
Synthesis of magnetic resonance images from computed tomography data using convolutional neural network with contextual loss function
Introduction
Magnetic resonance imaging (MRI) provides a wide range of soft-tissue contrast, such as T1-weighted images (T1WI) for anatomical structures and T2-weighted images (T2WI) for identifying lesions (1,2). Computed tomography (CT) has a lower discrimination between different soft tissues with less defined contours than MRI because of its relatively limited soft tissue contrast. Therefore, an MRI examination is important for an accurate diagnosis in medical scenarios where only CT images are available (3). Synthesis of MR images from CTs can assist clinicians in making medical decisions in such cases without MR images. In emergency treatment, MRI use is constrained by its limited availability and time cost (4). Real-time synthesis of MRI from CT images reduces the time cost for acquiring MRI data allowing timely diagnosis for acutely ill patients. Another limitation of MRI is its use in populations with metal implants or claustrophobia. Synthetic MRI eliminates the risk of displacement of pacemakers, joint prostheses, coronary stents, etc. (5), and avoids unnecessary discomfort in patients with claustrophobia during the actual process of MRI. More importantly, although CT is mainly employed for accurate target localization and dose calculation during radiotherapy treatment planning system (TPS), MRI may be beneficial for obtaining a more accurate picture of target structures for TPS compared to relying on CT images alone (6-8).
At present, most synthetic medical images are CT images derived from standard MR scans. There are generally three synthetic methods: image segmentation based on cluster statistics of different tissues (9-11), image registration based on associated MR/CT atlas (12,13), and deep learning based on convolutional neural network (CNN), including U-Net and generative adversarial networks (GAN) (14-18). The published studies cited here show that accurate synthetic CT images could be generated from single-sequence MR images in near real-time.
Compared with the published studies on the generation of synthetic CT images, there are few deep learning methods used to generate synthetic MR images (14,19,20). Among these studies, the network structure of CNN and loss function are two main factors that influenced the performance of synthetic MR images. For the former, Li et al. respectively used CycleGAN and U-Net to generate synthetic brain MR/CT images from their counterpart modality (14), the results of the quality of synthetic images by U-Net was higher than those of CycleGAN, it indicated that U-Net outperformed CycleGAN. In addition, synthesizing MR images from CT scans is currently limited because of the unclear boundary of soft tissues in CT (9) and the weakness of some loss functions in dealing with high-frequency information (21). There is high soft-tissue contrast in MR images compared to its CT counterparts (22), which means that more high-frequency soft-tissue information exists in MR images. For synthetic CT images from MRI, a structured-consistency loss function can be defined to process high-frequency data in MRI, but it is difficult to generate high-level synthetic MR images from low-level CT images (15). Therefore, the loss function is an important factor that can affect the performance of a CNN. There are two types of commonly used loss functions for optimizing the generated values of synthetic MR images: pixel-to-pixel loss function and global loss function (23). The former compares the predicted and actual values pixel-by-pixel under the same spatial coordinates to obtain characteristics such as mean square error (MSE) (17,20,24), binary cross-entropy (BCE) (25), etc. The global loss function can capture image features by comparing the statistics collected over the entire image. Specifically, the perceptual loss aims to synthesize T1WI planning CTs using deep learning (DL)-based frameworks, U-Net and CycleGAN (24). The contextual loss function has been used in a fully convolutional neural network (FCN) to generate pseudo-CTs from MRI, which confirms that it can improve the predicted performance of the CNN without changing the network architecture (26).
The main purpose of this study is to design a novel deep CNN, which is called Double ResNet-U-Net (DRUNet), with contextual loss by combining the strengths of both ResNet and U-Net. DRUNet is a general supervised learning system used for the synthesis of MRI. The synthetic MR images, including T1WI and T2WI from CT scans, achieved the desired results. More detailed comparisons and analyses of the proposed network and loss function are discussed in the following sections. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-846/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study passed the ethical approval of The First Affiliated Hospital of Shenzhen University’s Bioethics Committee, and the participants all signed the informed consent form. Data processing and model training for different procedures involve multiple systems and redundant processing. A general flowchart including the acquisition of multi-model images (CT, T1WI, and T2WI), image registration, image enhancement, image denoising, and model training is shown in Figure 1. The individual procedures are introduced in detail in the following sections.
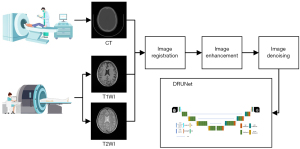
Data acquisition
The datasets came from Shenzhen Second People’s Hospital, China during the period from January 1, 2017 to December 30, 2019. The inclusion and exclusion criteria are as follows. The inclusion criteria: (I) age between 18 and 60 years old; (II) no hypertension and diabetes; (III) no trauma to the head. The exclusion criteria: (I) failure to complete MRI and CT examinations; (II) the presence of artifacts in the image leads to poor image quality; (III) presence of brain tumors, hemorrhage, infarct, and other diseases.
Initially, a total of 45 participants were selected, but five participants had hypertension or diabetes, two participants suffered severe head trauma in the past, one participant could not complete the MRI examination, one participant had poor quality brain CT or MRI images, and one participant had a brain tumor. Thus, 35 participants who underwent brain CT and MRI examinations at the same time were finally included.
For the specific acquisition parameters of MRI and CT, T1WI was acquired from 3D MPRAGE sequences of the transverse section using the following acquisition parameters: TR =1,600 ms [this refers to the MPRAGETR between two non-selective (180º) inversion pulses (27)], TE =3.37 ms, pixel size =1×1×1 mm3, image size =256×256, acquisition range =160 mm, turbo factor =125. T2WI was acquired from 3D SPACE sequences of the transverse section using the following acquisition parameters: TR =2,500 ms, TE =123 ms, pixel size =1×1×1 mm3, image size =256×256, acquisition range =160 mm, turbo factor =125. Both were acquired by GRAPPA with R-factor =2 on a 1.5T Avanto scanner (Siemens). CT images (120 kV, 330 mA, exposure time =500 ms, pixel size =0.5×0.5×1 mm3, image size =512×512) were acquired using SOMATOM Definition Flash (Siemens).
