
Attention-VGG16-UNet: a novel deep learning approach for automatic segmentation of the median nerve in ultrasound images
Introduction
Ultrasound technology provides real-time imaging of nerves, and has thus been widely applied in nerve diagnosis and treatment, for example, ultrasound-guided drug injection (1), peripheral nerve blockade (2), the diagnosis of traumatic nerve injuries (3,4), postoperative complications of nerve repair (5), inflammatory neuropathies (6), and nerve entrapment syndromes (7,8). The benefits of ultrasound imaging include the ability to depict nerve morphology, describe the degree of nerve injuries, quantify nerve size/pathology, uncover the underlying cause, and guide a likely intervention or forthcoming surgery; all of which share the common goal of localizing and segmenting the nerve. The nerve appears hyperechoic in ultrasound images with a honeycomb texture (9). However, nerve segmentation in ultrasound images is incredibly challenging due to the tiny and inconspicuous size of nerves, the low image quality, degradation of structure details induced by noise disturbance of the ultrasound waves, and blurred demarcation of anatomical tissues due to the low contrast with neighboring nerves.
Concerted efforts have previously been made for automatic detection and segmentation of the median nerve in medical ultrasound images. A machine learning framework was proposed to enable robust detection of the median nerve (10). Hafiane et al. (11) used deep learning combined with spatiotemporal information to robustly segment the nerve region. Ding et al. (12) proposed the BPMSegNet network for multiple instance segmentation in brachial plexus ultrasound images. Horng et al. (13) proposed a new convolutional neural network framework called DeepNerve for localization and segmentation of the median neve. Festen et al. (14) used a U-Net-shaped neural network for segmentation of the median nerve, and Wu et al. (15) evaluated the performance of the pretrained models using ultrasound images of the median nerve. However, in these studies, the ultrasound image of the median nerve was recorded at the level of wrist inlet. The segmentation contour of the median nerve displayed in a two-dimensional (2D) image lacks information of the morphology and travel direction when a section of the nerve needs to be evaluated from different perspectives. Important image features may be overlooked if only 2D images from the traditional sonogram are used for assessment (16). Therefore, this study aimed to propose a deep learning method for automatic segmentation of the median nerve in a sequence of ultrasound images and demonstrate the potential for 3D reconstruction of the median nerve based on the segmentation results.
Previous studies
The U-Net (17) is a commonly used model for segmentation in ultrasound images, and modified versions of U-Net (18-21) were proposed to detect and segment the brachial plexus. The Visual Geometry Group (VGG) network (22) proposed in ImageNet Large Scale Visual Recognition Challenge (ILSVRC) 2014 improves the recognition performance by increasing the depth of the convolutional neural network (CNN), in which the best network contains 16 weight layers. The VGG model is useful for object localization due to its tiny filter size-based design (23). The networks based on VGG architecture have been applied for detection and segmentation (24-28). Skip connection is a standard module in many convolutional architectures, and attention is a mechanism that can improve the performance of the model by incorporating an encoder and decoder.
Skip connections (i.e., shortcut connections), add an extra layer of connection from the network input to the output (skipping 1 or more layers) and have been analyzed in previous literature (29-31). The extra layer can be linear, such as identity mapping in ResNet (32), or non-linear with gating functions, as in Highway Networks (33). Skip connections are used to bypass the signal from 1 layer to another, which can help the algorithm avoid being attracted to spurious local optima and guide the algorithm to evolve towards a global optima (32,34), therefore solving the problem of gradient explosion and gradient vanishing during training in deep networks. Research has also shown that skip connections perform identity mapping and their inputs are added to the outputs of the stacked layers, forming the residual module (32,35-38).
Technically, summation and concatenation (copy and cut) of feature maps are the most popular operations in skip connections for feature fusion (39). In concatenation, only the number of channels for the features increases, while the information of each feature does not gain. Concatenation is often used to aggregate and merge the features extracted by multiple convolutional feature extraction frameworks. During summation, the information increases with the amount of the channel constant, which is beneficial to the classification of the final image. Furthermore, summation has been demonstrated to preserve the spatial information lost during the pooling operation and capture full resolution features (35).
The attention mechanism is a resource allocation scheme to allocate computing resources to more important tasks and solve the problem of information overload when computing power is limited. Previous research used the attention mechanism to connect deep layers to shallow layers, enabling the model to distinguish regions as a useful feature selection function in computer vision (40-42). The inputs of the attention mechanism are the outputs produced by the encoder. They are weighted and combined into the decoder at the current location to influence the output of the decoder. By weighting the outputs of the encoder, more context information from the raw data can be used while aligning the inputs with the outputs of the decoder.
