
Impact of point-spread function reconstruction on dynamic and static 18F-DOPA PET/CT quantitative parameters in glioma
Introduction
Gliomas represent approximately 80% of all malignant brain tumors and are associated with a high mortality rate (1). Besides contrast-enhanced magnetic resonance imaging (MRI), positron emission tomography (PET) using amino acids such as 3,4-dihydroxy-6-[18F]-fluoro-L-phenylalanine (18F-DOPA, FDOPA) plays a critical role in the initial clinical diagnosis, surgical treatment planning, and post-treatment follow-up (2). Recent studies showed that quantitative parameters extracted from FDOPA PET/computed tomography (CT) are useful for glioma assessment (3,4). According to European guidelines for PET imaging of glioma, visual assessment of tumor uptake is accompanied by quantitative assessment (2). Different quantification parameters of PET data using different methods can be employed. The static parameters are the most widely used for quantification in clinical practice due to its easiest feasibility, requiring a single-frame acquisition only. Among static parameters, the standardized uptake value (SUV), which is the tracer uptake normalized to the injected dose and to a normalization factor based on the subject’s anthropometric characteristics, is the most widely employed. The ratio of the SUV in the tumor region normalized to the SUV in a background region is the parameter the most used on PET imaging for glioma assessment (2). However, the use of PET kinetic parameters (needing a multi-frame acquisition), which add further knowledge regarding tumor behavior, has recently spread with newly developed PET systems (5-7). Quantitative parameters extracted from PET images can be influenced by the reconstruction algorithm used for image generation (8). Conventional iterative reconstruction methods lead to distortion at the edges of the field-of-view and PET images are subject to partial volume effect and spillover of signal between adjacent functional regions (9). The point-spread function (PSF) reconstruction technique recently available for PET imaging improved the image contrast to noise ratios and the spatial resolution, correcting for the photon mispositioning and increasing radial offset from the isocenter in PET systems (10-13). Studies using several PET tracers suggested that the implementation of PSF reconstruction increases static parameters values compared with non-PSF data (14-17). However, the influence of PSF on FDOPA quantification for glioma are missing. To the best of our knowledge, the impact of the PSF reconstruction on kinetic parameters quantification has not been studied yet.
The aim of the present study was to evaluate the impact of PSF reconstruction on quantitative FDOPA PET parameters in gliomas, especially for kinetic parameters.
We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/qims-21-742).
Methods
Patients
Fourteen patients from June 2018 to September 2019, from the “GLIROPA” clinical trial (NCT03525080) were analyzed. All patients were newly diagnosed for gliomas and selected for resective surgery in our center. Included patients had to be at least 18 years-old and covered by national health insurance; and neither be pregnant, nor in an emergency situation, nor be treated by carbidopa, catechol-O-methyl transferase inhibitor, haloperidol or reserpine medication. Sixteen patients were finally included. The dynamic acquisition was unsuccessful for 2 patients. All patients provided their written informed consent. This study has been performed in accordance with the Declaration of Helsinki (as revised in 2013) and approved by an independent national research ethics committee (IRB: CPPIDF1-2018-ND27-cat.2).
PET/CT imaging protocol
The patients were required to fast at least 4 h before undergoing the imaging protocol. Each patient underwent a CT scan without contrast agent injection, followed by a 40-min PET acquisition using list-mode acquisition with a single field of view centered on the brain (Siemens Healthcare Biograph mCT Flow, Knoxville, TN, USA). At the start of the PET scan, 2 MBq/kg of FDOPA were administered intravenously, without carbidopa premedication. PET data were reconstructed using Time of Flight (TOF) 3D ordered-subsets expectation maximization iterative algorithm (8 iterations, 21 subsets) with and without PSF, and smoothed with a gaussian filter at a full width at half-maximum of 4.0 mm. Corrections for random coincidences, normalization, dead-time losses, scatter, and attenuation were applied. Voxel size (XYZ) was 1×1×2 mm3. From the bolus arrival time, a 0–20-min static and dynamic (8×15 sec, 2×30 sec, 2×60 sec, 3×300 sec) series were reconstructed (18).
