficult problem, and typically faces three main challenges: (I) the difficulty in localizing the LV and distinguishing it from other features and artifacts; (II) the lack of edge information where contours are required; and (III) the shape variability of the endocardial and epicardial contours across slices, phases, and subjects.
In this work, we describe our method for the fast, robust, and fully automatic LV segmentation from short axis (SAX) cine MR images. The method localizes the LV and segments its major features including the endocardial and epicardial contours, and papillary and trabecular muscles in all phases. The segmentation algorithm was evaluated with a database of clinical cardiac MR studies with diverse pathology.
Materials and methods
Study population and imaging
For this study, data from two scanners were analyzed: GE (1.5 T, CV/i Excite, GE Healthcare, Milwaukee, WI), Philips (1.5 T, Achieva, Philips Medical Systems, Best, Netherlands). Cine steady state free precession (SSFP) MR-SAX images were obtained with imaging during 10-15 s breath-holds, with coverage from the atrioventricular ring to the apex. Patients with motion artifact, banding artifact or gross abnormality (e.g., due to a tumor) were excluded from the study.
GE scanner
Data from 133 patients were retrospectively analyzed (35 female, 98 male; age: 59.4±16.1 yrs). Imaging parameters were as follows: repetition/echo times, 3.7/1.6 ms; flip angle, 45º; matrix size, 256×256; field of view, 320 mm × 320 mm; contiguous sections, 10-14; section thickness were 8-10 mm; slice gap, 0 mm; and number of phases, 20. The patient datasets were classified into four groups representing diverse morphologies, based on the following clinical criteria: (I) heart failure with infarction (HFI) group had EF <40% and evidence of late gadolinium (Gd) enhancement; (II) heart failure with no infarction (HFNI) group had EF <40% and no late Gd enhancement; (III) LV hypertrophy (HYP) group had normal LV EF (>55%) and a ratio of LV mass/body surface area >83 g/m2 (16); and (IV) healthy (HEA) group had EF >55% and no hypertrophy. Patient selection ensured the group sizes were approximately equal (HFI N =34, HFNI N =30, HYP N =32, HEA N =37).
Philips scanner
Six datasets were analyzed (6 male; age: 61.5±12.4 yrs). Imaging parameters were as follows: repetition/echo times, 3.9/1.9 ms; flip angle, 60º; matrix size, 320×320; field of view, 330 mm × 330 mm; contiguous sections, 15-20; section thickness, 8 mm; slice gap, 0 mm; and number of phases, 25.
Manual contours
The endocardial and epicardial contours were drawn by three well-trained readers for all slices at the ED and ES phases, and confirmed by a cardiologist with 10 years of experience in cardiac imaging, and these manual contours were the evaluation reference standard. The inter- and intra-observer variability has been previously reported (17). The convention of including papillary muscles and trabeculations within the endocardial contour was followed (16). The most basal LV slice was defined as the one preceding the atrium having the ED blood pool ≥50% encircled by myocardium, and included the LV outflow tract (LVOT). The apical slice was defined as the ones that beyond the papillary muscles to the apex. The patients consented to research use of their imaging data, with approval by our institutional ethics review board.
Segmentation algorithm
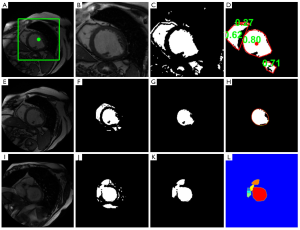
The calculation starts from the middle slice, and each image is processed sequentially in the basal direction, followed by sequential processing from the middle slice to the apex (18). The images for each cardiac phase are calculated independently. First, the myocardium is located by calculating the centroid of the LV blood pool on the middle slice at the specified phase based on a roundness metric technique (Figure 1A-D). Second, the endocardial and papillary muscles’ and trabeculations’ contours are detected by applying the optimal threshold method of Otsu (19) watershed, and 1D fast Fourier transform (FFT) (20) method (Figure 1E-I). Finally, the epicardial contour is calculated by mapping the pixels from Cartesian to polar coordinates, and using a multiple seed, region-growing method (Figure 2). Refer to the Supplementary Material for a detailed description of the image processing.