Our main aim in this work was to develop improved models for median nerve segmentation in ultrasound images and to reconstruct the 3D visualization of the median nerve based on the best segmentation results. Based on U-Net and VGG16, an improved network called VGG16-UNet was proposed. To enhance the improvement of the model performance, the attention mechanism and/or the residual module were added to the U-Net and VGG16-UNet to subsequently obtain the following models: Attention-UNet (A-UNet), Summation-UNet (S-UNet), and Attention-Summation-UNet (AS-UNet), Attention-VGG16-UNet (A-VGG16-UNet), Summation-VGG16-UNet (S-VGG16-UNet), and Attention-Summation-VGG16-UNet (AS-VGG16-UNet). Then, the performance of these models was evaluated by metrics including Dice, Jaccard, Precision, and Recall. Based on the weighted blending results of the median nerve image sequence and its corresponding best segmentation results, 3D visualization of the median nerve was reconstructed with the Visualization Toolkit (VTK) (43) to assist in clinical nerve diagnosis.
Contributions
In this study, we propose a deep learning approach to improve the accuracy of automatic segmentation of the median nerve in ultrasound image sequence. Specifically, we make the following contributions: (I) we propose a novel method by combining the VGG16 network and the architecture resembling the upsampling path of U-Net, aiming to improve the segmentation performance of U-Net model; (II) we apply the attention mechanism and residual module to the U-Net and VGG16-UNet and achieve better performance than the corresponding original models.
The proposed A-VGG16-UNet model provides the highest accuracy of automatic segmentation of the median nerve compared with other existing methods.
We present the following article in accordance with the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1074/rc).
Methods
Dataset description
In this work, a dataset of the median nerve of 20 healthy participants was collected by doctors from the Third Affiliated Hospital of Sun Yat-Sen University. The inclusion criteria were as follows: (I) aged 20–45 years; and (II) no history of peripheral nerve injuries or wrist injury. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University, and informed consent was provided by all individual participants.
For each participant, 1 or 2 ultrasound videos of the median nerve were acquired using an S-Nerve Ultrasound System (Fujifilm SonoSite Inc., Bothell, WA, USA) equipped with a HFL38x probe with a central frequency of 10 MHz. All participants were positioned with the palm facing upward, arm on the pillow, and wrist in neutral position. The probe was placed about 2 cm from the level of wrist inlet and then moved from the distal point to the proximal on the forearm, as illustrated in Figure 1. During scanning, the forearm and wrist remained stationary, and the sequence of transverse ultrasound images of the median nerve was recorded.
For the training set, 910 image frames were selected from 35 image sequences obtained from 19 participants to perform 5-fold cross-validation. The goal of selecting frames in a sequence of images was to capture the frames that were different from each other and make a final training set that covered as much diverse terrain as possible. Therefore, sequential images were chosen at an interval of 5 frames from the image sequence, and 26 image frames on average were selected for annotation from each acquired image sequence. Then, the image sequence of 207 frames collected from the remaining participants were used for the test set. The ground truth (GT) of the nerve segmentation of each frame was the corresponding binary mask generated from the manual delineation of the median nerve using an annotation tool LabelMe (44) under the guidance of an experienced doctor. In addition, each of the images in the dataset was cropped into 320×448 pixels.
Data augmentation
Deep neural networks often require a large amount of training data to achieve satisfactory performance. However, the available medical images are usually limited. Therefore, to reduce the impact of insufficient data, data augmentation is commonly used to increase the variability in the medical dataset. In this study, rotation, clip, zoom, translation, and horizontal flip were used in turn to augment the variability of the original training set in each iteration of training. Since the parameter values of each augmentation mentioned above were selected at random, different training sets could be obtained in different iteration batches. However, the number of images in the training set of each iteration remained constant.
Combination of attention mechanism and/or residual module with U-Net
Figure 2 displays the architectures of U-Net model and its variants, and the 3 blocks used in the models. The U-Net is a classical fully-convolutional network for classification, localization, and segmentation in ultrasound images (Figure 2A). It consists of an encoder (contracting path) and a decoder (expanding path). The output of the encoder is the feature map or vector that contains the information of the input. The decoder has the same structure as the encoder but takes feature maps as the input and provides a similar match to the actual input or intended output. Furthermore, concatenating feature maps from the contraction phase helps the expansion feature recover the information about the location of the respective object. The encoder process reduces the size of the input matrix by increasing the number of the feature maps. On the contrary, the decoder returns the matrix to its original size by minimizing the number of the feature maps. Therefore, the segmentation results can be compared with the ground truth (GT) in every pixel.
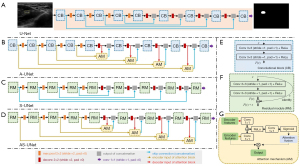
In this study, 3 variants of U-Net (A-UNet, S-UNet, and AS-UNet) were proposed by using the attention mechanism and/or the residual module based on U-Net. These 3 variants are described in the remainder of this section.
