
Prior information guided auto-contouring of breast gland for deformable image registration in postoperative breast cancer radiotherapy
Introduction
Breast cancer is the most diagnosed cancer and the leading cause of cancer death among women worldwide (1). Breast-conserving therapy (BCT), which involves a wide local excision followed by radiotherapy to the whole breast, has become the standard treatment for early-stage breast cancer (2). In postoperative breast cancer radiotherapy, accurate delineation of the breast tumor bed and its target volume is essential, and registering the image acquired before surgery to the image acquired after surgery can help to define target volumes. Recently, deformable image registration (DIR) has been introduced to define the breast tumor bed with different image modalities, including CT (3), contrast CT (4), MRI (5), and PET-CT (6). However, most of these are intensity-based methods, and no application of feature-based or hybrid methods has been reported in breast cancer radiotherapy.
As reported by many researchers, incorporating contour information with intensity-based DIR can achieve better registration accuracy (7,8). However, in the registration of breast CT, the region of interest (ROI) that could be used as constraints are almost unavailable except the unaffected breast gland. In this study, the contour of breast gland was only used for input of DIR algorithm and not for treatment planning purpose. The contour of breast gland is important for ROI-based image registration algorithms. It could not only be used as additional feature in conjunction with intensity-based DIR for finer searching of tumor bed and its target volume, but also serve as ROI for quantitative assessment of registration accuracy. However, contouring of breast gland is quite difficult in planning CT. For one reason breast gland do not have a unique CT number distribution in CT images. Thus, it cannot be segmented by simply using traditional segmentation methods based on CT numbers (such as gray-level thresholding and region growing) or shape-based methods (such as snakes and active shape model). For another reason, due to surgical interference (deformation and removal of glandular tissue) and post-surgical changes (seroma and edema, etc.) the density of breast gland regions may be affected considerably, which may potentially result in poorer contrast from surrounding tissue.
The manual contouring process is time-consuming and subject to considerable inter-and intra-observer variability (9). As an auxiliary tool, the automatic contouring of breast gland has mostly been investigated in the field of diagnostic CT with higher contrast. Zhou et al. (10,11) proposed a fully automated scheme for segmenting breast gland in non-contrast CT images. This scheme calculates each voxel’s probability of belonging to breast gland or chest muscle in CT images as the reference of the segmentation and identifies breast gland based on CT number automatically. The probability is estimated from the location of breast gland and chest muscle in CT images, and the location is investigated from a knowledge base that stores pre-recognized anatomical structures using different CT scans. Using the probabilistic atlas and density (CT number) estimation, the proposed method was efficient and robust for breast gland segmentation. However, there are challenges for planning CT with low-contrast ROIs. Firstly, the location and shape of breast gland vary considerably and it is difficult to construct a “universal atlas” for all patients. Secondly, surgical interference and post-surgical changes may compromise the accuracy of breast gland locations based on CT number.
Deep learning (DL) methods are capable of many computer vision tasks, and there has been increasing interest in their application to radiation therapy. In this study, a prior information guided DL network was used to contour breast gland automatically from planning CT. In Methods section, the prior information for prediction is first introduced. Then, the DL network for segmentation is described in detail. Next, the prediction model’s performance is evaluated quantitatively. In Results Section, the results of DL network for auto-contouring are summarized. Finally, the proposed method’s advantages and disadvantages are discussed, and the future work is prospected.
Methods
Datasets
Six left-sided breast cancer patients who underwent breast-conserving surgery and postoperative radiotherapy in our hospital were enrolled in this study. The mean age of patients was 50 years (range, 44–59 years), and the pathological diagnosis in all was invasive ductal carcinoma with a stage of T1–T2N0M0. All patients underwent a lumpectomy with sentinel lymph node dissection, and tumor-negative margins were ensured during a single operation. The breast volumes varied from 373 to 919 cm3, averaging 617 cm3. The study was approved by the local Ethics Committee and informed consent was waived in this retrospective study.
