
Automated knee cartilage segmentation for heterogeneous clinical MRI using generative adversarial networks with transfer learning
Introduction
Osteoarthritis (OA) is the most common cause of knee pain and disability among the patients over the age of 50 with an estimated annual treatment cost of more than $51 billion in the United States alone (1,2). Knee magnetic resonance imaging (MRI) is one of the best imaging modalities to determine the severity of OA. It has been shown to be predictive of outcomes after surgeries such as arthroscopic partial meniscectomy (3-5). Manual grading of cartilage disease using semi-quantitative grading systems, however, is time-consuming and suffers from inter-observer variability limiting its routine use in clinical practice (6). Similarly, quantitative MRI, i.e., compositional and morphologic techniques, which can detect early cartilage degeneration and serve as biomarkers for disease prognostication are not routinely used in clinical practice. One of the major hurdles preventing the adoption of quantitative imaging into clinical practice is the need for cartilage segmentation which, to date, requires significant manual effort and time. Manual or semi-automatic cartilage segmentation is not only laborious and time-consuming but also suffers from intra- and inter-reader variability (7,8). Researchers have attempted to build an end-to-end deep learning network to detect knee joint abnormalities without the segmentation step by using multi-sequence, multi-planar MR images as inputs (9). The diagnostic performance of the model, however, was significantly lower than that of humans. Alternative deep learning approaches including tissue segmentation as a separate step in the pipeline achieved diagnostic performance comparable to humans (10-12). These results suggested that the segmentation stage may still be a necessary step in the deep learning pipeline to maximize diagnostic performance of the models in the detection of early cartilage degeneration (13).
Efforts have been made recently to build fully automated segmentation models based on deep learning (14-23), with cartilage segmentation performance as good as 0.880 in Dice coefficient (21). These models, however, are typically trained and tested on homogeneous research datasets, e.g., the osteoarthritis initiative (OAI) dataset (http://nda.nih.gov/oai). It is not known how well these results will directly translate to the heterogeneous data sets found in clinical practice, since the images are typically obtained from an assortment of MR scanners with variations in imaging parameters, and image quality. To the best of our knowledge, no previous studies have provided an automated cartilage segmentation model that has been successfully applied to a heterogeneous set of clinical MR images or explored the ability to use transfer learning (TL) to improve model performance for cartilage segmentation on heterogeneous MR images collected from clinical routines with different field strengths, different MR systems, different coils, and different protocols etc. Our initial experience applying such models to clinical datasets resulted in poor segmentation performance (Table 1). To improve model performance, models could be trained directly from the clinical datasets. However, such training would require large training datasets that would have to be manually segmented to prevent overfitting which could occur if only a small dataset is used (24). Consequently, the enormous amount of annotated training data needed becomes prohibitive for direct model training. An alternate approach to improve model performance is to optimize a previously trained model with homogeneous data using transfer learning (22,25-34). The advantage of the transfer learning is that only small amount of manually segmented clinical data is needed.
Table 1
| Method | Overall | FC | LTC | MTC | PC | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DC | HD95 (mm) | ASSD (mm) | DC | HD95 (mm) | ASSD (mm) | DC | HD95 (mm) | ASSD (mm) | DC | HD95 (mm) | ASSD (mm) | DC | HD95 (mm) | ASSD (mm) | |||||
| OAI/R1 | 0.519 | 6.227 | 1.224 | 0.504 | 7.035 | 1.373 | 0.445 | 5.699 | 1.959 | 0.465 | 11.21 | 2.296 | 0.499 | 6.098 | 1.492 | ||||
| APM/R1 | 0.018 | 61.15 | 30.21 | 0.794 | 1.769 | 0.460 | 0.692 | 7.613 | 1.171 | 0.645 | 8.260 | 1.464 | 0.003 | 116.0 | 37.84 | ||||
| MIX/R1 | 0.741 | 2.361 | 0.447 | 0.741 | 2.195 | 0.412 | 0.703 | 3.705 | 0.729 | 0.586 | 5.082 | 0.937 | 0.836 | 1.387 | 0.269 | ||||
| TL/R1 | 0.819* | 1.463* | 0.350* | 0.812* | 0.978* | 0.386* | 0.823* | 1.754* | 0.317* | 0.750* | 3.546 | 0.647 | 0.874* | 1.240 | 0.210* | ||||
| TL/R2 | 0.785 | 1.895 | 0.417 | 0.774 | 2.037 | 0.490 | 0.769 | 2.071 | 0.417 | 0.712 | 2.663 | 0.614 | 0.859 | 1.069* | 0.225 | ||||