Image preprocessing
- Image registration. For the same patient, the CT/MR image pair (CT/T1WI or CT/T2WI) was aligned with a linear affine registration algorithm using FSL software (28). The affine registration simulates the global motion of viscera and improves visualization of soft tissues in the course of medical treatment (29,30).
- Image mask. Because large amounts of low-level noise exist in non-brain regions of CT images (Figure 2A), image masks for the brain region will reduce the noise impact on experiment performance. In this study, the watershed algorithm (31) conducted on the original CT images (Figure 2A) yielded masks (Figure 2B) to eliminate noise from non-brain regions for CT, T1WI and T2WI as shown in Figure 2C-2E.
- Image enhancement. The original CT images of brain soft tissue have low contrast, and image enhancement can increase it in soft tissues and benefit image computation. After truncating and adjusting the window width of CT images appropriately according to the Hounsfield unit (HU) values of different organs, the numerical difference of brain soft tissue in CT data was greatly increased, enhancing the contrast of regions of interest against bone windows. The HU histogram of the adjusted CT image provided more detailed information about soft tissues (Figure 2F).
DRUNet model
DRUNet combines U-Net (16) and ResNet (32) to generate synthetic MR images from CT images. The U-shaped model is usually used to generate pixel-level segmentation results based on the codec structure, that is, the encoder and decoder. Image segmentation is a special form of image generation. U-Net can be used for image generation under certain conditions, and it guarantees the optimal transfer of spatial information from input to output images (33). Two types of residual networks were constructed for the proposed U-shaped structure to take advantage of ResNet and U-Net compatibility.
When ResNet was designed as the encoder of the U-Net to extract complex features from the input CT images hierarchically, ResNet-18/34 used the basic blocks and ResNet-50/101/152 used the bottleneck block. There are two types of basic or bottleneck blocks, residual blocks with downsampling (Figure 3A,3B), and the residual blocks without downsampling (Figure 3C,3D). Specifically, Figure 3A,3C show bottleneck blocks; however, the former undergoes the process of downsampling by convolution (kernel =3, stride =2, padding =1) as shown by the dark grey trapezoid block in Figure 3A, and a residual block with a downsampling convolution (kernel =1, stride =2) as shown by the light grey trapezoid block in Figure 3A. Figure 3B,3D show basic blocks, but that in Figure 3B has an identical downsampling process, which is also illustrated by the light grey trapezoid blocks. To distinguish between them in the following design of the decoder, Figure 3A is called a down-bottleneck, Figure 3B is a down-basic block, Figure 3C is a bottleneck block, and Figure 3D is a basic block.
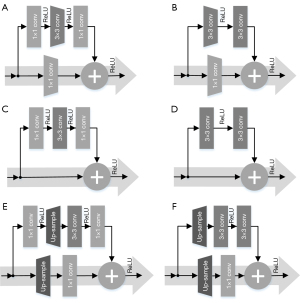
When ResNet was designed as the decoder of the U-Net, the decoding was transformed in a concatenated manner and the convolutional features from the decoder were fused with the corresponding features from the encoder. Then, the predicted MR images were gradually reconstructed from low to high resolution. To match the hierarchical structure of our encoder, the decoder uses a series of residual networks to generate synthetic images, reverse the entire convolutional component of the encoder, and replace the downsampling convolution of the encoder with nearest-neighbor interpolation upsampling to retrieve the output images where the corresponding changed residual blocks are called up-bottleneck or up-basic blocks. As shown in Figure 3E, the up-bottleneck block of the encoder was a variant of the down-bottleneck of the decoder, which substituted convolutional downsampling (kernel size =3, stride step =2, and padding =1) in Figure 3A with a combination of upsampling (scale =2) and convolution operation (kernel size =1, stride step =1), the operation of upsampling is labeled with the opposite trapezoid in Figure 3A. The same rule was applied to the basic block as shown in Figure 3F. Although deconvolution (i.e., transposed convolution) follows a similar procedure as upsampling, it generates checkerboard pattern artifacts if the stride step and kernel size are improper, thus resulting in uneven overlap during deconvolution (34). To avoid artifact formation, nearest-neighbor interpolation was used to double the output size of the last feature layer, and then a convolution (kernel size =1 and stride step =1) was performed. The integration of interpolation and convolution reduced the checkerboard artifacts. In addition, the skip connection, or concatenation, is a unique structure in U-Net, which ensures that the recovered feature maps can incorporate more low-level features from different scales (35). In the start phase of each scale block in the decoder, the concatenation of the last layer output and the corresponding same-scale output from the encoder doubled the feature size of this layer, so input channels were half the amount of output channels to match the feature size at the beginning of each scale layer. This is called the cate-bottleneck block.
Taking DRUNet-101 as an example (Figure 4), it has a symmetrical structure and four layers for both the encoder and decoder. The layers of equivalent hierarchy between the encoder and decoder have the same characteristic scale, and the scale between adjacent hierarchical layers has a double size relationship from top to bottom. The detailed feature scales of DRUNet-101, such as layer name, module name, input shape, and output shape, are described in Table 1. The convolutional layers of DOWN 4 and UP 1, for instance, have the main body structure of size 28×28, the former feature size is changed from 512×56×56 to 1,024×28×28. Inversely, the latter layer concatenated with upper layer is changed from 2,048×28×28 to 512×56×56. The DOWN 3 and UP 2 has a feature scale of 56×56, which is double of that in DOWN 4 and UP 1. D-CENTER and U-CENTER are the lowest-level structures that are responsible for handling high-dimensional features. Owing to the excellent performance of ResNet in image classification and recognition, the number of bottlenecks of ResNet in each layer was transferred to the designed CNN model. Thus, the blocks in each layer of DRUNet-101 have the numbers 3, 4, 23, 3, 3, 23, 4, and 3 in sequence, similar to the number of convolutional components of ResNet-101 (i.e., 3, 4, 23, 3), but with the addition of the mirror structures behind.