The image data originally consisted of six preoperative CT scans and six planning CT scans. The preoperative CT scan was contrast-enhanced and performed 1 week before breast conserving surgery (BCS), while the planning CT scan was non-contrast-enhanced and performed on average 10 weeks after BCS. Patients undergoing preoperative CT were placed in the supine position, with their hands naturally extended and placed on both sides of the head. Image acquisitions were performed according to the standard clinical protocol, 60 s after the injection of 100 cm3 intravenous contrast agent. The dimensions of preoperative CT volume varied from 512×512×51 to 512×512×60 in voxels, and the slice thickness was 5.0 mm. The in-plane image resolution varied from 0.68×0.68 mm to 0.94×0.94 mm. In postoperative CT simulation, the patients were in the supine position, immobilized on a breast bracket with no degree of incline and placed using arm support (with both arms above the head). The dimensions of planning CT volume varied from 512×512×41 to 512×512×53 in voxels, and the in-plane image resolution varied from 1.18×1.18 mm to 1.37×1.37 mm. The slice thickness was also 5.0 mm. The contour of breast gland in both preoperative CT and planning CT was delineated and peer-reviewed by two radiation oncologists in our department using the Pinnacle treatment planning system (Phillips Medical Systems). For any inconsistent contours, an agreement was reached based on their union. The breast gland contour in planning CT was defined as the ground truth label.
All preprocessing was performed in the 3D Slicer platform (12) (www.slicer.org). The volumes were all resampled to an isotropic resolution of 1×1×5 mm and then cropped to 256×256×32 pixels around the breast’s centroid. Data augmentation was implemented using the SlicerIGT extension (13). To realistically simulate human body deformation (equivalent to repeat scanning), a smoothly changing dense deformation field was applied on both image data and ground truth labels. For this purpose, two fiducial point lists (each including 57 points) were specified on one image. Two pairs of these points defined the LR-direction displacements (one chosen from 1/2/3 mm and the other chosen from 4/6/8 mm), which resulted in nine combinations, while the remaining pairs of points stayed at the same position. Thin-plate spline transform was then used to align each pair of points in the point list, and the resulting transform was then interpolated and smoothed (14). Each image data and ground truth label was deformed nine times using the resulting transform, resulting in 120 sets of CT scan and label images available for the model training. All CT scan and label images were resized to 240×240×152 to meet the input requirement of 3D U-Net, and the voxel size was changed to 1.07×1.07×1.05 mm after resizing.
Prior information
The overall workflow, which consists of training and testing stages, is shown in Figure 1. The dashed line indicates the data flow for model training, while the solid line indicates the data flow for model testing. The preoperative CT with its contour and the planning CT with its contour were exported from the treatment planning system. The affine registration between the preoperative CT and planning CT was first performed using Elastix registration software (15,16) integrated in 3D Slicer. The resulting transform was then applied to map the contour of breast gland in preoperative CT to its correspondence in planning CT. The affine registration and label transformation were all performed in 3D. This transformed contour provides a preliminary estimation of breast gland in planning CT and facilitates the searching of more accurate contour by the prediction model.
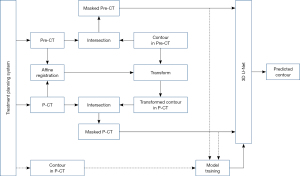
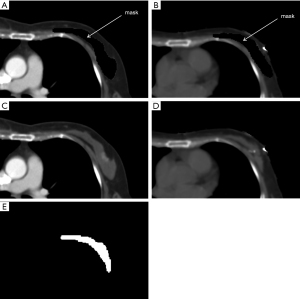
As shown in Figure 1, both preoperative and planning CT images with the masked contour of breast gland were generated via intersection operator and used as two input channels of the prediction model. The contour of breast gland in preoperative CT was masked, as shown in Figure 2A, and the transformed contour of breast gland in planning CT was used as prior information and masked in the same way as shown in Figure 2B. The CT number for both masked areas was set to −3,024, which is the cutoff value of CT image and easily identified from the surrounding tissue. For reference, the original preoperative CT and planning CT images without mask are shown in Figure 2C,2D, respectively. The ground truth labels for breast gland and non-gland in planning CT are represented by white and black colors, as shown in Figure 2E, which is the third input channel of the prediction model.