| R1/R2 | 0.782 | 1.766 | 0.505 | 0.763 | 1.730 | 0.524 | 0.774 | 1.766 | 0.556 | 0.728 | 2.396* | 0.605* | 0.865 | 1.083 | 0.335 | ||||
| TL/R1 (1.5T) | 0.797 | 1.691 | 0.331 | 0.795 | 1.418 | 0.326 | 0.742 | 2.652 | 0.664 | 0.734 | 2.616 | 0.510 | 0.844 | 1.522 | 0.327 | ||||
| TL/R1 (3T) | 0.832 | 1.565 | 0.364 | 0.823 | 1.482 | 0.406 | 0.802 | 2.163 | 0.627 | 0.768 | 2.864 | 0.694 | 0.896 | 0.739 | 0.350 | ||||
| Philips (1.5T) | 0.774 | 2.882 | 0.456 | 0.775 | 3.009 | 0.420 | 0.691 | 3.475 | 0.762 | 0.652 | 3.760 | 0.772 | 0.856 | 2.030 | 0.272 | ||||
*, Best performance values. The comparison between 1.5T and 3T scans are shown as well. OAI, the initial model trained on the OAI dataset; APM, model trained from scratch on the APM dataset; MIX, model trained from scratch on the OAI and APM mixed dataset; TL, the transfer learned model fine-tuned on the clinical dataset; Philips, TL model trained on the APM Siemens data and tested on the APM Philips data; R1, reader 1; R2, reader 2; FC, femoral cartilage; LTC, lateral tibial cartilage; MTC, medial tibial cartilage; PC, patellar cartilage; DC, Dice coefficient; HD95, 95 percentile Hausdorff distance; ASSD, average symmetric surface distance.
The purpose of this study was to build a model using a conditional generative adversarial network (cGAN) to automatically segment heterogeneous clinical MRI scans by optimizing a pre-trained model built from a homogeneous research dataset with transfer learning from a small dataset of heterogenous clinical MR scans. Specifically, we compared the performance of the model with transfer learning to that of the model without transfer learning and to the performance of manual segmentation by 2 radiologists. If successful, the automatic segmentation model could be used to facilitate qualitative and quantitative grading of OA which would not only allow for the adoption of these methods into routine clinical practice but for the development of models to guide patient management and predict patient outcomes.
We present the following article in accordance with the MDAR checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-459/rc).
Methods
Data
The datasets used in this study included two parts: a set of publicly available homogeneous MRIs from the OAI collected in a research setting, and a set of heterogeneous MRIs locally collected through clinical routines. OAI dataset (https://nda.nih.gov/oai/) was used to pre-train our model. It contained 176 sagittal knee MR images collected on Siemens 3T Trio scanner using the 3D sagittal double-echo steady state (DESS) sequence with the same coil model and acquisition parameter settings. Each image consisted of 160 slices (0.7 mm slice thickness) with fixed FOV of 14 cm2 and matrix size of 384×384 with cartilage manually segmented by iMorphics (35).
The clinical knee MR images used for transfer learning model training, validation and test were collected from the Cleveland Clinic OME cohort (36), a prospective orthopaedic surgical cohort within Cleveland Clinic health care system. The cohort was operationalized in 2015 and has collected more than 45,000 episodes of care representing over 40,000 patients at 16 Cleveland Clinic sites through the end of 2019. Most of the patients had gone through MRI at the baseline at different locations across the Cleveland Clinic health care system with different scanner models and various 2D/3D imaging parameters. To re-train our model using transfer learning, 25 sets of knee MRIs (15 1.5T and 10 3T) from 9 different locations scanned on 6 different Siemens scanner models were randomly selected from the cohort for patients underwent arthroscopic partial meniscectomy (APM). In addition, 5 more APM patients knee MRIs collected on a Philips 1.5T Achieva scanner were randomly selected for testing purpose. The patients had an age of 45 years and older with no prior orthopedic knee surgeries. Sagittal 2D fat-saturated proton density weighted (PDw) images were common to all the knee MRIs and used for the segmentations. Many institutions did not obtain 3D images as part of routine knee MRI and qualitative grading of cartilage was typically done using 2D images. These 30 sagittal PDw MRIs were a heterogeneous dataset obtained on a variety of scanner models from different vendors, acquired at different field strengths with varying coils, fields of view, spatial resolutions, slice thicknesses, image contrast, and image quality (see Table 2). The positioning of patient knees also varied, with lateral-medial offset from the isocenter ranging from 0 to 120 mm. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by our institutional review board with study number 19-005. Informed consent was waived.