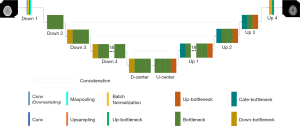
Table 1
| Layer name | Input shape | Output shape |
|---|---|---|
| DOWN 1 | 3×448×448 | 64×224×224 |
| DOWN 2 | 64×224×224 | 256×112×112 |
| DOWN 3 | 256×112×112 | 512×56×56 |
| DOWN 4 | 512×56×56 | 1,024×28×28 |
| D-CENTER | 1,024×28×28 | 2,048×14×14 |
| U-CENTER | 2,048×14×14 | 1,024×28×28 |
| UP 1 | 2,048×28×28 | 512×56×56 |
| UP 2 | 1,024×56×56 | 256×112×112 |
| UP 3 | 512×112×112 | 64×224×224 |
| UP 4 | 64×224×224 | 1×448×448 |
DRUNet-101, double ResNet-U-Net with double 101 convolutional layers.
Loss function
The loss function measures the similarity between the generated image and the target image during both medical image generation and reconstruction (36). It can be classified into two types: pixel-to-pixel loss functions and global loss functions. When compared in terms of appearance, pixel-to-pixel faces problems such as the lack of high-frequency information and over-smoothness in MSE and BCE loss (21). Contextual loss was initially proposed for super-resolution reconstruction of non-aligned data, which makes the generated image sharper and brighter (23,37). The contextual information in an image shows the importance of the proposed deep learning model (38). The key idea of contextual loss is to consider an image as a collection of features and apply a similarity measure to these features instead of measuring spatial location. In this study, contextual loss was compared with MSE loss and BCE loss to test individual performance. We describe these three loss functions in this section.
MSE loss
MSE loss is the squared L2-norm, also known as the least-squares error (LSE). It minimizes the sum of the squares of the differences between the real MR images IMR and the synthetic MR images from the CT scans SynMR(ICT). For the M×N images:
BCE loss
Here, the loss function was used by combining a “Sigmoid” layer with a “BCE loss” in one single class, which is more numerically stable than a plain “Sigmoid” followed by a “BCE loss”. The log-sum-exponent was calculated to evaluate the numerical stability of the BCE loss.
Contextual loss
Contextual loss calculates the similarity between the real MR images IMR and synthetic MR images SynMR(ICT) using the following mathematical formulas (23,26):
First, dij is the raw distance, which represents the cosine distance between xi and yj,
where si=fvgg[SynMR(ICT)] and rj=fvgg(TMR) are the feature points extracted from the layer conv5_4, which is the third convolution layer inside the fifth convolutional block, of VGG-19 for synthetic MR images and real MR images, and is the mean of the N feature points of the real MR images.
As shown in Figure 5, the blue circle represents the synthetic features of the generated MR images in Figure 5A, the yellow star represents the real features of the real MR images in Figure 5B, and CXij is the contextual similarity between the prediction and target in Figure 5C.

Then, the raw distance is normalized to get the relative distance:
where ε=1e−5.
The next variable is exponent distance, which is derived from shifting the relative distance to similarities by exponentiation, which can magnify the distance measure:
where h =0.5 is a bandwidth parameter.
Then, the contextual similarity between feature points was defined as the normalized similarities of the exponent distance.
As depicted in Figure 5, transforming data into a high-dimensional feature space of a pre-trained model (VGG19) is the key idea to guarantee the similarity measure of both aligned and non-aligned data. The similarity measure can then be directly applied to high-dimensional feature points.
Finally, negative logarithms are used as the final texture loss function
Evaluation of DRUNet
To evaluate the performance of the proposed DRUNet, peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and Tenenbaum gradient (Tenengrad) were calculated with the synthetic MRI images against the real MRI images for each patient. The definitions of PSNR, SSIM, and Tenengrad are briefly described as follows.
PSNR is often used to measure the quality of signal reconstruction, such as image synthesis, and is often defined simply by the MSE. For the real MR images IMR and the synthetic MR images from CT images SynMR(ICT) of size M×N with B bits, the MSE between them is defined as Eq. [1]; then,
SSIM is also a full reference image quality evaluation index that can measure image similarity in terms of brightness, contrast, and structure.
where μI and μS denote the mean of images IMR and SynMR(ICT), σI and σS indicate the variance of images IMR and SynMR(ICT), and σIS calculates the covariance of images IMR and SynMR(ICT). c1 and c2 are constants included to avoid division by 0.
Tenengrad is a gradient function used to evaluate image clarity without a reference image (39). It calculates the horizontal and vertical gradient values using the Sobel operator. There are more high-frequency signals in well-focused images and they have sharper edges and clearer details. The definition of image sharpness based on Tenengrad is as follows,
where T is the given threshold of edge detection, and Gi and Gj are the convolutions of Sobel operators of horizontal and vertical edge detection at pixel point (i,j) respectively.
Root mean squared error (RMSE) is commonly used for measuring the error rate of regression models. In there, RMSE was used to evaluate the performance of DRUNet on cross validation.
where SynMR(ICT) is the synthetic MR images from CT images, and IMR is the real MR images, i represent the pixel position, and m is the total number of pixels in both synthetic MR images and real MR images.
To ensure that differences in other factors are not masking or enhancing a significant difference in means, the independent-samples t-test was used to compare two groups for different loss functions (contextual loss, MSE, and BCE loss), and different deep learning models [different layers DRUNet and ordinary U-Net (17)] with a P value (P<0.001) and a confidence interval of difference.
Training details
The proposed DRUNet models were implemented using Pytorch 1.5.1, and all computations were performed using high-performance computing (HPC) with an NVIDIA GeForce 2070 GPU and Intel Core i7-8700 CPU. The proposed models had an increasing training time with increase of layers, that is, 18 h for DRUNet-34, 50 h for DRUNet-50, and 85 h for DRUNet-101, when finishing 100 epochs with a batch size of 5.