Image analysis
Spherical volumes-of-interest (VOI) of 1 cm3 on biopsy samples locations and on the contralateral cortex (background) were generated by a nuclear medicine physician with the Syngo.via software (Siemens Healthcare) on the 0–20-min static reconstruction without PSF. The background VOI was projected onto the 0–20-min static reconstruction with PSF. The 33 spherical tumor VOIs were drawn on the 0–20-min static PET reconstruction without PSF registered with the FLAIR weighted MR imaging used for the thirty-three MRI-guided brain biopsies. Then, the tumors VOIs drawn were projected onto the 0–20-min static PET reconstruction with PSF and projected onto each frame of the dynamic PET reconstruction with and without PSF. On the early PET image with the maximum blood pool activity of the dynamic reconstruction without PSF, a VOI was manually drawn within the middle cerebral artery and projected onto each frame of the dynamic reconstruction with and without PSF to estimate an imaging-derived input function (IDIF). FDOPA IDIF was obtained after corrections for metabolites and hematocrit for each patient. IDIF was fitted to the measured fractions of metabolites taken from the publication of Huang et al. (19). The SUV for each voxel was calculated using the following formula: SUV = tissue radioactivity concentration/[injected dose/patient weight]. SUVmax and SUVmean were respectively the maximum and the mean of the SUVs of the VOI. The reversible single-tissue compartment model with blood volume parameter (with K1 = rate constant from blood to tissue, k2 = rate constant from the tissue compartment to the arterial blood) was used to extract kinetic parameters (PMOD software version 3.8; PMOD Technologies; Zürich, Switzerland) (18).
Statistical analysis
Descriptive parameters were expressed as median, and range. Differences of quantitative parameters between both reconstruction algorithms were compared using the Wilcoxon signed-rank test for paired data. A P value <0.05 was considered as statistically significant, with false discovery rate adjustment for multiple comparisons. Statistical analysis was performed using IBM SPSS Statistics 25 (SPSS Ltd.).
Results
Patients
There were 9 men and 5 women, with a median age of 40 years (range, 23–66 years). The distribution of the 14 cases was as follows: 6 astrocytomas-IDH-mutant; 2 oligodendrogliomas/1p19q-codeleted-IDH-mutant; and 6 IDH-wild-type glioblastomas.
Impact of PSF reconstruction on quantitative parameters
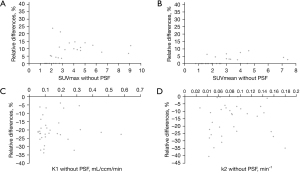
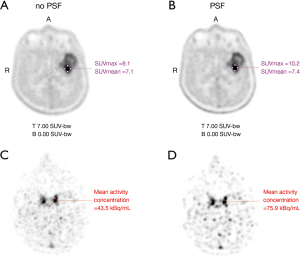
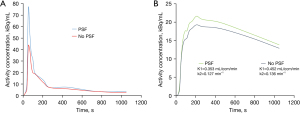
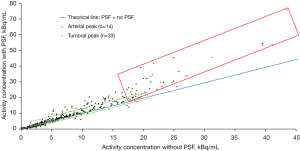
All data are set out in Table 1. Regarding static parameters, PSF images provided higher tumor SUVmax than non-PSF images for 23/33 VOIs [median SUVmax =3.0 (range, 1.4–10.2) with PSF vs. 2.7 (range, 1.4–9.1) without PSF; P<0.001] and higher tumor SUVmean for 12/33 VOIs [median SUVmean =2.0 (range, 0.8–7.6) with PSF vs. 2.0 (range, 0.8–7.4) without PSF; P=0.002]. Background SUVmax and SUVmean were not statistically affected by PSF reconstruction [respectively median SUVmax =1.7 (range, 1.3–2.0) with PSF vs. 1.7 (range, 1.3–1.9) without PSF; P=0.346 and median SUVmean =1.5 (range, 1.0–1.8) with PSF vs. 1.5 (range, 1.0–1.7) without PSF; P=0.371]. Applying the optimal cut-off