Table 2
| Model | Field strength (Tesla) | Coil | FOV (mm) | Matrix size | Slice thickness (mm) | Repetition time (ms) | Echo time (ms) | Flip angle | Number of slices | Number of scans |
|---|---|---|---|---|---|---|---|---|---|---|
| Siemens Aera | 1.5 | Siemens Tx/Rx 15-Channel Knee Coil | 140×140 | 384×384 | 3 | 2,790 | 15 | 150 | 27 | 2 |
| Siemens Avanto | 1.5 | Siemens CP Extremity | 140×140 | 320×320 | 3 | 2,660, 3,190 | 14, 15 | 180 | 25, 30 | 2 |
| Siemens Espree | 1.5 | Body | 140×140 | 384×384 | 3 | 2,700 | 11 | 180 | 30 | 1 |
| Siemens Symphony | 1.5 | Siemens CP Extremity | 140×140, 150×150, 160×160 | 256×256, 320×320 | 3 4 |
2,920, 3,000, 3,140, 3,260, 3,400, 3,500, 3,790, 3,920 | 14, 15, 17 | 180, 150 | 23, 24, 27, 28, 30, 31 | 10 |
| Siemens Trio | 3 | Siemens 8-Channel Knee Coil | 140×140 | 448×448 | 2.5 | 5,970 | 10 | 150 | 40 | 2 |
| Siemens Verio | 3 | Siemens 8-Channel Knee Coil | 134×140, 154×160 | 308×320 | 2.5 | 3,250, 3,550, 3,620 | 16 | 150 | 35, 38 | 8 |
| Philips Achieva | 1.5 | Philips SENSE-Knee-8; SENSE-Flex-M | 140×140, 150×150, 186×186 | 480×480, 528×528, 560×560, 704×704 | 3 | 3,312, 3,581, 3,583, 3,589, 3,988 | 15, 30 | 90 | 25, 27, 30 | 5 |
Two radiologist readers manually segmented the Siemens MRIs using an in-house segmentation tool under the supervision of an attending musculoskeletal radiologist with 13 years of experience. After setting the rule set for segmentation during a training session, each sagittal MRI image was segmented separately by the readers. The cartilage of the four different subregions [femoral condyle (FC), patella (PC), medial tibial plateau (MTC), and lateral tibial plateau (LTC)] was included, consistent with the OAI data cartilage segmentation. Each of the 25 Siemens cases contained approximately 23–40 sagittal MRI images for model training, validation, and test. Each case required around 2 hours of work, per radiologist. One radiologist (reader 1) segmented all the 25 Siemens cases. The other (reader 2) segmented the 5 Siemens test cases. Reader 1 further segmented the 5 Philips test cases.
Evaluation metrics
The quantitative metrics chosen to evaluate the cartilage segmentation performance included the Dice coefficient (DC) (37), the 95 percentile Hausdorff distance (HD95) (38,39), and the average symmetric surface distance (ASSD) (40). The DC focuses on assessing the overlap of two sets of segmentations. The Hausdorff distance calculates the maximum point distance between the segmentations, which is sensitive to outliers. The ASSD looks for averaged surface difference between the segmentations. Therefore, these three metrics are complementary to each other.
The Dice coefficient is defined as
where G is the ground truth segmentation and S is the segmentation to be evaluated. The DC is a widely accepted metric for knee tissue segmentation. It ranges from 0 to 1, with 0 indicating no overlap in segmentation and 1 indicating a perfect agreement in segmentation. Volumetric DC was calculated to accurately reflect model segmentation performance.
The Hausdorff Distance (HD) (41) between G and S is defined by
where the direct Hausdorff distance is given by
The HD is an informative metric since it is an indicator of the largest segmentation error. The HD is, however, known to be sensitive to outliers. The HD95 replaces the maximum in HD with the 95 percentile to reduce the impact of outliers on HD.
The average symmetric surface distance is the average of all distances from points on the boundary of S (∂S) to the boundary of (∂G) and from points on ∂G to ∂S:
where denotes the Euclidean norm and denotes the cardinality of a set. The smaller the ASSD, the better the segmentation boundaries agree to each other.
Model specifics
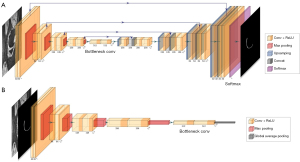
The architecture of the deep learning segmentation model was based on the cGAN (42), which was one variant of the generative adversarial networks (GAN) (43). As detailed in (21) (Figure 1), the U-Net (44) was used in place of the generative network or the generator in cGAN to generate four segmentation masks for the four cartilage compartments and a background channel. It contained an encoding path and a decoding path. Every step of the encoding path consisted of two repeated 3×3 unpadded convolutions with stride 2, each followed by batch normalization and a rectified linear unit (ReLU), and a 2×2 max pooling layer with stride 2 for downsampling. Each step of the decoding step consisted of a 2×2 upsampling convolutional layer and two repeated 3×3 convolutional layers followed by batch normalization and a ReLU. Each step in the encoding path was connected to the corresponding step in the decoding path with a skip connection. The number of feature channels was doubled in each step of the encoding path, and halved in each step of the decoding path. The output block contained a 1×1 convolutional layer and a softmax activation function to map the features to the probability maps for the four cartilage compartments channels and a background channel. A threshold of 0.5 was applied to each channel to obtain the binary segmentation masks of different cartilage compartments.