In this task of synthesizing MR images, a total of 35 participants were enrolled following the inclusion and exclusion criteria as described in Section 2.1, including 19 women (40.9±10.4 years old) and 16 men (39.4±12.0 years old) without hypertension, diabetes, trauma, brain tumors, hemorrhage, and other diseases. Among these participants, 30 patients were randomly selected for model training and validation, which accounted for 90% and 10%, respectively. The remaining 5 patients were used for the final test after completion of model training and validation. Originally, there were 130 to 190 slices for one patient’s MR or CT scan; thus, the total number of T1WI and T2WI was 5053 and 5081 slices, respectively, after image preprocessing. Augmenting the images by rotating each image by 90°, 180°, and 270°, the slice numbers of T1WI and T2WI were 20,212 and 20,324, respectively, which were sufficient for deep learning to train a model to synthesize MRI images. Because MR images present significant intensity variations across patients and their intensity has no fixed meaning, the MRI image was transformed from a single channel to three channels to perform transfer learning with the pre-trained ImageNet model before the proposed DRUNet model training. Then, we used channel means (0.485, 0.456, and 0.406) and channel standard deviations (0.229, 0.224, and 0.225) to standardize our dataset and eliminate the effects of scale differences.
Results
Three loss functions were applied to the proposed DRUNet model: MSE loss, BCE loss, and contextual loss. The synthetic T1WI and T2WI images from CT images under the different loss functions of DRUNet-101 are shown in the first and second rows of Figure 6, and the images from the first to third columns were synthesized by MSE loss, BCE loss, and contextual loss, respectively. The images in the last column are the actual T1WI and T2WI images. The synthetic MR images produced by contextual loss were intuitively closer to the actual ones, especially when there were more details and high-frequency components in the synthetic images after zooming into the regions of interest.
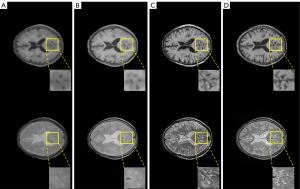
Except for the comparison of visual effects, PSNR, SSIM, and Tenengrad function were calculated to numerically verify the superiority of contextual loss. In Table 2, it is clear that the contribution of contextual loss is superior to the other functions because the larger PSNR and SSIM demonstrated that the synthetic images were closer to the actual images, and the larger Tenengrad function proved that the synthetic images have higher resolutions.
Table 2
| Loss function | PSNR | SSIM | Tenengrad | |||||
|---|---|---|---|---|---|---|---|---|
| T1WI | T2WI | T1WI | T2WI | T1WI | T2WI | |||
| MSE loss | 23.52±1.51 | 21.87±1.43 | 0.87±0.05 | 0.71±0.02 | 7.64±1.33 | 8.14±1.72 | ||
| BCE loss | 23.04±1.75 | 22.46±1.68 | 0.84±0.05 | 0.73±0.02 | 11.11±2.15 | 14.16±2.66 | ||
| Contextual loss | 34.25±2.06 | 33.50±1.08 | 0.97±0.03 | 0.98±0.05 | 17.03±2.75 | 19.76±3.54 | ||
DRUNet-101, double ResNet-U-Net with double 101 convolutional layers; PSNR, peak signal-to-noise ratio; SSIM, structural similarity; Tenengrad, Tenenbaum gradient; MSE, mean square error; BCE, binary cross-entropy. T1WI, T1-weighted images; T2WI, T2-weighted images.
To test how the ResNet layer affects the performance of DRUNet, three types of ResNet were constructed for the DRUNet, respectively, thus obtaining models like DRUNet-34, DRUNet-50, and DRUNet-101. The ordinary U-Net was also considered for the comparison experiment. There are few significant differences among the synthetic MR images generated by DRUNet-34, DRUNet-50, and DRUNet-101, as shown in Figure 7. The first four images in the first two rows are the synthetic T1WI, whereas those in last two rows are synthetic T2WI images, which were generated by DRUNet-34, DRUNet-50, DRUNet-101, and ordinary U-net, respectively. The pictures below them are the corresponding difference maps between synthetic and actual images. The real images in Figure 7E are located in the last column. In Figure 7A-7C generated using the proposed DRUNet, the differences between synthetic T2WI images and actual images are larger than those between synthetic T1WI images and actual images. Compared with the difference maps of T1WI images, the higher pixel differences of T2WI images are present in intracranial areas more than in other skull areas. For the images in Figure 7D generated by ordinary U-net (17), it was more obvious that their difference maps had outstanding defects, and the brain sulci and gyri in the synthetic T1WI had a reversed image rendering performance compared with actual T1WI, especially in the image of (D2). Although the synthetic T2WI had a significant image-rendering performance in brain areas, the entire synthetic image was full of checkerboard pattern artifacts, especially in the background of the synthetic T2WI. There were also checkerboard pattern artifacts in the foreground of the synthetic T1WI and T2WI when zooming in on these images. Thus, the spatial resolution of the synthetic images generated by DRUNet-101 was superior to that of the others and was more pronounced in the enlarged local areas of the image.
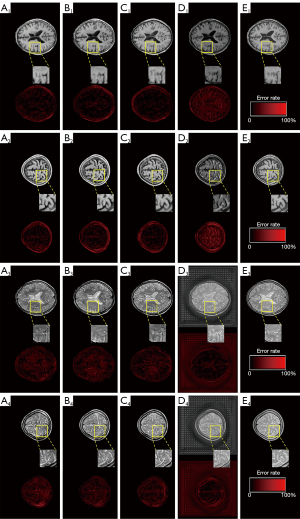
Table 3 illustrates the overall statistical analysis of the three quantitative metrics (PSNR, SSIM, and Tenengrad) for synthetic MR images by DRUNet-34, DRUNet-50, and DRUNet-101, and ordinary U-net. Although the differences in the aforementioned quantitative indices among these models were not obvious, the performance of DRUNet-101 was slightly superior to the other models, whereas the performance of ordinary U-net was slightly inferior to DRUNet. There is an upward tendency with an increase in the number of DRUNet layers in the PSNR and SSIM. Although Tenengrad function does not obey this rule, it is only a quality evaluation method without reference images, which reflects the image resolution but not the accuracy of the generated images. Therefore, DRUNet-101 has the expected performance and sharpness, although DRUNet-50 and DRUNet-34 have higher Tenengrad values in synthetic T1WI and T2WI.