3D U-Net architecture
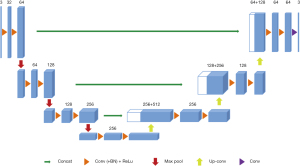
A 3D U-Net architecture previously used for brain tumor segmentation was employed in this study (17), as shown in Figure 3. The initial series of convolutional layers were interspersed with max pooling layers and successively decreased the input image’s resolution from 132 to 9 in the encoding process (left part of Figure 3). These layers were followed by a series of convolutional layers interspersed with upsampling operators, successively increasing the input image’s resolution from 9 to 132 in the decoding process (right part of Figure 3). A batch normalization layer was introduced before each rectified linear unit (ReLU) layer. In the original U-Net implementation, all convolution, max pooling, and upsampling operations were carried out in 2D (18). This was later extended to a 3D U-Net by Çiçek et al. (17).
The 3D U-Net was trained on image patches, and the test patches were finally stitched into a complete segmented test volume through an overlap-tile strategy. A random patch extraction datastore that contains the training image and pixel label data was used to feed the training data to the network. The patch size was 132×132×132 voxels, and 16 randomly positioned patches were extracted from each pair of volumes and labels during training. For prediction, the overlap-tile strategy was used to predict the labels for each test volume, which was then padded to make the input size a multiple of the network’s output size and compensated for the effects of valid convolution. The overlap-tile algorithm selected overlapping patches, predicted the labels for each patch, and then recombined the patches, which avoids border artifacts using the valid part of the convolution in the neural network (18).
Like the standard U-Net, the deep-learning network used in this study has an encoding path and a decoding path. In the encoding path, each layer contains two 3×3×3 convolutions, each followed by a ReLu, and then a 2×2×2 max pooling with strides of two in each dimension. In the decoding path, each layer consists of an up-convolution of 2×2×2 by strides of two in each dimension, followed by two 3×3×3 convolutions, each followed by a ReLu. Shortcut connections from layers of equal resolution in the analysis path provide the essential high-resolution features to the synthesis path. In the last layer, a 1×1×1 convolution reduces the number of output channels to the number of labels which was 2 in our case. The architecture has 1.907e7 parameters in total. Bottlenecks could be avoided by doubling the number of channels already before max pooling, as suggested in (19) and this scheme was adopted in the synthesis path.
Evaluations
A three times five-fold cross-validation procedure was applied to the 60 patient datasets. Each fold (containing 12 patients) was selected as the test data, and the remaining four folds (containing 48 patients) were used as training data. Averaging the prediction accuracy on five folds led to the overall accuracy estimate of the proposed method. The dice similarity coefficient (DSC), which is frequently used for training U-Nets (18), was used as the loss function. The prediction model’s performance with the input of prior information (as shown in Figure 2A,2B) was compared to the prediction model without the input of prior information (as shown in Figure 2A,2D). The average DSC, together with standard deviation of the prediction model with two types of input images were calculated, and the average Hausdorff distance (HD), together with standard deviation, were also calculated. Paired t-test was used to compare the results, and P<0.05 was considered statistically significant. All statistical analyses were performed in R statistical software (version 3.6.3).
The Adaptive moment estimation (Adam) was used to optimize the loss function. The initial learning rate was set as 0.0005, the learning rate drop factor as 0.95, and the validation frequency as 20. The network was trained with maximal 50 epochs and tested on a workstation equipped with NVIDIA Geforce GTX 1080 TI GPU with 11 GB of memory.
Results
The training time for 3D U-Net was approximately 30 hours, while the prediction time was 20 seconds per patient. In this binary segmentation, each pixel is labeled as breast gland or non-gland, as shown in Figure 2E. Without the input of prior information, the average DSC of breast gland was 0.775±0.065, while with the input of prior information, this was 0.830±0.038. The difference between the average DSC of breast gland resulting from the prediction models without and with the input of prior information was statistically significant (0.775 vs. 0.830, P=0.0014<0.05). Comparatively, the average DSC of breast gland was 0.70±0.01 using the traditional gray-level thresholding segmentation method. For fair comparison, without the input of prior information, the average HD of breast gland was 44.979±20.565, while with the input of prior information, this was 17.896±5.737. The difference between the average HD of breast gland resulting from the prediction models without and with the input of prior information was statistically significant (44.979 vs. 17.896, P=0.002<0.05). Comparatively, the average HD of breast gland was 55.898±25.345 using the traditional gray-level thresholding segmentation method.