The A-UNet (Figure 2B) combines 4 attention mechanisms with U-Net to better represent features learned by convolutional layers (Figure 2G). The 2 inputs of the attention mechanism are the feature maps extracted from the encoder and the decoder, which can restore the same spatial resolution as the input image. The output of the attention mechanism is concatenated to the result after the corresponding upsampling operation. The kernel size of the 3 convolutions was 1×1, and the size of the output of the attention fusion was the same as the size of the corresponding encoder feature. Therefore, the attention map was calculated with a pointwise operation.
The S-UNet (Figure 2C) can leverage features by adding residual modules based on U-Net. The architecture in Figure 2F can be a residual module, in which a summation operation of the skip connection was applied, adding the input of the first convolution layer to the output of the second convolution layer of the same module. Ultimately, there were 9 residual modules in S-UNet, which made better use of the information in the feature maps of different layers for model learning.
The AS-UNet (Figure 2D) combines the characteristics of A-UNet and S-UNet for better segmentation performance.
The proposed VGG16-UNet and its variants
Figure 3 displays the architectures of VGG16 and its variants. As shown in Figure 3A, the VGG16 network has 13 convolutional layers, 5 pooling layers, and 3 fully connected layers in the end of the network. The VGG16 network features a homogeneous architecture that only performs 3×3 convolution and 2×2 max pooling from the beginning to the end.
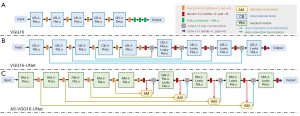
For the improved VGG16-UNet (Figure 3B), similar to the networks in (24-26), the final 3 fully connected layers of VGG16 (the green solid rectangles in Figure 3A) were replaced with architecture that resembled the decoding part of U-Net, which formed the expanding path with convolution layers and upsampling layers. Hence, the VGG16 without the final 3 fully connected layers was retained as the contracting path. Additionally, 3 more modifications were performed in this study. (I) For the original rectified linear unit (ReLu) activation function in 7 convolution layers (the final four convolutional blocks in Figure 3B), they were replaced with Leaky ReLu (α=0.1). (II) We used 4 skip connections (3 concatenations and a summation) to combine feature maps of different modules in contracting path and expanding path. (III) The size of the kernel for the upsampling operation was 4×4.
The AS-VGG16-UNet (Figure 3C) applies attention mechanisms and residual modules to VGG16-UNet. Meanwhile, A-VGG16-UNet and S-VGG16-UNet, which are constructed like A-UNet and S-UNet, were also used in this study.
3D nerve visualization using the VTK
More recently, 3D ultrasound image reconstruction based on 2D images has become a popular method to analyze abnormalities in some parts of the anatomy (45). The VTK toolbox is an open source, free software system for 3D computer graphics, image processing and visualization (43). It was originally designed for medical applications, so it has powerful capabilities for medical visualization (46). It encapsulates some common visualization algorithms, such as surface rendering used in Marching Cubes (MC) and volume rendering used in light projection (Ray-Casting), to reconstruct the 3D structure of objects from 2D images into a type of packaging in the form provided to the user.
The volume rendering method uses the transfer functions to convert volume data values into optical properties such as color, opacity, and gradient, which are then combined into pixels on the screen to form a 3D image. The VTK is also used for 3D reconstruction and visualization (45,47).
In this paper, 3D median nerve reconstruction and visualization were completed with the volume rendering method and by implementing vtkPiecewiseFunction and vtkColorTransferFunction classes as the design of transfer functions. The weighted blending results of the image sequence of the median nerve and the responding segmentation results were input as a volume into the system by using the function vtkJPGReader.
Evaluation metrics
To quantitatively measure the image segmentation performances of the 8 models, this study defined 2 region-based assessment metrics, Precision (Eq. [1]) and Recall (Eq. [2]), which were calculated based on the region overlap between manually and automatically segmented results. In addition, Dice similarity coefficient (Dice, Eq. [3]) and Jaccard similarity coefficient (Jaccard, Eq. [4]) were used to evaluate the general accuracy of the mass segmentation. The values of these 4 metrics are between 0 and 1. They are defined as follows:
where TP, FP, and FN denote the pixel numbers of true positives, false positives, and false negatives, respectively. The GT denotes the ground truth, SR denotes the segmented result, and |∙| denotes the region size.
Experimental setup
All the models used in this study were implemented using the Python 3.6 programming language with Tensorflow and Keras libraries, and all experiments were performed on NVIDIA TITAN X graphics processing unit (GPU). The training parameters and training strategy of the models are outlined in Table 1, in which each assignment is optimal for each model. Additionally, each model was trained with a training set after data augmentation.