Table 3
| Networks | PSNR | SSIM | Tenengrad | |||||
|---|---|---|---|---|---|---|---|---|
| T1WI | T2WI | T1WI | T2WI | T1WI | T2WI | |||
| DRUNet-34 | 34.19±2.07 | 33.43±1.11 | 0.96±0.04 | 0.96±0.06 | 17.85±2.87 | 19.82±3.67 | ||
| DRUNet-50 | 34.23±2.02 | 33.44±1.12 | 0.97±0.04 | 0.97±0.04 | 18.10±2.94 | 19.57±3.40 | ||
| DRUNet-101 | 34.25±2.06 | 33.50±1.08 | 0.97±0.03 | 0.98±0.05 | 17.03±2.75 | 19.76±3.54 | ||
| U-Net (17) | 34.08±2.16 | 27.61±0.06 | 0.90±0.06 | 0.73±0.12 | 13.62±2.36 | 21.79±0.70 | ||
DRUNet-X, double ResNet-U-Net with double X convolutional layers; PSNR, peak signal-to-noise ratio; SSIM, structural similarity; Tenengrad, Tenenbaum gradient; T1WI, T1-weighted images; T2WI, T2-weighted images.
Furthermore, statistical analyses with independent-samples t-tests were performed on the results (Tables 2,3) and are listed in Table 4, including the P value and confidence interval of difference. The middle two rows of the table are the statistical results obtained by comparing different loss functions under the same DRUNet-101, and the last row shows the statistical results when comparing different networks under the same contextual loss function. There is a significant difference (P<0.001) in the statistical results, and the narrower 95% confidence interval of difference demonstrated that DRUNet-101 with contextual loss outperforms the other models.
Table 4
| Experimental control | PSNR | SSIM | |||
|---|---|---|---|---|---|
| P value | Confidence interval of difference | P value | Confidence interval of difference | ||
| Contextual loss vs. MSE loss | <0.001 | 10.92, 11.45 | <0.001 | 0.20, 0.22 | |
| Contextual loss vs. BCE loss | <0.001 | 10.86, 11.40 | <0.001 | 0.18, 0.20 | |
| DRUNet-101 vs. U-Net (17) | <0.001 | 2.59, 3.47 | <0.001 | 0.14, 0.17 | |
DRUNet-101, double ResNet-U-Net with double 101 convolutional layers; PSNR, peak signal-to-noise ratio; SSIM, structural similarity; MSE, mean square error; BCE, binary cross-entropy.
To confirm the optimization of the aforementioned model parameters, the convergence rate of these models was investigated using loss curve graphs (Figure 8). Figure 8 reflect the loss curves when generating T1WI and T2WI images, respectively. With a decrease in layer number, the validating loss curves oscillate more drastically, especially for DRUNet-34 shown in Figure 8. In addition, the validating loss curves of the ordinary U-Net fluctuated drastically, which illustrates that the ordinary U-Net model has a weak convergence ability. More importantly, all curves gradually flattened out after 60 epochs of data training, which indicates that the overfitting problem was eliminated in the parameter setting and that accuracy gains were successfully obtained from increased depth. Therefore, DRUNet-101 exhibits considerably lower training loss and is generalizable to the validation data, which also fits well with the aforementioned conclusion.

To further evaluate the generalization performance of a DRUNet-101 on a given dataset, the performance of 5-fold cross validation through RMSE was conducted on the synthetic model of T1WI and T2WI as illustrated in Table 5. The differences among each fold were no significant, and the variances within each fold were also slight, all values of RMSE are within the acceptable ideal range. This eliminates the undesirable effects of unbalanced data division and proves the generalization performance of DRUNet on different datasets. Furthermore, all the RMSE values of T2WI were lower than that of T1WI, which was in keeping with the rule of validating loss curves of Figure 8 where the validation loss values of T2WI in Figure 8B were lower than that of T1WI in Figure 8A.
Table 5
| K-fold | T1WI | T2WI |
|---|---|---|
| 1 | 1.16±0.06 | 0.11±0.02 |
| 2 | 1.51±0.04 | 0.19±0.06 |
| 3 | 1.14±0.07 | 0.19±0.01 |
| 4 | 1.66±0.03 | 0.30±0.01 |
| 5 | 2.12±0.07 | 0.17±0.03 |
| Average | 1.52 | 0.19 |
RMSE, root mean squared error.
In this study, the MSE, BCE, and contextual loss functions were used to optimize the model parameters. The aforementioned results demonstrate that the performance of contextual loss is better than that of MSE and BCE loss. Visualization of the convolution filters and feature maps of CNN models is also very significant for analyzing the CNN mechanism as shown in Figure 9, where “JET” colormap is used to reflect the model weight, the higher the weight value, color tends to warm tone. The convolution filters can reflect part of the extracted features; thus, the visualization of the filters is very significant for the analysis of the CNN mechanism. MSE, BCE, and contextual algorithms were used as loss functions to optimize the model parameters. The experimental results demonstrated that the performance of contextual loss was better than that of BCE and MSE loss. Figure 9A-9C shows the heat maps of the 64 filters with a size of 7×7 from the first convolutional layer. If the filter detects an image region that is similar to its textural features, it will be activated to obtain a high value when striding over that area, and this feature of the image will be preserved in the feature maps. It can be seen that the vast majority of filters for contextual loss have more complicated textures, indicated by different shades of red, yellow, and blue visually, which ensures that more detailed features of the image can be fetched to the next layer for further high-dimensional feature extraction. In contrast, there were more red clustered areas in the filter maps generated by MSE loss, which demonstrated that more low-frequency features tended to be extracted in the feature maps. Hence, to a certain degree, the performance of the convolutional filters was in agreement with the experimental results: contextual loss > BCE loss > MSE loss.