Evaluation and statistical analysis
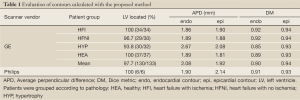
In order to quantitatively evaluate the automatically detected endocardial and epicardial contours of the ED and ES phases of all slices, four quantitative measures were assessed (17). Average perpendicular distance (APD) is the distance from the automatic contour to the corresponding manually drawn expert contour, averaged over all contour points. Dice metric (DM) (21) is a measure of contour overlap utilizing the area of automatically (A) and manually segmented (M) contours, and their intersection area (I) with formula DM = I/[0.5× (A+M)]. In addition, the percentage of patients where the blood pool is correctly localized and distinguished from other features was calculated (LV located).
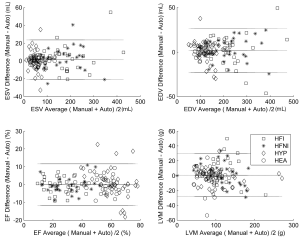
Paired sample, two-tailed Student’s t-test statistics (α=0.05) were computed to determine if the functional values calculated were significantly different when using automatic contours instead of manual contours. The end systolic volume (ESV), end diastolic volume (EDV), EF and left ventricular mass (LVM) were evaluated. The manually and automatically determined ESV, EDV, EF and LVM were also compared by means of linear regression to determine the correlations. Bland-Altman plots were calculated to assess the bias and limits of agreement between manual and automatic contours.
To investigate the impact of methodological difference on clinical care, EF used for patient classification in this study (HFNI: EF <40%, HFI: EF <40%, HYP: EF >55%, HEA: EF >55%) were applied to the corresponding patient group to evaluate whether clinical classification would differ based on manual and automated results. The categorical variable of agreement between the manual and automated results was assessed with Cohen’s Kappa statistics.
The computation time was tested on consumer hardware (2×2.8 GHz Quad-core Intel Xeon Mac Pro, Apple) with a non-optimized Matlab code (Mathworks) implementation.
To provide a direct basis of comparison for our results, the APD, DM and EF were calculated with commercial, automatic segmentation software [QMass® MR 7.5, Medis Medical Imaging Systems (22), Leiden] using our GE data. We adopted a threshold of APD >10 mm to eliminate clearly erroneous contours detected by QMass. For these cases, the automatic contour was replaced by the corresponding manual contour to simulate the expected usage of the automatic contours and subsequent calculation of APD, DM and EF.
Results
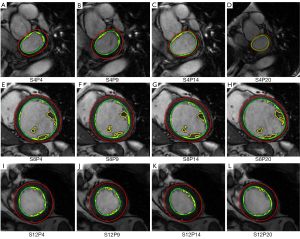
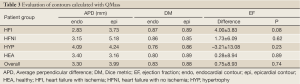
The contour evaluation results averaged across all subjects, for all slices at the ED and ES phases of each group, are shown in Table 1. Representative segmentation contours for one dataset are shown in Figure 3. For three of the 133 datasets, the segmentation algorithm failed to locate the LV automatically, thus manual corrections were applied. The HYP group had the largest APD for both endo- and epicardial contours, and the smallest DM for the endocardial contours. The APD was small indicating good agreement between individual contours. The APD for GE data was 2.08 and 1.92 mm (1.7 and 1.6 pixels) for endo- and epi-cardial contours, and the APD for Philips data was 1.9 and 2.14 mm (1.5 and 1.8 pixels), respectively.
Full Table
The paired sample t-test and linear regression results for ESV, EDV, EF and LVM between manual and the automatic methods are shown in Table 2. There were no significant differences found between automatically and manually determined ESV, EDV, EF and LVM. The automatically determined ESV and EDV for individual patient groups showed excellent correlations (R≥0.89) with those derived from manual contours. The EF and LVM of the automatically and manually analyzed studies correlated very well generally, with lower correlations evident for the HYP and HEA groups. Grouping together all subjects and pathological categories, the coefficient of determination (R2) between the manual and automatic methods was high for all functional parameters (ESV: 0.98, EDV: 0.98, EF: 0.90, and LVM: 0.88).