Table 1
| Methods | Batch size | Training epochs | No. of trainable variables | Loss function | Optimizer | Learning rate |
|---|---|---|---|---|---|---|
| U-Net | 2 | 30 | 31,031,685 | Cross entropy | Adam | 0.00001 |
| A-UNet | 2 | 30 | 31,380,805 | Cross entropy | Adam | 0.00001 |
| S-UNet | 2 | 30 | 32,427,333 | Cross entropy | Adam | 0.00001 |
| AS-UNet | 2 | 30 | 32,242,885 | Cross entropy | Adam | 0.00001 |
| VGG16-UNet | 2 | 30 | 38,337,473 | Cross entropy | SGD | 0.01 |
| A-VGG16-UNet | 2 | 30 | 40,493,377 | Cross entropy | SGD | 0.01 |
| S-VGG16-UNet | 2 | 30 | 39,290,177 | Cross entropy | SGD | 0.01 |
| AS-VGG16-UNet | 2 | 30 | 41,880,769 | Cross entropy | SGD | 0.01 |
SGD, stochastic gradient descent.
Results
In this study, we proposed and constructed 8 models based on U-Net and VGG. For each model, 5-fold cross-validation was performed on the training set, and the test set was used to illustrate the effectiveness of the trained models. The evaluation metrics mentioned in the Methods section were used to compare the segmentation results of the 8 models and ground truths. The metrics were calculated for each image in the test set, and all values of the evaluation metrics were presented as mean ± standard deviation. Paired t-tests were performed on the metrics’ values of different models. We also compared our proposed methods with 3 other methods, including ResUNet (48), MultiResUNet (49), and UNet++ (50).
Evaluation of segmentation using U-Net and the proposed U-Net variants
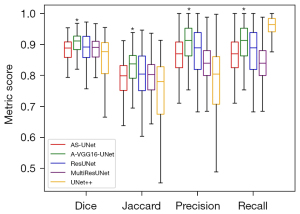
Table 2 shows the 4 evaluation metrics used to evaluate and analyze the segmentation performance of the U-Net and its 3 variants. The 3 variants of U-Net had higher scores than U-Net on Dice, Jaccard, and Precision. The A-UNet had significantly higher values of Dice, Jaccard, and Recall (0.869±0.054, 0.772±0.082, and 0.932±0.040, P<0.01, respectively) and outperformed S-UNet; AS-UNet had significantly higher values of Dice, Jaccard, and Precision (0.881±0.038, 0.789±0.059 and 0.865±0.065, P<0.01, respectively) and performed better than A-UNet. The quantitative segmentation results suggested that AS-UNet had the best performance.
Table 2
| Methods | Metrics | |||
|---|---|---|---|---|
| Dice | Jaccard | Precision | Recall | |
| U-Net | 0.846±0.048 | 0.739±0.071 | 0.788±0.072 | 0.926±0.036 |
| A-UNet | 0.869±0.054+ | 0.772±0.082+ | 0.819±0.084 | 0.932±0.040+ |
| S-UNet | 0.855±0.056 | 0.751±0.084 | 0.871±0.085 | 0.845±0.067 |
| AS-UNet | 0.881±0.038* | 0.789±0.059* | 0.865±0.065* | 0.901±0.043 |
| VGG16-UNet | 0.868±0.047 | 0.769±0.072 | 0.792±0.077 | 0.965±0.028 |
| A-VGG16-UNet | 0.904±0.035** | 0.826±0.057** | 0.905±0.061** | 0.909±0.061 |
| S-VGG16-UNet | 0.891±0.044 | 0.806±0.071 | 0.834±0.077 | 0.962±0.033 |
| AS-VGG16-UNet | 0.893±0.038 ++ | 0.810±0.061 ++ | 0.879±0.061 ++ | 0.912±0.046 |
+, the statistically significant improvement at P<0.01 vs. S-UNet; *, the statistically significant improvement at P<0.01 vs. A-UNet; ++, the statistically significant improvements at P<0.01 vs. S-VGG16-UNet; **, the statistically significant improvement at P<0.01 vs. the other three VGG16-UNet models.
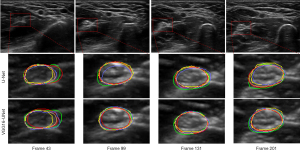
As shown in the second row of Figure 4, the segmentation contours produced by A-UNet (red), S-UNet (purple), and AS-UNet (orange) were closer to the ground truths (yellow), compared to U-Net (green). The results demonstrated the significance of the attention mechanism and the residual module to improve the performance of the U-Net model.