Details of the network architecture are illustrated in Figure 2A. The discriminative network or the discriminator followed the architecture of a typical convolutional neural network as shown in Figure 2B. It contained 5 convolutional blocks followed by average pooling, 1×1 unpadded convolution, and sigmoid activation. Each convolutional block consisted of two repeated 3×3 unpadded convolutional layers, each followed by batch normalization and ReLU activation, and a 2×2 max pooling layer. The objective loss function was a combination of the averaged DC segmentation loss and the feature matching loss for the generator, and the binary cross entropy loss for the discriminator, as defined in (21).
Model training, validating, and testing
The OAI dataset knee MRIs were randomly split into 70%, 20%, 10% for training, validation, and test respectively for model pre-training. The 25 sets of clinical PDw Siemens knee MRIs were randomly divided into 15, 5, 5 for training, validation, and test respectively for transfer learning using the OAI pretrained model. The clinical PDw MRIs were further augmented by counterclockwise 90-degree rotations and mirroring for training and validation. All scans and segmentation masks were interpolated with bi-cubic and nearest neighbor interpolation respectively to 384×384 to fit the input size of the pretrained OAI model. The discriminator and the generator were trained alternatively, with the discriminator trained once for each generator training iteration. No layers were frozen during transfer learning. The ADAM optimizer was used for all model training with default moment values, an initial learning rate of 1e-3, and a decay rate of 0.9. The batch size was set to 10 due to GPU memory limitation. The maximum number of iterations was set to 90,000. Early stopping was imposed when the learning rate reached 1e-8 to avoid potential overfitting.
Four models were trained and applied to the clinical test cases for comparison: (I) an OAI pretrained model; (II) a model trained from scratch on the APM dataset; (III) a model trained from scratch on the OAI and APM combined dataset; and (IV) a TL model transferred from the OAI pretrained model to the APM dataset. The dependence of the TL model performance on the training set size was further investigated by comparing the model performance trained on different number of training sets (training sizes =1, 5, 10 and 15 respectively) and tested on the 5 Siemens test cases. The TL model performance on scanners from a different vendor (Philips 1.5T Achieva) was also tested. The training, validation, and test of the models were realized with the Python deep learning frameworks Keras (45) and Tensorflow (46) on a NVIDIA Titan Xp GPU.
Inter reader variation
Inter reader variation was assessed on 5 Siemens test cases from one random draw that were not included in the model training and validation. The model trained with segmentation from reader 1 as ground truth was evaluated on the test cases and compared with both readers using DC, HD95, and ASSD. The agreement between the manual segmentation of the two readers was also evaluated.
Cross-validation
Due to the small size of the test set, the transfer learning model for the clinical knee MRIs was further tested for robustness using cross-validation. The 25 sets of Siemens MRIs were randomly partitioned into 5 groups with each group containing 5 sets of MRIs. A 5-fold cross validation was performed on the 5 groups, each time using 4 groups for training and validation and one for testing. In addition, a 4-fold cross-validation was applied to the 4 groups of each training and validation step, using 3 for training and one for validation. The models with best DC on the validation sets through the 4-fold cross-validations were chosen. The 22 test cases from the 5-fold cross validation, excluding the cases with flow artifacts or failed fat saturation, were used to further examine the TL model performance difference between the 1.5T and 3T scans.
Results
The time for one instance of transfer learning model training on the APM dataset was approximately 26 h on the NVIDIA Titan Xp GPU. Once trained, however, the mean time cost per case for automated cartilage segmentation on the test cases was under 2 seconds.