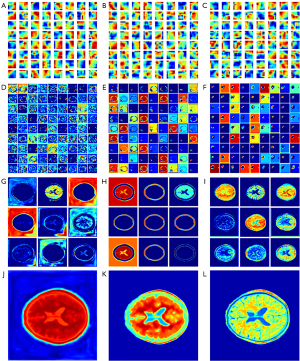
In addition, feature maps extracted from the proposed models were used to further illustrate the superiority of contextual loss. The most convincing feature maps were selected to analyze the performance variations among the MSE loss, BCE loss, and contextual loss. As shown in Figure 9, the feature maps generated by MSE loss, BCE loss, and contextual loss were aligned in three columns from left to right: Figure 9D,9G,9J are the feature maps of the lowest layer, the penultimate convolutional layer, and the final output generated by MSE loss, respectively; Figure 9E,9H,9K are the feature maps of the lowest layer, the penultimate convolutional layer, and the final output generated by BCE loss, respectively; Figure 9F,9I,9L are the feature maps of the lowest layer, the penultimate convolutional layer, and the final output generated by contextual loss, respectively. Aside from this, we used random zooming in pictures to distinguish texture or frequency information in the feature map. Specifically, the 81 images of the lowest layer were randomly selected from a total of 2,048 feature maps and were arranged with a grid size of 9×9 as shown in Figure 9D-9F. Similarly, the nine images of the penultimate layer were randomly selected from a total of 64 feature maps and were arranged with a grid size of 9×9 as shown in Figure 9G-9I.
The highest-dimensional feature maps of the lowest layer in DRUNet-101 are in the first row, and it is clear that the features generated by contextual loss are sparser and more abstract than the corresponding maps from the other two loss functions, which demonstrates that the features extracted from contextual loss are clearer and more definite for the concretization of features (40). The feature maps of the penultimate convolutional layer of the decoder are shown in the third row, and the last row presents the final feature maps to the output. The details of the MR images are depicted more clearly in each feature map generated by contextual loss, and the final output feature map also reflects more high-frequency information in the synthetic MR images due to contextual loss. Specifically, there are sparser signals in Figure 9F and denser signals in Figure 9D. The former is far more representative of high-dimensional characteristics. Although there are also sparse signals in Figure 9E, it focuses more on the skull area, which is not the region of interest. High-frequency signals inside the skull appear more richly in Figure 9I, including the gray matter, white matter, cerebrospinal fluid, sulcus, and gyrus. In contrast, there are more details about the skull in Figure 9H. The signals of the cerebral tissue are mixed with noise, as shown in Figure 9G. Figure 9I appears to have a blurred background noise. The detailed information in the brain regions is unclear despite noise removal from unrelated areas in Figure 9H. Figure 9L, generated by contextual loss, has the best rendering effects of MR images compared with Figure 9J,9K generated by MSE and BCE loss. In summary, contextual loss, a loss function originally used for hyper-resolution image reconstruction, can achieve the desired results in the medical image transfer task from CT to MR images.
Discussion
As a common generated network, CycleGAN and U-Net are typically used for medical images generation (41). Therefore, Li et al. (14,20) compared the performance of U-Net and CycleGAN to transform brain MR/CT images to their counterpart modality. The SSIM and PSNR of synthetic CT by U-Net and CycleGAN were 0.972 vs. 0.955, 28.84 vs. 26.32, respectively, and these identical figures of synthetic MRI were 0.946 vs. 0.924, 32.35 vs. 30.79, respectively. The quantitative results indicated that the U-Net method outperformed the CycleGAN method. Therefore, the U-Net was mainly adopted as the DRUNet in the research to synthesize MR images from CT scans. DRUNet is a general synthesis system for supervised learning and is different from some seemingly similar deep learning models that are limited to the application of a few residual blocks to the U-shaped model (38,42,43). Based on LinkNet and D-LinkNet, the whole ResNet-18 and ResNet-34 were set as their encoders, respectively (44,45), which enables full exploitation of the advantages of ResNet, and the decoder is designed based on ResNet. The final experimental results, including PSNR and SSIM, verified that ResNet integrated with U-Net outperformed several residual blocks inserted into U-Net on the synthetic scene.
The introduction of contextual loss from image super-resolution enriched the high-frequency information that existed in the synthesis of MR images through the evaluation indicator Tenengrad. The use of contextual information was initially investigated using a deep learning model, which proved the importance of contextual information in an image (38). The emergence of gram loss prompted the use of CNN to represent textures and images and synthesize new ones (46). This network uses the gram matrix to activate the texture feature of the VGG-19 layer. Then, perceptual loss (47,48) was proposed to apply a gram matrix to penalize differences in colors, textures, and exact shapes when outputs deviated from the target images. Few studies have applied global loss functions to medical image synthesis before. Dar et al. (49) used perceptual loss to improve the synthesis performance of T1WI and T2WI. In contrast to the aforementioned global loss functions in aligned images, contextual loss (23,37) aims to tackle image transformation for non-aligned data based on both context and semantics. Although contextual loss function was used in the FCN to generate synthetic CTs from MRI, this was only a simple application of contextual loss. Furthermore, the availability of contextual loss used for the synthesis of medical images (26) was confirmed by detailed analyses such as loss curves and feature maps.
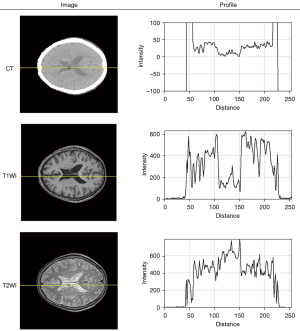
There is a nearly inverse tendency of contextual loss curves when generating synthetic T1WI and T2WI in Figure 8, which was possibly associated with the larger differences between CT and T2 images than between CT and T1 images. Some attempts have been made to explain this performance by plotting a profile across cerebral spinal fluid on the same position as CT, T1WI, T2WI in Figure 10. It can be clearly seen that there is a more parallel tendency between the profile of CT and T1WI than the counterparts of CT and T2WI. An obvious valley of the curve occurred in the distance ranging from 100 to 150 for the CT and T1WI, whereas the curve of T2WI has a visible convexity in the same position. In addition to feature maps, the superiority of the contextual loss function in processing high-frequency information was carried out by the visualization of the convolutional filter and heat map, as shown in Figure 9.