Evaluation of Segmentation using the proposed VGG16-UNet and its variants
The experiment results of the proposed VGG16-UNet and its variants were quantitatively evaluated with evaluation metrics. As shown in Table 2, all 3 variants of VGG16-UNet performed better compared with VGG16-UNet. Similar to U-Net and its variants, the results reconfirmed that the attention mechanism and the residual module can enhance the performance of the deep learning model for segmentation. The AS-VGG16-UNet model had significantly higher values of Dice, Jaccard, and Precision (0.893±0.038, 0.810±0.061, and 0.879±0.061, P<0.01) and performed better than S-VGG16-UNet. Interestingly, the segmentation results of A-VGG16-UNet obtained the highest results (Dice of 0.904±0.035, Jaccard of 0.826±0.057, and Precision of 0.905±0.061; P<0.01).
The segmentation results produced by VGG16-UNet and its variants are illustrated in the third row in Figure 4. Compared to A-VGG16-UNet (red), S-VGG16-UNet (purple), and AS-VGG16-UNet (orange), the segmentation contours of VGG16-UNet (green) expressed the largest difference with the ground truths (yellow). This is consistent with the quantitative results shown in Table 2.
Evaluation of architectures of U-Net and VGG16-UNet
The results in Table 2 indicate that VGG16-UNet and its variants performed better than the corresponding U-Net and its variants. With more convolutional layers, VGG16-UNet could extract higher dimensional image representations by processing local information layer by layer in comparison with U-Net.
Interestingly, both A-UNet and S-UNet improved the Precision, but S-UNet greatly reduced the Recall. The reason might be that the U-Net used in this study could not perform well on feature extraction for the details inside the median nerve. Adding the attention mechanism or the residual module to U-Net improved the Precision, and the Precision of S-UNet increased more than that of A-UNet. This indicated that the residual module could supplement the missing information due to insufficient network depth and could extract more details of features. However, doing so resulted in under-segmentation to some extent, which increased false negatives and therefore decreased Recall. Furthermore, since VGG16-UNet could already extract the features of the internal details of the median nerve, adding the residual module to VGG16-UNet made little difference to its original feature extraction ability and then had a limited impact on the Recall.
Comparison of AS-UNet and A-VGG16-UNet with other methods
The test set of the median nerve image sequence was used to test different methods. Figure 5 shows the comparison of evaluation results of AS-UNet, A-VGG16-UNet, ResUNet, MultiResUNet, and UNet++. The metric scores of Dice, Jaccard, and Precision of A-VGG16-UNet were significantly improved in comparison with the other four methods (P<0.05). Recall of A-VGG16-UNet was significantly improved in comparison with AS-UNet, ResUNet, and MultiResUNet (P<0.05).
3D reconstruction and visualization using VTK based on the segmentation results
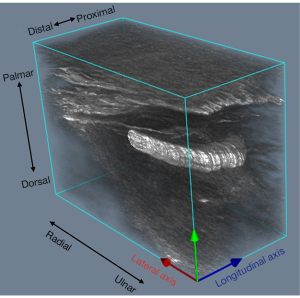
Since A-VGG16-UNet achieved the best performance, the segmentation results of the image sequence in the test set were produced by the trained A-VGG16-UNet model and then used for 3D reconstruction. The segmentation results and the corresponding image sequence were blended with different weights to highlight the general shape and travel direction of the median nerve in 3D visualization. The blending results were input as a volume into the system using the function vtkJPGReader. Therefore, 3D median nerve reconstruction and visualization were obtained by using the volume rendering method in VTK.
As shown in Figure 6, the significantly highlighted area illustrates the general shape and travel direction of the median nerve from the distal to the proximal on the forearm (Figure 1). The 3D reconstruction was displayed from different perspectives to view the morphological features of the median nerve.
Discussion
Deep learning methods based on VGG and U-Net have been used for medical or natural image segmentation (24-26,51,52), but seldom for nerve segmentation in ultrasound images. The main challenges are the low contrast of ultrasound images, the tiny and inconspicuous size of the nerve, and imaging noise. Since VGG16 is quite simple and highlight for having only small convolutional filters (23), and U-Net possesses the ability of precise pixel-level localization, we proposed a novel model named VGG16-UNet based on VGG16 and U-Net. Meanwhile, the attention mechanism and the residual module were applied to U-Net and VGG16-UNet, to produce their corresponding variants (A-UNet, S-UNet and AS-UNet, A-VGG16-UNet, S-VGG16-UNet and AS-VGG16-UNet). The spatial attention mechanism can prevent missing pixel-level information and improve the accuracy of feature extraction (38). The residual module can prevent the vanishing gradient problem by applying identity mapping to facilitate the training process (36).