Applying the trained transfer learning model to the holdout test sets yielded an overall averaged DC of 0.819 (±0.039), an averaged HD95 of 1.463 (±0.827) mm, and an averaged ASSD of 0.350 (±0.114) mm. For comparison, an averaged DC of 0.519 (±0.088), an averaged HD95 of 6.227 (±2.751) mm, and an averaged ASSD of 1.224 (±0.465) mm were obtained on the holdout test set when applying the pretrained OAI model without transfer learning. The model trained from scratch using the APM dataset yielded an averaged DC of 0.018 (±0.005), an averaged HD95 of 61.15 (±5.944) mm, an averaged ASSD of 30.21 (±2.212) mm. The model trained from scratch on the mixed dataset with OAI and APM datasets combined produced an averaged DC of 0.741 (±0.083), an averaged HD95 of 2.361 (±0.988) mm, an averaged ASSD of 0.447 (±0.151) mm. Moreover, a comparison between the segmentation of the test sets from two readers showed an averaged DC of 0.782 (±0.046), an averaged HD95 of 1.766 (±0.321) mm, and an averaged ASSD of 0.505 (±0.033) mm. A more detailed breakdown comparison, including a comparison between 1.5T and 3T scans, for the four sub-compartments was shown in Table 1. The TL model performance on test images from a different vendor (Philips) was also shown in Table 1.
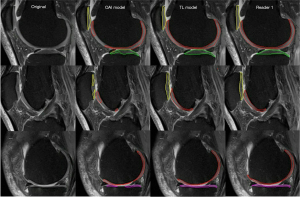
Figure 3 showed a sample comparison between the automatic and manual cartilage segmentation on lateral, central, and medial sides of sagittal fat-saturated PDw MR image slices collected on a Siemens 3T Verio scanner with FOV of 134×140 mm2, matrix size of 308×320, and the number of slices being 38.
The dependence of the TL model performance on the number of training cases was plotted in Figure 4. Specifically, Figure 4A-4C showed the dependency plots of the TL model performance on the training sizes 1, 5, 10 and 15 in terms of DC, HD95, and ASSD respectively. For each evaluation metric, both the overall model performance and the compartment-wise model performance were plotted.

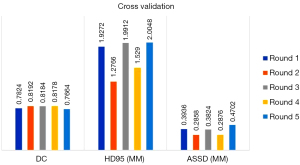
The results of the TL model performance on the respective holdout test sets of the 5-fold cross validation were shown in Figure 5. The overall mean (± SD) DC, HD95 and ASSD for all 25 test cases of the 5-fold cross validation were 0.801 (±0.051), 1.746 (±0.944) mm, and 0.364 (±0.149) mm, respectively. Compartment-wise violin plots for the TL model performance in DC, HD95, and ASSD on all 25 test cases of the 5-folder cross validation were depicted in Figure 6 respectively.

Discussion
In this study, we observed that the model trained on the homogeneous OAI dataset did not perform well on the heterogeneous clinical dataset (overall DC of 0.519). However, when the OAI model was used as a pre-trained model for transfer learning to the clinical dataset, the TL model showed a markedly improved performance, even using only a small training dataset. Moreover, the transfer learning model performance was similar to that of the 2 radiologists (overall DC of 0.819). Specifically, the trained TL model achieved overall a Dice coefficient of 0.819, an HD95 of 1.463 mm, and an ASSD of 0.350 mm on a randomly drawn and holdout clinical test dataset with various MR scanner models, field strengths, image resolution, contrast, and quality. It clearly outperformed its counterpart pre-trained on the homogenous OAI dataset collected in a research setting in all three evaluation metrics for all four sub-compartments as shown in Table 1. The TL model trained with reader 1’s segmentation was also tested for inter-reader agreement on the holdout test set by comparing to reader 2’s segmentation, which yielded a similar agreement as that between reader 1 and reader 2.
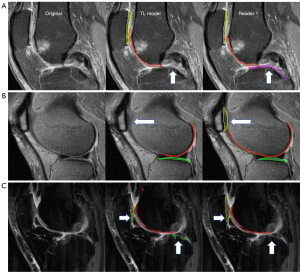
Compartment-wise, the TL model performance on PC was the best considering all the three metrics as shown in Table 1; the performance on MTC was the worst. As shown in Figure 7A, in the central part of the knee joint close to the tibial eminence, the MTC was not segmented by the TL model but was segmented by reader 1 according to experience in this case. There was however inconsistency in manual segmentation for this difficult area, which may have caused confusion to the model. Focusing on the weight bearing area only may help remediate this problem and improve the model performance without impacting clinical diagnosis.
We also examined the model performance when trained from scratch using the APM dataset and the OAI and APM combined dataset. As shown in Table 1, the performance of the APM model on all compartments was worse compared to the TL model. In particular, it failed on PC, which led to the worst overall performance among all the models. This could be due to the relatively small number of slices available containing PC to train the model. The model performance on the rest compartments, however, was better than directly applying the OAI model. The mixed model performed better than the OAI model and improved the PC results of the APM model, but still worse than the TL model.
Furthermore, there was a clear trend of the TL model performance with regard to the training size as shown in Figure 4. Specifically, the overall DC increased as the number of training cases increased (Figure 4A); the overall HD95 and ASSD decreased as the number of training cases increased (Figure 4B,4C). Moreover, the curves flattened when using 15 training cases. Compartment-wisely, FC and PC showed similar trend. LTC and MTC, however, may benefit from more training data.