ratio to discriminate between low- and high-grade gliomas reported by Bund et al. (20), three tumor locations classified as low-grade on PET images without PSF were classified as high-grade on PET images. Regarding kinetic parameters, PSF showed significantly lower tumors K1 and k2 than non-PSF (respectively median K1 =0.077 (range, 0.043–0.445) mL/ccm/min with PSF vs. 0.101 (range, 0.055–0.578) mL/ccm/min without PSF; P<0.001 and median k2 =0.070 (range, 0.025–0.146) min–1 with PSF vs. 0.081 (range, 0.027–0.180) min–1 without PSF; P<0.001). All lesions showed a lower K1 and k2 when using PSF reconstruction compared to non-PSF. Applying the optimal cut-off for k2 to discriminate between low- and high-grade gliomas found in our previous study (17), three tumor locations classified as high-grade on PET images without PSF were classified as low-grade on PET images. Percentage differences in quantitative parameters using PSF and non-PSF reconstruction are displayed in Figure 1. Examples of higher static parameters and lower kinetic parameters using PSF images compared to non-PSF images in a 59-year-old man with a left temporal IDH1-wild-type glioblastoma are shown in the Figure 2 and Figure 3. Comparing activity concentrations extracted from each frame of the corrected arterial time-activity curves and tumor time-activity curves with PSF as a function of the activity concentrations without PSF, the present results show that highest activity concentrations are more impacted by the PSF reconstruction than lowest activity concentrations (Figure 4).
Table 1
| Parameters | Without PSF | With PSF | P value | Relative differences (%) |
|---|---|---|---|---|
| Tumoral VOI | ||||
| SUVmax | 3.4/2.7 (2.0) (1.4–9.1) | 3.6/3.0 (2.2) (1.4–10.2) | <0.001* | +7.4/+6.7 (6.8) (0.0/+23.8) |
| SUVmean | 2.6/2.0 (1.6) (0.8–7.4) | 2.7/2.0 (1.6) (0.8–7.6) | 0.002* | +1.8/0.0 (2.6) (0.0/+8.7) |
| K1 (mL/ccm/min) | 0.153/0.101 (0.121) (0.055–0.578) | 0.122/0.077 (0.097) (0.043–0.445) | <0.001* | –20.5/–21.9 (8.0) (–4.2/–34.2) |
| k2 (min–1) | 0.086/0.081 (0.039) (0.027–0.180) | 0.073/0.070 (0.033) (0.025–0.146) | <0.001* | –16.0/–12.2 (10.9) (–0.1/–40.5) |
| Background VOI | ||||
| SUVmax | 1.6/1.7 (0.2) (1.3–1.9) | 1.6/1.7 (0.3) (1.3–2.0) | 0.346 | +1.2/0.0 (2.3) (0.0/+5.6) |
| SUVmean | 1.4/1.5 (0.2) (1.0–1.7) | 1.4/1.5 (0.3) (1.0–1.8) | 0.371 | +0.9/0.0 (2.2) (0.0/+6.3) |
*, P value <0.05 was considered statistically significant. Mean/median values displayed with their respective standard deviation and range. PSF, point-spread function; SUV, standardized uptake value; VOI, volumes-of-interest.
Discussion
In this study, we investigated the influence of PSF on the quantitative static and kinetic features extracted from FDOPA PET/CT for glioma assessment at diagnosis. Other radiotracers have been synthetized for glioma assessment (other amino-acid radiotracers like 11C-methionine and [18F]-fluoroethyl-tyrosine or fibroblast activation protein ligands). Several studies have shown that these different amino-acid radiotracers performed equally well in the assessment of gliomas (21-23). However, the use of 11C-methionine is limited because carbon 11’s half-life is 20 min and the [18F]-fluoroethyl-tyrosine radiotracer is less commonly available amino acid radiotracer than FDOPA in Europe, and particularly in France (24).