Full Table
The Bland-Altman analysis (Figure 4) confirmed the good agreement between the automatic and manual contours with negligible biases (1.51 mL, 1.69 mL, –0.02%, –0.66 g for ESV, EDV, EF and LVM, respectively). The limits of agreement (±1.96 SD) were as follows: ±22.1 mL (ESV), ±11.6% (EF), ±25.0 mL (EDV), and ±28.8 g (LVM).
The patient classification agreement between automated and manual contours were good (HFNI: Kappa statistic 0.84, P-value <0.001, HFNI: Kappa statistic 1, P-value 0, HYP: Kappa statistic 0.57, P-value 0.004, HEA: Kappa statistic 0.51, P-value 0.002). The agreement is highly significant.
The average computation time of LV location was 0.096 s per subject. The average computation time for segmentation was 0.8 s per image. Moreover, the algorithm segmented the basal slice with LVOT by a fast watershed technique (0.027 s per image). Manual analysis for EF calculation (ES and ED phases) required 10-15 min per subject.
The QMass contour evaluation results were calculated for comparison and shown in Table 3. All contours were detected. The average APD for endocardial and epicardial contours was higher (3.30, 3.99 mm) than for the proposed algorithm. The EF difference (QMass EF- manual EF) shows that QMass overestimated the EF (except for the hypertrophy patient group), although this was statistically insignificant for all groups.
Full Table
Discussion
This study demonstrates that the proposed image driven segmentation algorithm rapidly and accurately quantifies clinically relevant parameters with minimal user input. This method is based on the assumptions that (I) the heart is approximately in the centre of the original image, and (II) the left ventricular blood pool is approximately circular. These assumptions are routinely satisfied in cine SSFP MR imaging, and therefore the method should be suitable for datasets with a wide range of anatomy, function, and image contrast as required for clinical use.
Novel aspects of this algorithm include that it obviates initialization by manually drawn contours, prior statistical shape model, or gray-level appearance model. In addition, the algorithm locates the LV automatically by utilizing a roundness metric that is fast (0.096 s per subject) and robust (97.7% located), which compares favorably to another automatic LV location technique (23).
The algorithm detects not only endocardial contours and epicardial contours, but also the papillary muscles’ and trabeculations’ contours. Clinical studies have employed different quantification methods for calculation of LV volume, mass, and EF by including or excluding papillary muscles and trabeculations’ in the ventricular cavity. Recent studies have shown that the papillary muscles and trabeculations’ have a significant impact on calculation of LV volume, mass (24) and EF (25). Therefore the proposed method provides additional important options for daily clinical application.
A difficult challenge of LV segmentation is the accurate delineation of the epicardial contours. The typical problem is the ballooning of epicardial contours at the junction between myocardium and lung parenchyma and subdiaphragmatic tissues, due to the small intensity differences between these tissues. Following transformation from Cartesian to polar coordinates, the multiple-seed region-growing method was found to provide a robust solution that accounted for intensity gradients at edges that differed around the myocardium
Accurate assessment of quantitative LV parameters is essential for all imaging techniques. For instance, current guidelines for the implantation of cardioverter defibrillators use a selection criterion based upon EF, mandating that an accurate quantitative method be used to determine EF (26). Furthermore, reducing the quantity of patients required for clinical research studies depends upon increasing the interstudy reproducibility. CMR currently has the best interstudy reproducibility of any imaging technique for both mass and volume in the left and right ventricles, and is considered superior to that of two dimensional echocardiography (1). Interstudy reproducibility, determined from the standard deviation of the differences in two measurements of a parameter over a reasonable time period wherein no clinical change is expected, is currently influenced by inter- and intra-observer variability (27,28). Automatic segmentation algorithms will further reduce interstudy variability, therefore reducing patient numbers required. This advantage together with the time efficiency for quantitation by study readers will reduce the costs of clinical trials while improving data quality (2).
