
Accuracy of individualized 3D modeling of ossicles using high-resolution computed tomography imaging data
Introduction
Improvements in modern imaging technology, computer-aided design (CAD), and rapid prototyping have resulted in significant repair and regenerative medicine progress. The development of clinical medicine has also taken a turn toward highly individualized and precise diagnoses and treatments. CAD 3D printing technology, which comprehensively integrates different areas of material science, and tissue engineering, has become some of the most promising research areas in biomedical science (1,2). Based on the development of imaging technology and high-resolution computed tomography (HRCT) scanning, multiplanar reformation (MPR) and 3D volume rendering (3D VR) can be applied to predefine the status of the ossicular chain in patients (3-5). However, whether 3D printing technology can accurately replicate individualized ossicles based on HRCT data requires further analysis.
The present study aimed to investigate the individualized 3D modeling of ossicles and carry out an accuracy study based on HRCT imaging data. The findings are intended to provide a theoretical foundation for realizing 3D printing individualized ossicles based on image data.
Methods
Auditory ossicular bone image data collection
Subjects
A total of 24 patients with sudden deafness sought consultation at the Department of Otorhinolaryngology Head and Neck Surgery at the Sixth Medical Center of People’s Liberation Army General Hospital (Beijing, China) between October 2013 and June 2014 were enrolled in the study.
The study was conducted following the Declaration of Helsinki and was approved by the Ethics Committee of the Sixth Medical Center of People’s Liberation Army General Hospital. All participants signed informed consent forms before participation.
HRCT data
All participants underwent a 256-slice spiral HRCT temporal bone axial scan, yielding a series of Digital Imaging and Communications in Medicine (DICOM) documents. HRCT scan data of normal temporal bones were collected before data conversion was done using Mimics 16.0 interactive medical image processing software (Materialise NV, Leuven, Belgium), computer-aided modeling, and 3D visualization of ossicles were carried out. Restoration authenticity and accuracy were verified, and the feasibility of 3D printing individualized ossicular prostheses was investigated.
The Philips Brilliance iCT 256-slice spiral CT (Phillips, the Netherlands) was used to obtain ossicular DICOM data of individualized ossicles through conventional continuous cross-sectional scanning of axial images and the use of bone algorithms. The slice thickness was 0.67 mm.
Medical imaging conversion software
HRCT image post-processing was carried out using the Philips Extended Brilliance Workspace (EBW) 4.0 workstation (Philips, the Netherlands). The 3D visualization, segmentation, extraction, and CAD of DICOM data were carried out using Mimics 16.0 interactive medical image processing software.
MPR and 3D reconstruction of ossicular HRCT data
In addition to the axial positions, 2D images of the other planes in the scanning data were taken from the MPR reconstruction. The axial images were processed into ossicular MPR and 3D VR data via the EBW 4.0 workstation. To generate sharper edges, thereby facilitating the determination of fine structural landmarks, the Y-sharp bone algorithm was used for the reconstruction of slices at an interval of 0.34 mm. In normal adults, 128–140 slices were obtained, and coronal-plane images of the ossicular chain’s structure were retained and displayed. To display the ossicles’ multiple landmark structures and facilitate measurement, we selected slices of the malleus, incus, stapes, and the malleus–incus parallel plane that could display joints on the same slice.
CAD processing of DICOM data
Mimics 16.0 interactive medical image processing software was selected for the processing of the DICOM data. The ossicular reconstruction process and parameter settings are described below and in Figure 1.
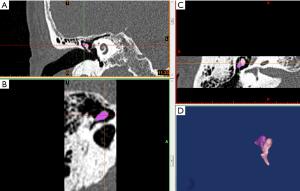
Image input
Mimics 16.0 was used to read coronal-plane temporal bone HRCT scan images (in DICOM format). This software automatically reads the value of the original HRCT scan. Slice thickness was 0.2 mm, slice interval was 0.2 mm, and pixel lattice was 0.195 mm, with a total of 75 slices.
Threshold segmentation
After the images’ quadratic differential processing, the threshold average of the ossicular tissues was defined as 1,200–4,000 HU. The threshold conversion method was used to further separate skeletal tissue data after the gradient had been adjusted.