The experimental segmentation results of the median nerve dataset showed that, for both U-Net and VGG16-UNet, the models constructing with the attention mechanism and/or the residual module performed better than their original models. This demonstrated that the 2 additions can leverage more learned features between the layers to improve the performance of the models. The attention mechanism can transform the original image’s spatial information into another space while retaining essential information or properties, and the residual module can preserve the spatial information lost during the pooling operation to alleviate the disparity between the encoder-decoder features. Furthermore, the model with the attention mechanism was superior to that with the residual module for both U-Net and VGG16-UNet. This showed that the attention mechanism can integrate features from the encoder and decoder, while the residual module uses identity mapping to add the input to the output of the stacked layers.
The proposed VGG16-UNet combines the characteristics of U-Net and VGG, which improves the performance of U-Net. Furthermore, the variants of VGG16-UNet outperformed the corresponding variants of U-Net, respectively, which confirmed the effectiveness of the proposed VGG16-UNet in median nerve ultrasound image segmentation.
Interestingly, this study found that AS-VGG16-UNet outperformed AS-UNet, which indicated that VGG16 as the encoder can improve feature extraction for segmentation. However, the performance of AS-VGG16-UNet was slightly lower than that of A-VGG16-UNet. The possible reason might be that the fusion of features in different scales in the encoder degrades the representation of the originally extracted features. It has been demonstrated that the features in different scales extracted by VGG-like architecture greatly represent the characteristics of the corresponding level (53). Therefore, adding residual modules to the encoder of A-VGG16-UNet may influence its primarily extracted features, and consequently affect the performance of A-VGG16-UNet.
Finally, with a trained A-VGG16-UNet model, the automatic segmentation of the median nerve in the image sequence was obtained and used for 3D reconstruction. The morphology and travel direction of the median nerve was displayed in 3D visualization. Therefore, the corresponding alternations induced by nerve trauma or carpal tunnel syndrome could be detected. From this, the swelling or compression and the nerve continuity can be evaluated and provide information for diagnosis and follow-up rehabilitation (54,55).
Previous studies paid more attention to the median nerve in the carpal tunnel, and the images were acquired with the probe positioned at the carpal tunnel inlet. Many models, such as U-Net-shaped, FPN, Mask-R-CNN, and DeepNerve, were used for segmentation of the median nerve. Few studies have applied VGG models in median nerve segmentation. Additionally, although the image sequences of the median nerve were originally acquired in (13-15), the segmentation results were displayed in 2D image frames without providing the 3D information of the nerve. In this study, we scanned the median nerve of the forearm by moving the probe from the distal to the proximal. We built VGG16-UNet models with/without the attention mechanism and/or the residual module for automatic segmentation of the median nerve in an image sequence, and then a 3D image of the median nerve was created to visualize morphology and the location of the median nerve from different perspectives.
Although the results were promising, this study had limitations in accuracy and performance of the proposed networks. The data for model training was limited and only came from healthy participants, which might have resulted in overfitting of the model for less-represented features. The model would be less effective in the segmentation of ultrasound data from unhealthy participants. Therefore, expanding the dataset and including the data from a wider range of patients is required to improve the practicability of the models. In addition, only the ultrasound images of the median nerve on the forearm were used in the experiments, so it might be difficult to generalize these findings to other types of nerve. Therefore, the performance of the proposed networks should be evaluated and assessed with more types of healthy and injured nerves in future work.
Acknowledgments
Funding: This work was supported in part by the Guangdong Science and Technology Program (No. 2016A020216017), the Program of Pearl River Young Talents of Science and Technology in Guangzhou (No. 201610010011), and the National Natural Science Foundation of China (No. 81371560).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1074/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1074/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University and informed consent was provided by all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fuse H, Sumiya H, Ishii H, Shimazaki J. Treatment of hemospermia caused by dilated seminal vesicles by direct drug injection guided by ultrasonography. J Urol 1988;140:991-2. [Crossref] [PubMed]