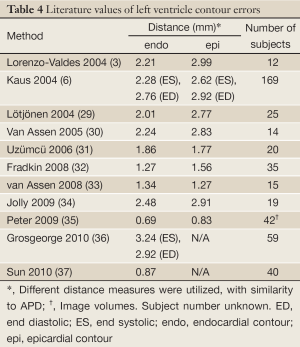
Among the four groups, the results of the hypertrophy group were least successful for both the endocardial and epicardial contours because the concomitant hypertrophy of the papillary muscles obscured the target contours. Table 2 shows that there were good correlations for all of the four groups between automatically and manually determined clinical parameters, as expected from the small APD and large DM. These results compare favorably to recent literature describing automatic or semi-automatic segmentation methods (Table 4). Although dataset specifics are not uniform and thus hinder direct comparison, this meta-analysis provides additional confidence in our method.
Full Table
The mean difference shown in Table 2 compares well to interobserver variation of manually drawn contours in previous reports (17,38). The Bland-Altman plots demonstrate negligible biases for ESV, EDV, EF and LVM, and the limits of agreements were reasonable considering the image quality heterogeneity of the datasets. While the proposed method is fully automatic, setting and adjusting the parameters interactively by checking the contours visually could further improve results.
In conclusion, the proposed fully automated segmentation technique is fast, robust and effective for the quantification of cine cardiac MR in clinical practice.
Acknowledgements
The authors thank the Canadian Foundation for Innovation (CFI) and the Canadian Institutes of Health Research (CIHR) for their grant support, and Circle Cardiovascular Imaging for licensing the technology.
Disclosure: The authors declare no conflict of interest.
Supplementary Material
LV location
This section presents a method based on a roundness metric to automatically locate the LV blood pool’s centroid on the middle slice at the specified phase. This procedure consists of five steps (refer to Figure 1A-D):
- Choose the middle (normally in the mid-cavity level) slice image in a given phase (e.g., ED) as the target image;
- Specify a centered, fixed rectangular region of interest (ROI) on the target image. The size of the rectangular is 110×110 pixels (Figure 1A);
- Apply the optimal threshold method of Otsu (19) to convert the ROI to a binary image (Figure 1C);
- Remove all objects smaller than a predefined threshold (40 pixels) and compute the convex hull of the surviving objects;
- Compute the roundness metric
of each surviving convex-hulled object, where A is area and P is perimeter length. R =1 for a circle. The object with the largest roundness metric is recognized as the LV blood pool (Figure 1D), and a 11×11 mask is defined centered on the centroid of the LV blood pool for the subsequent segmentation.
LV endocardial contour detection
The endocardial contour is detected by the following steps (refer to Figure 1E-L):
- Specify a ROI dilated from the previously identified LV blood pool;
- Apply the optimal threshold method of Otsu to convert the ROI to a binary image (Figure 1F);
- The LV blood pool object is identified by choosing the object that has maximum overlap with the predefined mask centered on the LV blood pool centroid (Figure 1G);
- Compute the convex hull of the refined blood pool (Figure 1H);
- Smooth the convex hull’s contour by applying the 1D fast Fourier transform (FFT) (20). We first compute the FFT of the x coordinate of the contour point index, multiply the result by a low pass filter transfer function (retaining the four lowest frequency components), then take the inverse transform to produce the smoothed x coordinate. Repeat for y coordinates (Figure 1H);
- Identify and segment basal slice with LVOT. If the ratio of current contour’s major axis length (L) to preceding contour’s L is larger than a predened threshold (1.2, in this work), basal slice with LVOT is identified. Then, the blood pool is separated from the LVOT by the following steps: Calculate the Euclidean distance transform of the binary object, i.e., compute the distance between each object pixel and its nearest background pixel. Then calculate the watershed regions of the distance image (Figure 1I-L). Then compute the smoothed contour (as in step V).
Detection of contour delineating LV papillary muscles and trabeculations
Black pixels in the smoothed LV blood pool (Figure 1H) are detected as papillary muscles and trabeculations.