Redundancy elimination
After segmented ossicular regional data were input, the ossicles were used as the center to identify the boundaries of the epitympanum, mesotympanum, and posterior tympanum. Redundant data at the sides and surrounding tissues on the X, Y, and Z axes were removed.
Ossicular calculation
Morphology-opening and region-growing algorithms were used to obtain ossicular data.
3D segmentation
3D segmentation was carried out according to the ossicles selected from grayscale screening and the ossicular structure obtained from opening algorithms, segmented independently and displayed.
The assessment method for reconstructed ossicles
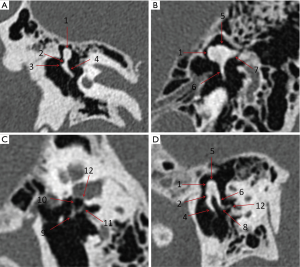
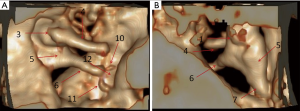
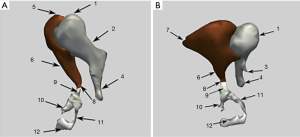
Three experienced doctors (2 otologists and 1 doctor from the imaging department) were responsible for carrying out independent observation of the images. A total of 12 anatomical landmarks in the ossicular chain were assessed: the caput mallei (label 1), collum mallei (label 2), processus lateralis mallei (label 3), manubrium mallei (label 4), corpus incudis (label 5), crus longum incudis (label 6), crus breve incudis (label 7), lenticular process (label 8), caput stapedis (label 9), crus anterius stapedis (label 10), crus posterius stapedis (label 11), and stapes footplate (label 12). A 3-point grading scheme was used to score the landmarks: 2 points for a clear and complete anatomical structure; 1 point for a visible anatomical structure, but unclear or incomplete display; and 0 points for a non-visible anatomical structure (6). When the structure assessment scores from the 3 doctors were the same, that score was taken as the final score of the structure. When the scoring was not consistent, the same score from 2 doctors was taken as the final score of that structure.
Statistical analysis
SPSS 18.0 software (SPSS, Chicago, IL, USA) was used for statistical processing, and the kappa consistency test was used. As middle-ear diseases were excluded from the study, it was assumed that the ossicular chain structures would be normal and that the scores could be used to calculate the sensitivity percentage of the 3 methods for displaying the ossicular structure. As it has already been shown that MPR and 3D VR reconstruction achieves relatively good restoration of the ossicular chain’s morphology, the Mimics 16.0 3D ossicular reconstruction’s consistency was tested against that of MPR and 3D VR reconstruction. The 3D VR reconstruction images were also used as a gold standard for evaluating the degree of restoration of the Mimics 16.0 3D ossicular reconstruction in the 12 anatomical landmarks.
In the kappa consistency test, a κ-value of >0.75 indicated very good consistency, a κ-value of 0.4–0.75 indicated good consistency, and a κ-value of <0.4 indicated poor consistency. P<0.05 was considered to be statistically significant.
Results
General characteristics
A total of 24 patients with sudden deafness (36 ears), who sought consultation at the Department of Otorhinolaryngology Head and Neck Surgery between October 2013 and June 2014, were enrolled in the study. Fourteen patients were male, and 10 were female.
Ossicular images obtained from MPD, 3D VR, and Mimics 16.0 software
Obtaining MPR ossicular reconstruction images
To display the ossicles’ multiple landmark structures and facilitate measurement, we selected slices of the malleus, incus, stapes, and the malleus-incus parallel plane, which can display joints at the same slice (Figure 2).
Obtaining 3D VR-reconstructed ossicular dynamic images
Dynamic images were obtained after reconstruction, which enabled omnidirectional observation. Two observation directions were selected for image display: the lateral–medial observation of the external auditory canal and the anterior observation of the posterior tympanum (Figure 3).
Obtaining mimics 16.0-restored visualizable ossicular images
Mimics 16.0 software was used to generate 3D visualizations from data in DICOM format, and target data were obtained from segmentation. Rapid prototyping output was in STL format, which is currently universally used for 3D printing. For the observation, evaluation, and design of the objects, the 3D images were used to synthesize images in PNG format to obtain independent ossicular images (Figure 4).