Firstly, regarding static parameters, our results showed that tumor SUVmax and SUVmean obtained with PSF reconstruction were significantly higher than without PSF reconstruction. These results are consistent with data from previous studies assessing the impact of PSF on these parameters (11-14). Regarding the SUVmean, the results of the current study showed smaller deviations compared to the SUVmax. These observations are in line with results of recent studies (11,12), confirming that SUVmax is more sensitive to reconstruction parameters than SUVmean (25). In our study, the mean percentage change after PSF implementation is considerably lower than in most reports assessing the impact of PSF (12,14,26-28). This could be linked to the fact that the current study compared images reconstructed with and without PSF but with TOF algorithm, which already provides higher SUV than SUV obtained with reconstruction using non-TOF algorithm (9,13). The PET images showed essentially low tumors FDOPA uptake in our study. This could be the reason why the percentage changes were low. In the current study, no significant difference of the static parameters extracted with and without PSF was observed for background VOI. Furthermore, comparing activity concentrations from each frame of the arterial and tumor time-activity curves used for the kinetic analysis, the present results confirm that differences between PSF and non-PSF reconstruction decrease with lower tracer uptake, in line with previous published data (11,26,27). Indeed, PSF algorithm produces edge overshoot called Gibbs artifact, resulting in an artificially increased contrast of hot structures and hyperresolution of focal uptake artifacts, especially if combined with TOF algorithm (29). At low contrast, these are essentially absent but are present at high contrast. Authors showed that the visual lesion detectability is improved by the edge elevations (30,31).
Secondly, regarding kinetic parameters, PSF showed significantly lower K1 and k2 than non-PSF. This is due the fact that the increase of activity concentration measured using PSF is more prominent for the arterial activity concentrations than the tumoral activity concentrations. To the best of our knowledge, this study is the first analyzing the impact of the PSF reconstruction on quantitative parameters extracted with dynamic acquisition.
Munk et al. showed that recovery coefficients were non-monotonic as function of lesion size and less reproducible for PSF reconstructed images of small lesions using clinically relevant phantom (32). So, the tumor and blood pool VOI sizes should be considered for quantitative analysis with PSF. Moreover, Bowen et al. demonstrated that the choice of PSF modelling in the reconstruction can have a significant effect on kinetic parameters (33). Due to these Gibbs artefacts and quantitative errors, this method is not recommended for FDOPA PET/CT quantification in glioma (2). Therefore, in our institution, both PSF and non-PSF reconstructions are routinely used for FDOPA PET/CT performed for glioma assessment. PSF reconstruction is used for visual assessment and the non-PSF reconstruction is used to extract quantitative parameters.
The main limitation of this study is that we focused only on small VOIs. It is necessary to examine the effects of PSF on lesions with larger size. The low number of patients analyzed is another limitation. Further investigations analyzing the impact of PSF for the different glioma subgroups are needed.
The present study confirms that PSF significantly modifies quantitative features from tumor, especially for kinetic parameters. The FDOPA uptake quantification seems to be useful for the management of glioma, so the extraction of quantitative parameters should be performed with caution when using PSF algorithms.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/qims-21-742