Epicardial contour detection
The epicardial contour is calculated by the following steps (refer to Figure 2A-F):
- Map the pixels from Cartesian to approximately polar coordinates, as suggested previously (18). An outer boundary is calculated by dilation of the endocardial contour. The two contours are interpolated to the same number of points, and paired to derive scan lines, each of a predefined length (20 pixels) (Figure 2A). The result is a rectangular image that extends from the endocardial contour (top row) outward (bottom row) (Figure 2B);
- Use each top-row pixel as a region growing seed, with all grown regions summed and converted to a binary image (Figure 2C). For region growing, intensities are normalized by the original image’s maximum;
- Fill image holes by morphological operations (Figure 2D);
- The end point of each column’s grown region determines an edge point (Figure 2E);
- Inverse transform the edge point coordinates to the original (Cartesian) coordinate space to determine the epicardial contour (Figure 2F);
- Smooth the contour by applying the 1D fast Fourier transform (Figure 2F).
References
- Grothues F, Smith GC, Moon JC, et al. Comparison of interstudy reproducibility of cardiovascular magnetic resonance with two-dimensional echocardiography in normal subjects and in patients with heart failure or left ventricular hypertrophy. Am J Cardiol 2002;90:29-34. [PubMed]
- Keenan NG, Pennell DJ. CMR of ventricular function. Echocardiography 2007;24:185-93. [PubMed]
- Lorenzo-Valdés M, Sanchez-Ortiz GI, Elkington AG, et al. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med Image Anal 2004;8:255-65. [PubMed]
- Kurkure U, Pednekar A, Muthupillai R, et al. Localization and segmentation of left ventricle in cardiac cine-MR images. IEEE Trans Biomed Eng 2009;56:1360-70. [PubMed]
- Rezaee MR, van der Zwet PJ, Lelieveldt BP, et al. A multiresolution image segmentation technique based on pyramidal segmentation and fuzzy clustering. IEEE Trans Image Process 2000;9:1238-48. [PubMed]
- Kaus MR, von Berg J, Weese J, et al. Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal 2004;8:245-54. [PubMed]
- Cordero-Grande L, Vegas-Sánchez-Ferrero G, Casaseca-de-la-Higuera P, et al. Unsupervised 4D myocardium segmentation with a Markov Random Field based deformable model. Med Image Anal 2011;15:283-301. [PubMed]
- Mitchell SC, Lelieveldt BP, van der Geest RJ, et al. Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. IEEE Trans Med Imaging 2001;20:415-23. [PubMed]
- O’Brien SP, Ghita O, Whelan PF. A novel model-based 3D +time left ventricular segmentation technique. IEEE Trans Med Imaging 2011;30:461-74. [PubMed]
- Paragios N. A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans Med Imaging 2003;22:773-6. [PubMed]
- Ben Ayed I, Lu Y, Li S, et al. Left ventricle tracking using overlap priors. Med Image Comput Comput Assist Interv 2008;11:1025-33. [PubMed]
- Lin X, Cowan B, Young A. Model-based Graph Cut Method for Segmentation of the Left Ventricle. Conf Proc IEEE Eng Med Biol Soc 2005;3:3059-62. [PubMed]
- Cocosco CA, Niessen WJ, Netsch T, et al. Automatic image-driven segmentation of the ventricles in cardiac cine MRI. J Magn Reson Imaging 2008;28:366-74. [PubMed]
- Codella NC, Weinsaft JW, Cham MD, et al. Left ventricle: automated segmentation by using myocardial effusion threshold reduction and intravoxel computation at MR imaging. Radiology 2008;248:1004-12. [PubMed]
- Petitjean C, Dacher JN. A review of segmentation methods in short axis cardiac MR images. Med Image Anal 2011;15:169-84. [PubMed]
- Alfakih K, Plein S, Thiele H, et al. Normal human left and right ventricular dimensions for MRI as assessed by turbo gradient echo and steady-state free precession imaging sequences. J Magn Reson Imaging 2003;17:323-9. [PubMed]
- Connelly KA, Detsky J, Graham J, et al. Multicontrast late gadolinium enhancement imaging enables viability and wall motion assessment in a single acquisition with reduced scan times. J Magn Reson Imaging 2009;30:771-7. [PubMed]
- Lu Y, Radau P, Connelly K, et al. Segmentation of Left Ventricle in Cardiac Cine MRI: An Automatic Image-Driven Method. Lect Notes Comput Sci 2009;5528:339-47.