- Barrington MJ, Kluger R. Ultrasound guidance reduces the risk of local anesthetic systemic toxicity following peripheral nerve blockade. Reg Anesth Pain Med 2013;38:289-99. [Crossref] [PubMed]
- Zeidenberg J, Burks SS, Jose J, Subhawong TK, Levi AD. The utility of ultrasound in the assessment of traumatic peripheral nerve lesions: report of 4 cases. Neurosurg Focus 2015;39:E3. [Crossref] [PubMed]
- Bilgici A, Cokluk C, Aydın K. Ultrasound neurography in the evaluation of sciatic nerve injuries. J Phys Ther Sci 2013;25:1209-11. [Crossref] [PubMed]
- Fantoni C, Erra C, Fernandez Marquez EM, Ortensi A, Faiola A, Coraci D, Piccinini G, Padua L. Ultrasound Diagnosis of Postoperative Complications of Nerve Repair. World Neurosurg 2018;115:320-3. [Crossref] [PubMed]
- Goedee HS, van der Pol WL, Hendrikse J, van den Berg LH. Nerve ultrasound and magnetic resonance imaging in the diagnosis of neuropathy. Curr Opin Neurol 2018;31:526-33. [Crossref] [PubMed]
- Chang KV, Mezian K, Naňka O, Wu WT, Lou YM, Wang JC, Martinoli C, Özçakar L. Ultrasound Imaging for the Cutaneous Nerves of the Extremities and Relevant Entrapment Syndromes: From Anatomy to Clinical Implications. J Clin Med 2018;7:457. [Crossref] [PubMed]
- Chang KV, Kim SB. Editorial: Use of Ultrasound in Diagnosis and Treatment of Peripheral Nerve Entrapment Syndrome. Front Neurol 2020;10:1348. [Crossref] [PubMed]
- Smistad E, Iversen DH, Leidig L, Lervik Bakeng JB, Johansen KF, Lindseth F. Automatic Segmentation and Probe Guidance for Real-Time Assistance of Ultrasound-Guided Femoral Nerve Blocks. Ultrasound Med Biol 2017;43:218-26. [Crossref] [PubMed]
- Hadjerci O, Hafiane A, Conte D, Makris P, Vieyres P, Delbos A. Computer-aided detection system for nerve identification using ultrasound images: a comparative study. Inform Med Unlocked 2016;3:29-43. [Crossref]
-
Hafiane A Vieyres P Delbos A Deep learning with spatiotemporal consistency for nerve segmentation in ultrasound images. Available online: https://arxiv.org/abs/1706.05870 -
Ding Y Yang Q Wu G Zhang J Qin Z. Multiple Instance Segmentation in Brachial Plexus Ultrasound Image Using BPMSegNet. Available online: https://arxiv.org/abs/2012.12012 - Horng MH, Yang CW, Sun YN, Yang TH. DeepNerve: A New Convolutional Neural Network for the Localization and Segmentation of the Median Nerve in Ultrasound Image Sequences. Ultrasound Med Biol 2020;46:2439-52. [Crossref] [PubMed]
- Festen RT, Schrier VJMM, Amadio PC. Automated Segmentation of the Median Nerve in the Carpal Tunnel using U-Net. Ultrasound Med Biol 2021;47:1964-9. [Crossref] [PubMed]
- Wu CH, Syu WT, Lin MT, Yeh CL, Boudier-Revéret M, Hsiao MY, Kuo PL. Automated Segmentation of Median Nerve in Dynamic Sonography Using Deep Learning: Evaluation of Model Performance. Diagnostics (Basel) 2021;11:1893. [Crossref] [PubMed]
- Lee CY, Chang TF, Chou YH, Yang KC. Fully automated lesion segmentation and visualization in automated whole breast ultrasound (ABUS) images. Quant Imaging Med Surg 2020;10:568-84. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, 2015.
- Kakade A, Dumbali J, editors. Identification of nerve in ultrasound images using u-net architecture. Mumbai, India: 2018 International Conference on Communication information and Computing Technology (ICCICT), 2018.
- Rubasinghe I, Meedeniya D. Ultrasound nerve segmentation using deep probabilistic programming. Journal of ICT Research and Applications 2019;13:241-56. [Crossref]
- Wang Y, Geng J, Zhou C, Zhang Y, editors. Segmentation of Ultrasound Brachial Plexus Based on U-Net. 2021 International Conference on Communications, Information System and Computer Engineering (CISCE), 2021.
- Wu H, Liu J, Wang W, Wen Z, Qin J, editors. Region-aware Global Context Modeling for Automatic Nerve Segmentation from Ultrasound Images. Vancouver, Canada: Proceedings of the AAAI Conference on Artificial Intelligence, 2021.
-
Simonyan K Zisserman A Very deep convolutional networks for large-scale image recognition. Available online: https://arxiv.org/abs/1409.1556 -
Inan MSK Alam FI Hasan R Deep Integrated Pipeline of Segmentation Leading to Classification for Automated Detection of Breast Cancer from Breast Ultrasound Images. Available online: https://arxiv.org/abs/2110.14013 -
Iglovikov V Shvets A. Ternausnet: U-net with vgg11 encoder pre-trained on imagenet for image segmentation. Available online: https://arxiv.org/abs/1801.05746 - Pravitasari AA, Iriawan N, Almuhayar M, Azmi T, Fithriasari K, Purnami SW, Ferriastuti W. UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation. Telkomnika 2020;18:1310-8. [Crossref]
-
Balakrishna C Dadashzadeh S Soltaninejad S Automatic detection of lumen and media in the IVUS images using U-Net with VGG16 Encoder. Available online: https://arxiv.org/abs/1806.07554 - Haque MF, Lim HY, Kang DS, editors. Object detection based on VGG with ResNet network. Auckland, New Zealand: 2019 International Conference on Electronics, Information, and Communication (ICEIC), 2019.