The TL model performance difference between 1.5T and 3T scans was also examined as shown in Table 1. The overall DC of the TL model on 3T scans was significantly higher than that on 1.5T scans. The differences in the overall distance metrics, however, were not significant. Similar story was observed among different compartments. One of the potential reasons for the increased DC on 3T scans could be the increased signal to noise ratio compared to 1.5T scans. Studies with larger dataset are needed to better understand the model performance difference between 1.5T and 3T scans.
In addition, the TL model performance on MR scans from a different vendor was also tested. Specifically, the TL model trained on the APM Siemens scans were further tested on 5 APM patient scans collected from a Philips 1.5T Achieva scanner with Philips coils and various acquisition parameters (Table 2). As shown in Table 1, the TL model showed good overall as well as compartment-wise performance in all three metrics on the Philips test cases. The performance was slightly worse than that on the Siemens test cases, which was expected. Including scans from a different vendor in the training dataset should improve the TL model performance on the test cases from the vendor.
To test the robustness of the TL model on the small sized test data set, the best performed TL model was chosen for each round of the 5-fold cross-validation based on 4-fold cross validations among the training and validation sets, which is a standard practice for deep learning model training. Figure 5 showed the robustness of the TL models across the 5 rounds of cross validation when comparing the automatic segmentations with the manual segmentation references. Specifically, the DCs had minimal variation while the HD95s and ASSDs had some fluctuations. This was expected since the DC served as a general measure for checking segmentation overlaps, while the HD95 and the ASSD looked for segmentation boundary discrepancies. The HD95 captured the worst-case boundary discrepancy between the segmentations, and the ASSD evaluated the averaged boundary discrepancy. That being said, the fluctuations in HD95 and ASSD are comparable to the literature reported values for articular cartilage segmentation (19,47).
The compartment-wise robustness of the model on all 25 test cases was shown in violin plots (Figure 6) with DC, HD95, and ASSD as the metric respectively. The distribution of the DCs shown in Figure 6A for all 4 compartments was reasonably good except for PC, where there was a clear failure with a DC of 0.444. The cause for this failure was that the fat-saturation in the corresponding case failed during the clinical scan as shown in Figure 7B, which made it difficult for the model to distinguish between cartilage and other fat tissues. Including more failed fat-saturation cases in the model may help remediate the problem. For HD95 shown in Figure 6B, a clear outlier of HD95 of 36.283 mm was calculated for LTC. A close look at the case as shown in Figure 7C suggested that this was due to the heavy flow artifact collected during the clinical scan. The flow artifact was also one of the major contributors to the spikes in ASSD for LTC and PC shown in Figure 6C. Training the model on more images with non-ideal fat saturation or flow artifacts may help improve the model performance to clinical images with sub-optimal image quality.
One of the major potential problems of applying deep learning models to small training data set is model overfitting. Transfer learning has been recognized as a solution to this problem. In this study, we have in addition employed other remedies including a variable learning rate for early stopping, data augmentation with 90-degree rotations and mirroring, cross validation, and regularization. We observed an averaged DC of 0.875 on the 5 validation sets, compared to the averaged DC of 0.819 on the test sets, which indicated that a slight overfitting problem still existed. Inclusion of dropout layers in the TL model and usage of more sophisticated augmentation such as finer angle rotation, translation, and deformation may help to further reduce the overfitting problem and improve the model performance (48,49).
This study has several limitations. First, although the robustness of our TL model has been validated through cross validation, the small size of the test set is still one limitation. Evaluation on larger sized test dataset will be conducted before its implementation into clinical workflow. Second, we did not compare our current TL model with other model structures on this clinical dataset. Some initial comparisons have been made in the literature (21). We will extend this comparison with existing models (16,17) in future studies. We will also explore the possibility to build the generator using U-Net with VGG or ResNet backbones (50). In addition, only 2D networks were used in our current cGAN model. Multi-planer or fully 3D networks, while requiring more CPU/GPU computing resources, could incorporate more 3D information to improve segmentation accuracy and efficiency over 2D models (51-53). Third, although results in the DC, HD95, and ASSD were reported in this study, the loss function was solely based on the DC. Model performance of incorporating different metrics or a hybrid of them into the loss function needs to be investigated in the future. Fourth, single contrast PDw images were used in this study. Training and testing the TL model on multi-contrast images need to be further investigated. Another limitation is that all training images were collected with scanners from one vendor, although with different models, and all within the Cleveland Clinic system. In the future, we will develop and test models using multi-site and multi-vendor data.