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-742). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by an independent national research ethics committee (CPPIDF1-2018-ND27-cat.2). All individual participants consented to the use of their medical records for research purposes.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet 2012;205:613-21. [Crossref] [PubMed]
- Law I, Albert NL, Arbizu J, Boellaard R, Drzezga A, Galldiks N, la Fougère C, Langen KJ, Lopci E, Lowe V, McConathy J, Quick HH, Sattler B, Schuster DM, Tonn JC, Weller M. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and 18FFDG: version 1.0. Eur J Nucl Med Mol Imaging 2019;46:540-57. [Crossref] [PubMed]
- Somme F, Bender L, Namer IJ, Noël G, Bund C. Usefulness of 18F-FDOPA PET for the management of primary brain tumors: a systematic review of the literature. Cancer Imaging 2020;20:70. [Crossref] [PubMed]
- Xiao J, Jin Y, Nie J, Chen F, Ma X. Diagnostic and grading accuracy of 18F-FDOPA PET and PET/CT in patients with gliomas: a systematic review and meta-analysis. BMC Cancer 2019;19:767. [Crossref] [PubMed]
- Palard-Novello X, Blin AL, Bourhis D, Garin E, Salaün PY, Devillers A, Querellou S, Bourguet P, Le Jeune F, Saint-Jalmes H. Comparison of choline influx from dynamic 18F-Choline PET/CT and clinicopathological parameters in prostate cancer initial assessment. Ann Nucl Med 2018;32:281-7. [Crossref] [PubMed]
- Takesh M. The Potential Benefit by Application of Kinetic Analysis of PET in the Clinical Oncology. ISRN Oncol 2012;2012:349351. [Crossref] [PubMed]
- Girard A, Le Reste PJ, Metais A, Chaboub N, Devillers A, Saint-Jalmes H, Jeune FL, Palard-Novello X. Additive Value of Dynamic FDOPA PET/CT for Glioma Grading. Front Med (Lausanne) 2021;8:705996. [Crossref] [PubMed]
- Cheebsumon P, Yaqub M, van Velden FH, Hoekstra OS, Lammertsma AA, Boellaard R. Impact of 18FFDG PET imaging parameters on automatic tumour delineation: need for improved tumour delineation methodology. Eur J Nucl Med Mol Imaging 2011;38:2136-44. [Crossref] [PubMed]
- Soret M, Bacharach SL, Buvat I. Partial-volume effect in PET tumor imaging. J Nucl Med 2007;48:932-45. [Crossref] [PubMed]
- Tong S, Alessio AM, Kinahan PE. Noise and signal properties in PSF-based fully 3D PET image reconstruction: an experimental evaluation. Phys Med Biol 2010;55:1453-73. [Crossref] [PubMed]
- Conti M. Focus on time-of-flight PET: the benefits of improved time resolution. Eur J Nucl Med Mol Imaging 2011;38:1147-57. [Crossref] [PubMed]
- Akamatsu G, Mitsumoto K, Taniguchi T, Tsutsui Y, Baba S, Sasaki M. Influences of point-spread function and time-of-flight reconstructions on standardized uptake value of lymph node metastases in FDG-PET. Eur J Radiol 2014;83:226-30. [Crossref] [PubMed]
- Prieto E, Martí-Climent JM, Morán V, Sancho L, Barbés B, Arbizu J, Richter JA. Brain PET imaging optimization with time of flight and point spread function modelling. Phys Med 2015;31:948-55. [Crossref] [PubMed]
- Rogasch JM, Steffen IG, Hofheinz F, Großer OS, Furth C, Mohnike K, Hass P, Walke M, Apostolova I, Amthauer H. The association of tumor-to-background ratios and SUVmax deviations related to point spread function and time-of-flight F18-FDG-PET/CT reconstruction in colorectal liver metastases. EJNMMI Res 2015;5:31. [Crossref] [PubMed]
- Rogasch JMM, Albers J, Steinkrüger FL, Lukas M, Wedel F, Amthauer H, Furth C. Point Spread Function Reconstruction for Integrated 18F-FET PET/MRI in Patients With Glioma: Does It Affect SUVs and Respective Tumor-to-Background Ratios? Clin Nucl Med 2019;44:e280-5. [Crossref] [PubMed]
- Armstrong IS, Kelly MD, Williams HA, Matthews JC. Impact of point spread function modelling and time of flight on FDG uptake measurements in lung lesions using alternative filtering strategies. EJNMMI Phys 2014;1:99. [Crossref] [PubMed]