- Geng L, Zhang S, Tong J, Xiao Z. Lung segmentation method with dilated convolution based on VGG-16 network. Comput Assist Surg (Abingdon) 2019;24:27-33. [Crossref] [PubMed]
- Bishop CM. Neural networks for pattern recognition. Oxford, England: Oxford University Press, 1995.
- Ripley BD. Pattern recognition and neural networks. Cambridge, England: Cambridge University Press, 2007.
- Venables WN, Ripley BD. Modern applied statistics with S-PLUS. Springer Science & Business Media, 2013.
- He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Las Vegas, Nevada, USA: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016.
-
Srivastava RK Greff K Schmidhuber J Training very deep networks. Available online: https://arxiv.org/abs/1507.06228 -
Liu T Chen M Zhou M Du SS Zhou E Zhao T Towards understanding the importance of shortcut connections in residual networks. Available online: https://arxiv.org/abs/1909.04653 - Singadkar G, Mahajan A, Thakur M, Talbar S. Deep Deconvolutional Residual Network Based Automatic Lung Nodule Segmentation. J Digit Imaging 2020;33:678-84. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J, editors. Identity mappings in deep residual networks. European conference on computer vision. Springer, 2016.
- Oyedotun OK, Aouada D, Ottersten B, editors. Training very deep networks via residual learning with stochastic input shortcut connections. International Conference on Neural Information Processing. Springer, 2017.
- Ni Y, Xie Z, Zheng D, Yang Y, Wang W. Two-stage multitask U-Net construction for pulmonary nodule segmentation and malignancy risk prediction. Quant Imaging Med Surg 2022;12:292-309. [Crossref] [PubMed]
- Guo S, Jin Q, Wang H, Wang X, Wang Y, Xiang S. Learnable gated convolutional neural network for semantic segmentation in remote-sensing images. Remote Sensing 2019;11:1922. [Crossref]
-
Oktay O Schlemper J Folgoc LL Lee M Heinrich M Misawa K Mori K McDonagh S Hammerla NY Kainz B Attention u-net: Learning where to look for the pancreas. Available online: https://arxiv.org/abs/1804.03999 - Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, editors. Attention is all you need. Advances in Neural Information Processing Systems. Curran Associates, Inc., 2017.
- Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, Rueckert D. Attention gated networks: Learning to leverage salient regions in medical images. Med Image Anal 2019;53:197-207. [Crossref] [PubMed]
- Schroeder WJ, Avila LS, Hoffman W. Visualizing with VTK: a tutorial. IEEE Comput Graph Appl 2000;20:20-7. [Crossref]
- Russell BC, Torralba A, Murphy KP, Freeman WT. LabelMe: a database and web-based tool for image annotation. Int J Comput Vis 2008;77:157-73. [Crossref]
- Hafizah M, Kok T, Spriyanto E, editors. 3D ultrasound image reconstruction based on VTK. Catania, Italy: Proceedings of the 9th WSEAS International Conference on SIGNAL Processing, 2010.
- Brown A, Wilson G. The Architecture of Open Source Applications: Elegance, Evolution, and a Few Fearless Hacks. Lulu.com, 2011.
- Xu LQ, Pu F, Li SY, Li D, Fan YB, editors. Three-dimensional reconstruction and analysis of human central sulcus based on visualization toolkit. Shanghai, China: 2008 2nd International Conference on Bioinformatics and Biomedical Engineering, 2008.
- Liang S. Research on Breast Ultrasound Image Segmentaion Based on Residual U-shaped Convolution Neural Network [Master’s dissertation]. Guangzhou: South China University of Technology, 2018.
- Ibtehaz N, Rahman MS. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw 2020;121:74-87. [Crossref] [PubMed]
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans Med Imaging 2020;39:1856-67. [Crossref] [PubMed]
- Ghosh S, Chaki A, Santosh KC. Improved U-Net architecture with VGG-16 for brain tumor segmentation. Phys Eng Sci Med 2021;44:703-12. [Crossref] [PubMed]
- Kadry S, Rajinikanth V, Taniar D, Damaševičius R, Valencia XPB. Automated segmentation of leukocyte from hematological images—a study using various CNN schemes. J Supercomput 2021; [Crossref]
- Ding X, Zhang X, Ma N, Han J, Ding G, Sun J, editors. RepVGG: Making VGG-style convnets great again. Nashville, TN, USA: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2021.
- Wijntjes J, Borchert A, van Alfen N. Nerve Ultrasound in Traumatic and Iatrogenic Peripheral Nerve Injury. Diagnostics (Basel) 2020;11:30. [Crossref] [PubMed]
- Schminke U. Ultrasonography of peripheral nerves–Clinical significance. Perspect Med 2012;1:422-6. [Crossref]