Conclusions
In this study, we observed that the model trained on the publicly available homogeneous OAI dataset did not perform well on the heterogeneous clinical dataset. As a remedy, we created a transfer learning model using a small training data set. This TL model was able to automatically segment knee cartilage, with performance comparable to human, for a wide range of knee MR images chosen to reflect the realities of clinical practice, i.e., different field strengths, scanner models, coils, imaging parameters, and image quality. In addition, we showed the robustness of the proposed model through cross validation. We found it feasible to perform fully automated segmentation of clinical knee MR images, which would enable the clinical application of quantitative MR imaging technique and other prediction models for improved patient treatment planning.
Acknowledgments
The authors would also like to acknowledge NVIDIA for the donation of the Titan Xp GPU used in this study.
Funding: This study was partially supported by NIH/NIAMS R01AR073512, R01AR075422 and R01AR077452.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-459/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-459/coif). XL reports support from NIH/NIAMS R01AR075422 and R01AR077452. NS reports support from NIH R01-AR073512. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by our institutional review board with study number 19-005. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jinks C, Jordan K, Croft P. Measuring the population impact of knee pain and disability with the Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC). Pain 2002;100:55-64. [Crossref] [PubMed]
- Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, Gabriel S, Hirsch R, Hochberg MC, Hunder GG, Jordan JM, Katz JN, Kremers HM, Wolfe FNational Arthritis Data Workgroup. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 2008;58:26-35. [Crossref] [PubMed]
- Katz JN, Meredith DS, Lang P, Creel AH, Yoshioka H, Neumann G, Fossel AH, de Pablo P, Losina E. Associations among preoperative MRI features and functional status following arthroscopic partial meniscectomy. Osteoarthritis Cartilage 2006;14:418-22. [Crossref] [PubMed]
- Kijowski R, Woods MA, McGuine TA, Wilson JJ, Graf BK, De Smet AA. Arthroscopic partial meniscectomy: MR imaging for prediction of outcome in middle-aged and elderly patients. Radiology 2011;259:203-12. [Crossref] [PubMed]
- Cantrell WA, Colak C, Obuchowski NA, Spindler KP, Jones MH, Subhas N. Radiographic evaluation of knee osteoarthritis in predicting outcomes after arthroscopic partial meniscectomy. Knee 2020;27:1238-47. [Crossref] [PubMed]
- Colak C, Polster JM, Obuchowski NA, Jones MH, Strnad G, Gyftopoulos S, Spindler KP, Subhas N. Comparison of Clinical and Semiquantitative Cartilage Grading Systems in Predicting Outcomes After Arthroscopic Partial Meniscectomy. AJR Am J Roentgenol 2020;215:441-7. [Crossref] [PubMed]
- Carballido-Gamio J, Bauer J, Lee KY, Krause S, Majumdar S. Combined image processing techniques for characterization of MRI cartilage of the knee. Conf Proc IEEE Eng Med Biol Soc 2005;2005:3043-6. [Crossref] [PubMed]
- Carballido-Gamio J, Bauer JS, Stahl R, Lee KY, Krause S, Link TM, Majumdar S. Inter-subject comparison of MRI knee cartilage thickness. Med Image Anal 2008;12:120-35. [Crossref] [PubMed]
- Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 2018;15:e1002699. [Crossref] [PubMed]
- Liu F, Zhou Z, Samsonov A, Blankenbaker D, Larison W, Kanarek A, Lian K, Kambhampati S, Kijowski R. Deep Learning Approach for Evaluating Knee MR Images: Achieving High Diagnostic Performance for Cartilage Lesion Detection. Radiology 2018;289:160-9. [Crossref] [PubMed]
- Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging 2019;49:400-10. [Crossref] [PubMed]
- Tolpadi AA, Lee JJ, Pedoia V, Majumdar S. Deep Learning Predicts Total Knee Replacement from Magnetic Resonance Images. Sci Rep 2020;10:6371. [Crossref] [PubMed]
- Kijowski R, Liu F, Caliva F, Pedoia V. Deep Learning for Lesion Detection, Progression, and Prediction of Musculoskeletal Disease. J Magn Reson Imaging 2020;52:1607-19. [Crossref] [PubMed]
- Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 2013;16:246-53.
- Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80:2759-70. [Crossref] [PubMed]
- Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018;79:2379-91. [Crossref] [PubMed]
- Norman B, Pedoia V, Majumdar S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology 2018;288:177-85. [Crossref] [PubMed]
- Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H, editors. Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 4-7 April 2018.