The predicted labels of breast gland with the input of prior information overlaid on the planning CT images with the ground truths for all six patients are shown in Figure 4, with the predicted labels in white and the ground truths in black. The segmentation results are displayed in three orthogonal views. It showed that the majority of both contours were highly similar. The predicted contour had smoother boundary than the contour which was manually delineated by radiation oncologist.

Discussion
In this study, a prior information guided DL network was developed to contour breast gland automatically from planning CT. To the best of our knowledge, no similar study has been reported in the field of postoperative breast cancer radiotherapy. The preliminary results showed that the introduction of prior information with affine transform successfully improves the prediction accuracy of breast gland contour. This improvement may be attributed to the geometrical information provided by the preliminary estimation of breast gland in planning CT, which facilitates the searching of more accurate contour in subsequent step by the DL model.
An early reported DL based method for segmentation of breast was specified for diagnostic CT with higher contrast between anatomical structures (20). The advantage of the DL based method in comparison with the regional intensity based or overall histogram-based unsupervised learning methods (21) is obvious. It depends not only on the voxel intensities but also morphological variations and gradients within the CT image. However, this method is based on 2D U-Net, which is insufficient in learning spatial information between adjacent slices. Also, it could fail when planning CT with low-contrast ROIs are presented. In our study, the prior information was introduced to lend preliminary geometrical distribution of breast gland to the training model and guide the searching of more accurate and reliable contour.
Both preoperative CT and planning CT images (without mask) were initially used for the input of network training but it didn’t work well (average DSC =0.33). It looks like there were few high-contrast features in breast region which made less attention to network training. Therefore, the contour of breast gland in preoperative CT was masked with the cutoff value of CT image to enhance the contrast and it did work well (average DSC =0.78). Furthermore, the transformed contour of breast gland (presented as prior information) in planning CT was masked in the same way, and its effectiveness was validated by experiments (average DSC =0.83). It may be possible to provide an additional channel for prior information, although this will cost more resource of network. This would be a good attempt and will be tested in the future.
In this study only diagnostic CT and planning CT were used. There could be more input image modalities such as CBCT, ultrasound, and MRI. With more imaging information, the prediction accuracy of this model could be further improved. Besides, prior information with affine transform is less accurate. In our study, the affine registration only provided a preliminary estimation of breast gland in planning CT. It would be fine for patients with significant contour overlapping and could fail otherwise. For those patients without significant overlapping, more advanced non-rigid (deformable) transform would be introduced, which bring more accurate prior information to the training model.
There are several limitations of this study. First, the training set is limited even after data augmentation. In the data augmentation step, the deformation field was introduced to simulate real human body motion, which could be regarded as serial CT images acquired from a patient over multiple days. The augmented data could correlate with the original data moderately, and the correlation could be a factor causing bias in the network training. However, for validation of the effectiveness of prior information, its effect would be less influential. Second, massive cross-validation is needed to ensure the training’s stability due to the relatively small sample size. In the future, more data will be collected to make the model more reliable. Finally, the contouring of breast gland was manually performed on preoperative CT, and it would be better to accomplish this by automatic methods (21,22). In the future, traditional auto-contouring tools will be employed to accelerate this process.
Conclusions
A significant improvement in the prediction accuracy of breast gland contour was achieved with the introduction of prior information. This prior information provided an initial estimation of geometrical distribution of breast gland in planning CT and facilitated the subsequent searching of the refined contour by the prediction model. This method provides an effective way of identifying low-contrast ROIs in planning CT for postoperative breast cancer radiotherapy.
Acknowledgments
Funding: This work was partially supported by the National Natural Science Foundation of China (11875320, 11975312), Beijing Hope Run Special Fund of Cancer Foundation of China (LC2018A08), and Beijing Municipal Natural Science Foundation (7202170).