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 1979;9:62-6.
- Brigham EO, Morrow RE. The fast Fourier transform. IEEE Spectrum 1967;4:63-70.
- Van Leemput K, Maes F, Vandermeulen D, et al. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging 1999;18:897-908. [PubMed]
- List of Publications. Medis medical imaging systems. Available online: http://www.medis.nl/Products/PublicationLists/QMassMRPublications.htm, accessed January 18, 2011.
- Pednekar AS, Muthupillai R, Lenge VV, et al. Automatic identification of the left ventricle in cardiac cine-MR images: dual-contrast cluster analysis and scout-geometry approaches. J Magn Reson Imaging 2006;23:641-51. [PubMed]
- Papavassiliu T, Kühl HP, Schröder M, et al. Effect of endocardial trabeculae on left ventricular measurements and measurement reproducibility at cardiovascular MR imaging. Radiology 2005;236:57-64. [PubMed]
- Weinsaft JW, Cham MD, Janik M, et al. Left ventricular papillary muscles and trabeculae are significant determinants of cardiac MRI volumetric measurements: effects on clinical standards in patients with advanced systolic dysfunction. Int J Cardiol 2008;126:359-65. [PubMed]
- Moss AJ, Multicenter Automatic Defibrillator Implantation Trial. MADIT-II and its implications. Eur Heart J 2003;24:16-8. [PubMed]
- Bellenger NG, Burgess MI, Ray SG, et al. Comparison of left ventricular ejection fraction and volumes in heart failure by echocardiography, radionuclide ventriculography and cardiovascular magnetic resonance; are they interchangeable? Eur Heart J 2000;21:1387-96. [PubMed]
- Bellenger NG, Davies LC, Francis JM, et al. Reduction in sample size for studies of remodeling in heart failure by the use of cardiovascular magnetic resonance. J Cardiovasc Magn Reson 2000;2:271-8. [PubMed]
- Lötjönen J, Kivistö S, Koikkalainen J, et al. Statistical shape model of atria, ventricles and epicardium from short- and long-axis MR images. Med Image Anal 2004;8:371-86. [PubMed]
- van Assen HC, Danilouchkine MG, Frangi AF, et al. SPASM: a 3D-ASM for segmentation of sparse and arbitrarily oriented cardiac MRI data. Med Image Anal 2006;10:286-303. [PubMed]
- Uzümcü M, van der Geest RJ, Swingen C, et al. Time continuous tracking and segmentation of cardiovascular magnetic resonance images using multidimensional dynamic programming. Invest Radiol 2006;41:52-62. [PubMed]
- Fradkin M, Ciofolo C, Mory B, et al. Comprehensive segmentation of cine cardiac MR images. Med Image Comput Comput Assist Interv 2008;11:178-85. [PubMed]
- van Assen HC, Danilouchkine MG, Dirksen MS, et al. A 3-D active shape model driven by fuzzy inference: application to cardiac CT and MR. IEEE Trans Inf Technol Biomed 2008;12:595-605. [PubMed]
- Jolly MP, Xue H, Grady L, et al. Combining registration and minimum surfaces for the segmentation of the left ventricle in cardiac cine MR images. Med Image Comput Comput Assist Interv 2009;12:910-8. [PubMed]
- Peters J, Ecabert O, Meyer C, et al. Optimizing boundary detection via Simulated Search with applications to multi-modal heart segmentation. Med Image Anal 2010;14:70-84. [PubMed]
- Grosgeorge D, Petitjean C, Caudron J, et al. Automatic cardiac ventricle segmentation in MR images: a validation study. Int J Comput Assist Radiol Surg 2011;6:573-81. [PubMed]
- Sun H, Frangi AF, Wang H, et al. Automatic cardiac MRI segmentation using a biventricular deformable medial model. Med Image Comput Comput Assist Interv 2010;13:468-75. [PubMed]
- Danilouchkine MG, Westenberg JJ, de Roos A, et al. Operator induced variability in cardiovascular MR: left ventricular measurements and their reproducibility. J Cardiovasc Magn Reson 2005;7:447-57. [PubMed]