- You H, Sanli Y, Subramaniam RM. Impact of Point-Spread Function Reconstruction on 68Ga-DOTATATE PET/CT Quantitative Imaging Parameters. AJR Am J Roentgenol 2019;213:683-8. [Crossref] [PubMed]
- Girard A, Saint-Jalmes H, Chaboub N, Le Reste PJ, Metais A, Devillers A, Le Jeune F, Palard-Novello X. Optimization of time frame binning for FDOPA uptake quantification in glioma. PLoS One 2020;15:e0232141. [Crossref] [PubMed]
- Huang SC, Barrio JR, Yu DC, Chen B, Grafton S, Melega WP, Hoffman JM, Satyamurthy N, Mazziotta JC, Phelps ME. Modelling approach for separating blood time-activity curves in positron emission tomographic studies. Phys Med Biol 1991;36:749-61. [Crossref] [PubMed]
- Bund C, Heimburger C, Imperiale A, Lhermitte B, Chenard MP, Lefebvre F, Kremer S, Proust F, Namer IJ. FDOPA PET-CT of Nonenhancing Brain Tumors. Clin Nucl Med 2017;42:250-7. [Crossref] [PubMed]
- Lapa C, Linsenmann T, Monoranu CM, Samnick S, Buck AK, Bluemel C, Czernin J, Kessler AF, Homola GA, Ernestus RI, Löhr M, Herrmann K. Comparison of the amino acid tracers 18F-FET and 18F-DOPA in high-grade glioma patients. J Nucl Med 2014;55:1611-6. [Crossref] [PubMed]
- Kratochwil C, Combs SE, Leotta K, Afshar-Oromieh A, Rieken S, Debus J, Haberkorn U, Giesel FL. Intra-individual comparison of 18F-FET and 18F-DOPA in PET imaging of recurrent brain tumors. Neuro Oncol 2014;16:434-40. [Crossref] [PubMed]
- Becherer A, Karanikas G, Szabó M, Zettinig G, Asenbaum S, Marosi C, Henk C, Wunderbaldinger P, Czech T, Wadsak W, Kletter K. Brain tumour imaging with PET: a comparison between 18Ffluorodopa and 11Cmethionine. Eur J Nucl Med Mol Imaging 2003;30:1561-7. [Crossref] [PubMed]
- Verger A, Metellus P, Sala Q, Colin C, Bialecki E, Taieb D, Chinot O, Figarella-Branger D, Guedj E. IDH mutation is paradoxically associated with higher 18F-FDOPA PET uptake in diffuse grade II and grade III gliomas. Eur J Nucl Med Mol Imaging 2017;44:1306-11. [Crossref] [PubMed]
- Boellaard R, Krak NC, Hoekstra OS, Lammertsma AA. Effects of noise, image resolution, and ROI definition on the accuracy of standard uptake values: a simulation study. J Nucl Med 2004;45:1519-27. [PubMed]
- Andersen FL, Klausen TL, Loft A, Beyer T, Holm S. Clinical evaluation of PET image reconstruction using a spatial resolution model. Eur J Radiol 2013;82:862-9. [Crossref] [PubMed]
- Gellee S, Page J, Sanghera B, Payoux P, Wagner T. Impact of the point spread function on maximum standardized uptake value measurements in patients with pulmonary cancer. World J Nucl Med 2014;13:128-31. [Crossref] [PubMed]
- Lasnon C, Hicks RJ, Beauregard JM, Milner A, Paciencia M, Guizard AV, Bardet S, Gervais R, Lemoel G, Zalcman G, Aide N. Impact of point spread function reconstruction on thoracic lymph node staging with 18F-FDG PET/CT in non-small cell lung cancer. Clin Nucl Med 2012;37:971-6. [Crossref] [PubMed]
- Rapisarda E, Bettinardi V, Thielemans K, Gilardi MC. Image-based point spread function implementation in a fully 3D OSEM reconstruction algorithm for PET. Phys Med Biol 2010;55:4131-51. [Crossref] [PubMed]
- Kadrmas DJ, Casey ME, Conti M, Jakoby BW, Lois C, Townsend DW. Impact of time-of-flight on PET tumor detection. J Nucl Med 2009;50:1315-23. [Crossref] [PubMed]
- Schaefferkoetter J, Casey M, Townsend D, El Fakhri G. Clinical impact of time-of-flight and point response modeling in PET reconstructions: a lesion detection study. Phys Med Biol 2013;58:1465-78. [Crossref] [PubMed]
- Munk OL, Tolbod LP, Hansen SB, Bogsrud TV. Point-spread function reconstructed PET images of sub-centimeter lesions are not quantitative. EJNMMI Phys 2017;4:5. [Crossref] [PubMed]
- Bowen SL, Byars LG, Michel CJ, Chonde DB, Catana C. Influence of the partial volume correction method on (18)F-fluorodeoxyglucose brain kinetic modelling from dynamic PET images reconstructed with resolution model based OSEM. Phys Med Biol 2013;58:7081-106. [Crossref] [PubMed]