Footnote
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1141). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. JD served as the unpaid Guest Editor of the special issue. The authors report grants from the National Natural Science Foundation of China, Beijing Hope Run Special Fund of Cancer Foundation of China, and Beijing Municipal Natural Science Foundation during the study. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Informed consent was waived in this retrospective study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Litière S, Werutsky G, Fentiman IS, Rutgers E, Christiaens MR, Van Limbergen E, Baaijens MH, Bogaerts J, Bartelink H. Breast conserving therapy versus mastectomy for stage I-II breast cancer: 20 year follow-up of the EORTC 10801 phase 3 randomised trial. Lancet Oncol 2012;13:412-9. [Crossref] [PubMed]
- Wodzinski M, Skalski A, Ciepiela I, Kuszewski T, Kedzierawski P, Gajda J. Improving oncoplastic breast tumor bed localization for radiotherapy planning using image registration algorithms. Phys Med Biol 2018;63:035024 [Crossref] [PubMed]
- Kirova YM, Servois V, Reyal F, Peurien D, Fourquet A, Fournier-Bidoz N. Use of deformable image fusion to allow better definition of tumor bed boost volume after oncoplastic breast surgery. Surg Oncol 2011;20:e123-5. [Crossref] [PubMed]
- Yu T, Li JB, Wang W, Xu M, Zhang YJ, Shao Q, Liu XJ, Xu L. A comparative study based on deformable image registration of the target volumes for external-beam partial breast irradiation defined using preoperative prone magnetic resonance imaging and postoperative prone computed tomography imaging. Radiat Oncol 2019;14:38. [Crossref] [PubMed]
- Cho O, Chun M, Oh YT, Kim MH, Park HJ, Heo JS, Noh OK. Can initial diagnostic PET-CT aid to localize tumor bed in breast cancer radiotherapy: feasibility study using deformable image registration. Radiat Oncol 2013;8:163. [Crossref] [PubMed]
- Motegi K, Tachibana H, Motegi A, Hotta K, Baba H, Akimoto T. Usefulness of hybrid deformable image registration algorithms in prostate radiation therapy. J Appl Clin Med Phys 2019;20:229-36. [Crossref] [PubMed]
- Kubota Y, Okamoto M, Li Y, Shiba S, Okazaki S, Komatsu S, Sakai M, Kubo N, Ohno T, Nakano T. Evaluation of Intensity- and Contour-Based Deformable Image Registration Accuracy in Pancreatic Cancer Patients. Cancers (Basel) 2019;11:1447. [Crossref] [PubMed]
- van Mourik AM, Elkhuizen PH, Minkema D, Duppen JC, van Vliet-Vroegindeweij C. Multiinstitutional study on target volume delineation variation in breast radiotherapy in the presence of guidelines. Radiother Oncol 2010;94:286-91. [Crossref] [PubMed]
- Zhou X, Han M, Hara T, Fujita H, Sugisaki K, Chen H, Lee G, Yokoyama R, Kanematsu M, Hoshi H. Automated segmentation of mammary gland regions in non-contrast X-ray CT images. Comput Med Imaging Graph 2008;32:699-709. [Crossref] [PubMed]
- Zhou X, Kan M, Hara T, Fujita H, Sugisaki K, Yokoyama R, Lee G, Hoshi H. Automated segmentation of mammary gland regions in non-contrast torso CT images based on probabilistic atlas - art. no. 65123O. Proceedings of SPIE - The International Society for Optical Engineering 2007;6512.
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Ungi T, Lasso A, Fichtinger G. Open-source platforms for navigated image-guided interventions. Med Image Anal 2016;33:181-6. [Crossref] [PubMed]
- Gobbi DG, Peters TM. Generalized 3D nonlinear transformations for medical imaging: an object-oriented implementation in VTK. Comput Med Imaging Graph 2003;27:255-65. [Crossref] [PubMed]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Front Neuroinform 2014;7:50. [Crossref] [PubMed]
- Çiçek Ö, Abdulkadir A, Lienkamp S, Brox T, Ronneberger O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. 2016.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015.
- Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. 2015.
- Ghazi P, Hernandez AM, Abbey C, Yang K, Boone JM. Shading artifact correction in breast CT using an interleaved deep learning segmentation and maximum-likelihood polynomial fitting approach. Med Phys 2019;46:3414-30. [Crossref] [PubMed]
- Caballo M, Boone JM, Mann R, Sechopoulos I. An unsupervised automatic segmentation algorithm for breast tissue classification of dedicated breast computed tomography images. Med Phys 2018;45:2542-59. [Crossref] [PubMed]
- Pike R, Sechopoulos I, Fei B. A minimum spanning forest based classification method for dedicated breast CT images. Med Phys 2015;42:6190-202. [Crossref] [PubMed]