- Liu F. SUSAN: segment unannotated image structure using adversarial network. Magn Reson Med 2019;81:3330-45. [PubMed]
- Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med Image Anal 2019;52:109-18. [Crossref] [PubMed]
- Gaj S, Yang M, Nakamura K, Li X. Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks. Magn Reson Med 2020;84:437-49. [Crossref] [PubMed]
- Kessler DA, MacKay JW, Crowe VA, Henson FMD, Graves MJ, Gilbert FJ, Kaggie JD. The optimisation of deep neural networks for segmenting multiple knee joint tissues from MRIs. Comput Med Imaging Graph 2020;86:101793. [Crossref] [PubMed]
- Wang Y, Zhang Y, Wen Z, Tian B, Kao E, Liu X, Xuan W, Ordovas K, Saloner D, Liu J. Deep learning based fully automatic segmentation of the left ventricular endocardium and epicardium from cardiac cine MRI. Quant Imaging Med Surg 2021;11:1600-12. [Crossref] [PubMed]
- Caruana R, Lawrence S, Giles L. Overfitting in neural nets: backpropagation, conjugate gradient, and early stopping. Proceedings of the 13th International Conference on Neural Information Processing Systems; Denver, CO: MIT Press; 2000. p. 381-7.
- Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering 2010;22:1345-59. [Crossref]
- Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. [Crossref] [PubMed]
- Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. [Crossref] [PubMed]
- Hesamian MH, Jia W, He X, Kennedy P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging 2019;32:582-96. [Crossref] [PubMed]
- Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019;29:102-27. [Crossref] [PubMed]
- Chen KT, Schürer M, Ouyang J, Koran MEI, Davidzon G, Mormino E, Tiepolt S, Hoffmann KT, Sabri O, Zaharchuk G, Barthel H. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging 2020;47:2998-3007. [Crossref] [PubMed]
- Raghu M, Zhang C, Kleinberg J, Bengio S, editors. Transfusion: Understanding Transfer Learning for Medical Imaging. Advances in Neural Information Processing Systems; 2019.
- Karimi D, Warfield SK, Gholipour A. Transfer learning in medical image segmentation: New insights from analysis of the dynamics of model parameters and learned representations. Artif Intell Med 2021;116:102078. [Crossref] [PubMed]
- Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med 2021;128:104115. [Crossref] [PubMed]
- Wacker J, Ladeira M, Nascimento JEV, editors. Transfer Learning for Brain Tumor Segmentation. International MICCAI Brainlesion Workshop; 2020.
- Williams TG, Holmes AP, Bowes M, Vincent G, Hutchinson CE, Waterton JC, Maciewicz RA, Taylor CJ. Measurement and visualisation of focal cartilage thickness change by MRI in a study of knee osteoarthritis using a novel image analysis tool. Br J Radiol 2010;83:940-8. [Crossref] [PubMed]
- Cleveland OME. Clinic Orthopaedics. Implementing a Scientifically Valid, Cost-Effective, and Scalable Data Collection System at Point of Care: The Cleveland Clinic OME Cohort. J Bone Joint Surg Am 2019;101:458-64. [Crossref] [PubMed]
- Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology 1945;26:297-302. [Crossref]
- Fedorov A, Billet E, Prastawa M, Gerig G, Radmanesh A, Warfield SK, et al., editors. Evaluation of Brain MRI Alignment with the Robust Hausdorff Distance Measures2008; Berlin, Heidelberg: Springer Berlin Heidelberg.
- Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, et al. Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal 2014;18:359-73. [Crossref] [PubMed]
- Heimann T, van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 2009;28:1251-65. [Crossref] [PubMed]
- Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Transactions on Pattern Analysis and Machine Intelligence 1993;15:850-63. [Crossref]
- Mirza M, Osindero S. Conditional Generative Adversarial Nets2014 November 01, 2014:[arXiv:1411.784 p.]. Available online: https://ui.adsabs.harvard.edu/abs/2014arXiv1411.1784M.
- Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2; Montreal, Canada: MIT Press; 2014. p. 2672-80.
- Ronneberger OF, Philipp; Brox, Thomas. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv. 2015:1505.04597.
- Keras CF. GitHub repository. 2015.
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems2016 March 01, 2016:[arXiv:1603.04467 p.]. Available online: https://ui.adsabs.harvard.edu/abs/2016arXiv160304467A.
- Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J. Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Phys Med Biol 2014;59:7245-66. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1; Lake Tahoe, Nevada: Curran Associates Inc.; 2012. p. 1097-105.
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929-58.
- Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham) 2019;6:014006. [Crossref] [PubMed]
- Milletari F, Navab N, Ahmadi S, editors. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV); 2016 25-28 Oct. 2016.
- Çiçek Ö, Abdulkadir A, Lienkamp S, Brox T, Ronneberger O, editors. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. MICCAI; 2016.
- Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61-78. [Crossref] [PubMed]