In this study, synthetic MRIs were generated from CT scans for obtaining a more accurate target volume in medical scenarios where MRI examination is not available. Because of the application of the proposed method to radiotherapy, it is important to know whether the synthetic MR images can accurately determine the planning target volume and delineate the lesions. The dataset of CT and MRI images was initially screened according to the exclusion criteria. Therefore, the results cannot provide a reasonable interpretation that synthetic MRI can serve a more accurate target volume. Considering this deficiency, this article is a preliminary attempt to apply synthetic MR images to TPS, which provides the theoretical feasibility of MRI-assisted TPS and is beneficial for the establishment of tumor radiotherapy model in future research. In addition, the experiment was conducted on pairs of brain MRI and CT images, it has not been verified whether the model can be applied to medical images of other body parts. Next, images of various tissues and organs coming from clinical work will be used to train a common model for the synthesis of MRI. In addition, DRUNet is a 2D deep learning network, which has a potential inter-slice discontinuity problem while generating synthetic images. In future studies, 3D networks will be used to investigate the differences between 2D and 3D networks.
Conclusions
In this paper, a new DCNN model, DRUNet, was proposed based on ResNet and U-Net for cross-modality medical image synthesis, which synthesizes T1WI and T2WI from CT. Three loss functions (i.e., BCE loss, MSE loss, and contextual loss) were introduced to optimize the parameters of DRUNet until it converged to a stable state. The loss curves and feature maps illustrated that the contextual loss preserved the high-frequency information of the intracranial tissues and removed the low-frequency background noise. Comparing DRUNet models with different layer numbers (i.e., 34, 50, and 101) and ordinary U-Net using PSNR, SSIM, and Tenengrad, it was concluded that DRUNet was superior to the ordinary U-Net model and that DRUNet-101 had the optimal performance for both synthetic T1WI and T2WI. In summary, DRUNet-101 with contextual loss can be a useful model for synthesizing MR images from CT images of the brain. This can be valuable for further evaluation in future studies concerning clinical applications
Acknowledgments
Funding: This research was funded by the National Natural Science Foundation of China (No. 12075011, No. 82071280, and No. 61901008), the Natural Science Foundation of Beijing (No. 7202093), and Key Research & Development Program of Science and Technology Planning Project of Tibet Autonomous Region, China (No. XZ202001ZY0005G).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-846/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-846/coif). XH reports that this research was funded by the National Natural Science Foundation of China (No. 61901008). SG reports that this research was funded by the National Natural Science Foundation of China (No. 12075011) and the Natural Science Foundation of Beijing (Grant No. 7202093). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study passed the ethical approval of The First Affiliated Hospital of Shenzhen University’s Bioethics Committee, and the participants all signed the informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jonsson JH, Karlsson MG, Karlsson M, Nyholm T. Treatment planning using MRI data: an analysis of the dose calculation accuracy for different treatment regions. Radiat Oncol 2010;5:62. [Crossref] [PubMed]
- Oldendorf W. Chapter 8: advantages and disadvantages of MRI. Basics of Magnetic Resonance Imaging. Boston: Martinus Nijhoff Publishing, 1988:125-38.
- Zhao C, Carass A, Lee J, He Y, Prince JL, editors. Whole brain segmentation and labeling from CT using synthetic MR images. 2017 Machine Learning in Medical Imaging. Cham: Springer International Publishing, 2017.
- Yu HS, Gupta A, Soto JA, LeBedis C. Emergency abdominal MRI: current uses and trends. Br J Radiol 2016;89:20150804. [Crossref] [PubMed]
- Ditkofsky NG, Singh A, Avery L, Novelline RA. The role of emergency MRI in the setting of acute abdominal pain. Emerg Radiol 2014;21:615-24. [Crossref] [PubMed]
- Schmidt MA, Payne GS. Radiotherapy planning using MRI. Phys Med Biol 2015;60:R323-61. [Crossref] [PubMed]
- Glide-Hurst CK, Low DA, Orton CG. Point/Counterpoint. MRI/CT is the future of radiotherapy treatment planning. Med Phys 2014;41:110601. [Crossref] [PubMed]
- Fox T, Elder E, Crocker I. Chapter 3: image registration and fusion techniques. In: Paulino AC, Teh BS, editors. PET-CT in Radiotherapy Treatment Planning. Philadelphia: Elsevier, 2008:35-51.
- Hsu SH, Cao Y, Huang K, Feng M, Balter JM. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Phys Med Biol 2013;58:8419-35. [Crossref] [PubMed]
- Lei Y, Harms J, Wang T, Tian S, Zhou J, Shu HK, Zhong J, Mao H, Curran WJ, Liu T, Yang X. MRI-based synthetic CT generation using semantic random forest with iterative refinement. Phys Med Biol 2019;64:085001. [Crossref] [PubMed]
- Hsu SH, Dupre P, Peng Q, Tomé WA. A technique to generate synthetic CT from MRI for abdominal radiotherapy. J Appl Clin Med Phys 2020;21:136-43. [Crossref] [PubMed]
- Uh J, Merchant TE, Li Y, Li X, Hua C. MRI-based treatment planning with pseudo CT generated through atlas registration. Med Phys 2014;41:051711. [Crossref] [PubMed]
- Dowling JA, Lambert J, Parker J, Salvado O, Fripp J, Capp A, Wratten C, Denham JW, Greer PB. An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int J Radiat Oncol Biol Phys 2012;83:e5-11. [Crossref] [PubMed]
- Li Y, Li W, Xiong J, Xia J, Xie Y. Comparison of Supervised and Unsupervised Deep Learning Methods for Medical Image Synthesis between Computed Tomography and Magnetic Resonance Images. Biomed Res Int 2020;2020:5193707. [Crossref] [PubMed]
- Jin CB, Kim H, Liu M, Han IH, Lee JI, Lee JH, Joo S, Park E, Ahn YS, Cui X. DC2Anet: generating lumbar spine MR images from CT scan data based on semi-supervised learning. Appl Sci 2019;9:2521. [Crossref]
- Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Cham: Springer International Publishing, 2015:234-41.
- Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. [Crossref] [PubMed]
- Yang H, Sun J, Carass A, Zhao C, Lee J, Prince JL, Xu Z. Unsupervised MR-to-CT Synthesis Using Structure-Constrained CycleGAN. IEEE Trans Med Imaging 2020;39:4249-61. [Crossref] [PubMed]
- Jin CB, Kim H, Liu M, Jung W, Joo S, Park E, Ahn YS, Han IH, Lee JI, Cui X. Deep CT to MR Synthesis Using Paired and Unpaired Data. Sensors (Basel) 2019;19:2361. [Crossref] [PubMed]
- Li W, Li Y, Qin W, Liang X, Xu J, Xiong J, Xie Y. Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy. Quant Imaging Med Surg 2020;10:1223-36. [Crossref] [PubMed]
- Jiang Y, Li J. Generative adversarial network for image super-resolution combining texture loss. Appl Sci 2020;10:1729. [Crossref]
- Aisen AM, Martel W, Braunstein EM, McMillin KI, Phillips WA, Kling TF. MRI and CT evaluation of primary bone and soft-tissue tumors. AJR Am J Roentgenol 1986;146:749-56. [Crossref] [PubMed]
- Mechrez R, Talmi I, Zelnik-Manor L, editors. The contextual loss for image transformation with non-aligned data. 2019 European Conference on Computer Vision. Cham: Springer International Publishing, 2018.
- Kalantar R, Messiou C, Winfield JM, Renn A, Latifoltojar A, Downey K, Sohaib A, Lalondrelle S, Koh DM, Blackledge MD. CT-Based Pelvic T1-Weighted MR Image Synthesis Using UNet, UNet++ and Cycle-Consistent Generative Adversarial Network (Cycle-GAN). Front Oncol 2021;11:665807. [Crossref] [PubMed]
- Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, Shen D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. Med Image Comput Comput Assist Interv 2017;10435:417-25.
- van Stralen M, Zhou Y, Wozny P, Seevinck P, Loog M, editors. Contextual loss functions for optimization of convolutional neural networks generating pseudo CTs from MRI. SPIE Medical Imaging 2018. Houston: Image Processing, 2018.
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 2010;49:1271-81. [Crossref] [PubMed]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012;62:782-90. [Crossref] [PubMed]
- Rivest-Hénault D, Dowson N, Greer PB, Fripp J, Dowling JA. Robust inverse-consistent affine CT-MR registration in MRI-assisted and MRI-alone prostate radiation therapy. Med Image Anal 2015;23:56-69. [Crossref] [PubMed]
- Oguro S, Tuncali K, Elhawary H, Morrison PR, Hata N, Silverman SG. Image registration of pre-procedural MRI and intra-procedural CT images to aid CT-guided percutaneous cryoablation of renal tumors. Int J Comput Assist Radiol Surg 2011;6:111-7. [Crossref] [PubMed]
- Gonzalez RC, Woods RE. Digital image processing. NJ, USA: Prentice-Hall, Inc., 2002.
- He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Las Vegas, NV, USA: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- Esser P, Sutter E, editors. A bariational U-Net for conditional appearance and shape generation. Salt Lake City, USA: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018.
- Odena A, Vincent D, Olah C. Deconvolution and checkerboard artifacts. Distill 2016;1:e3. [Crossref]
- Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C, editors. The importance of skip connections in biomedical image segmentation. Deep Learning and Data Labeling for Medical Applications. Cham: Springer International Publishing, 2016.
- Chen Y, Liu L, Phonevilay V, Gu K, Xia R, Xie J, Zhang Q, Yang K. Image super-resolution reconstruction based on feature map attention mechanism. Appl Intell 2021;51:4367-80. [Crossref]
- Mechrez R, Talmi I, Shama F, Zelnik-Manor L, editors. Maintaining natural image statistics with the contextual loss. 2018 Asian Conference on Computer Vision. Cham: Springer International Publishing, 2019.
- Trimpl MJ, Boukerroui D, Stride EPJ, Vallis KA, Gooding MJ. Interactive contouring through contextual deep learning. Med Phys 2021;48:2951-9. [Crossref] [PubMed]
- Yeo TTE, Ong SH. Jayasooriah, Sinniah R. Autofocusing for tissue microscopy. Image and Vision Computing 1993;11:629-39. [Crossref]
- Eakins JP. Automatic image content retrieval - are we getting anywhere? Milton Keynes: De Montfort University, 1996:123-35.
- Cheng Z, Wen J, Huang G, Yan J. Applications of artificial intelligence in nuclear medicine image generation. Quant Imaging Med Surg 2021;11:2792-822. [Crossref] [PubMed]
- Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham) 2019;6:014006. [Crossref] [PubMed]
- Nasrin S, Alom MZ, Burada R, Taha TM, Asari VK, editors. Medical image denoising with recurrent residual U-Net (R2U-Net) base auto-encoder. Dayton, OH: 2019 IEEE National Aerospace and Electronics Conference (NAECON), 2019.
- Chaurasia A, Culurciello E, editors. LinkNet: Exploiting encoder representations for efficient semantic segmentation. St. Petersburg, FL, USA: 2017 IEEE Visual Communications and Image Processing (VCIP), 2017.
- Zhou L, Zhang C, Wu M, editors. D-LinkNet: LinkNet with pretrained encoder and dilated convolution for high resolution satellite imagery road extraction. Salt Lake City, USA: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 2018.
- Snelgrove X, editor. High-resolution multi-scale neural texture synthesis. SIGGRAPH Asia 2017 Technical Briefs. Bangkok, Thailand: Association for Computing Machinery, 2017.
- Johnson J, Alahi A, Fei-Fei L, editors. Perceptual losses for real-time style transfer and super-resolution. 2016 European Conference on Computer Vision. Cham: Springer International Publishing, 2016.
- Xu X, Ye Y, Li X. Joint demosaicing and super-resolution (JDSR): network design and perceptual optimization. IEEE Trans Comput Imaging 2020;6:968-80. [Crossref]
- Dar SU, Yurt M, Karacan L, Erdem A, Erdem E, Cukur T. Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Trans Med Imaging 2019;38:2375-88. [Crossref] [PubMed]