Consistency comparison
When comparing the Mimics 16.0-restored visualizable ossicles with MPR ossicular reconstruction, both methods had a display rate of >90% for the first 7 landmarks, and both had κ-values of >0.75. The display rate of the 8th landmark was 56.9% in Mimics 16.0 and 75% in MPR, with a κ-value of 0.566. The display rate of the 9th landmark was 61.1% in Mimics 16.0 and 90.3% in MPR, with a κ-value of 0.616. The display rate of the 10th landmark was 54.1% in Mimics 16.0 and 76.3% in MPR, with a κ-value of 0.842. The display rate of the 11th landmark was 62.5% in Mimics 16.0 and 84.7% in MPR, with a κ-value of 0.593. The display rate of the 12th landmark was 25% in Mimics and 76.4% in MPR, with a κ-value of 0.317. All kappa tests were P<0.05, denoting statistical significance (Table 1).
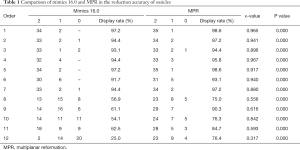
Full table
When comparing Mimics 16.0-restored individualized ossicles with 3D VR ossicular reconstruction, both methods had a display rate of >90% for the first 7 landmarks, and both had κ-values of >0.75. The display rate of the 8th landmark was 56.9% in Mimics 16.0 and 54.1% in 3D VR, with a κ-value of 0.818. The display rate of the 9th landmark was 61.1% in Mimics 16.0 and 80.1% in 3D VR, with a κ-value of 0.489. The display rate of the 10th landmark was 54.1% in Mimics 16.0 and 63.9% in 3D VR, with a κ-value of 0.798. The display rate of the 11th landmark was 62.5% in Mimics 16.0 and 81.9% in 3D VR, with a κ-value of 0.767. The display rate of the 12th landmark was 25% in Mimics 16.0 and 52.8% in 3D VR, with a κ-value of 0.298. All kappa tests were P<0.05, denoting statistical significance (Table 2).
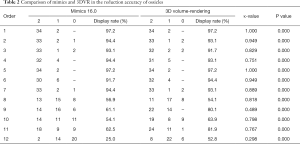
Full table
These results show that ossicular morphology converted by Mimics 16.0 software had a display rate of >90% when used to display 7 of the landmarks (the caput mallei, collum mallei, processus lateralis mallei, manubrium mallei, corpus incudis, crus longum incudis, and crus breve incudes). The Mimics 16.0-restored images showed excellent matching with the ossicular images from MPR and 3D VR reconstruction. In displaying the lenticular process, stapedis caput, crus anterius stapedis, and crus posterius stapedis, the display rate was around 60%, with κ-values of >0.4, showing good matching with the ossicular images from MPR and 3D VR reconstruction. However, the display rate of the stapes footplate was only 25% and showed greater differences, with 76.4% in MPR and 52.8% in 3D VR.
Discussion
The temporal bone is the most complex in the human body in terms of anatomical structure, and its ossicles are located deep within it. The ossicular chain that is formed of the 3 ossicles comprises the most delicate and complex joint connections in the human body. In a normal middle-ear structure, due to the vibration area’s ratio and the length of the lever arm, when sound waves are transmitted to the vestibular window through vibrations in the tympanum and the activity of the ossicular chain, sound pressure is amplified up to 22 times. Therefore, ossicular chain lesions may be severely detrimental to this amplification effect. Clinically, hearing loss caused by ossicular chain lesions is usually at around 30 decibels, reaching up to 50–60 decibels in severe cases. If moderate or severe hearing loss is due to a break in one of the links in the mechanical transduction device of the middle ear, tympanoplasty and ossicular chain reconstruction can be carried out to bridge this air-bone gap and restore connectivity in the ossicular chain, or even achieve precise anatomical reduction (7,8).
Ossicles have a delicate structure and are deeply located, meaning they cannot be directly observed by routine examination. Only temporal bone imaging can provide relatively intuitive information. However, due to the scanning slice thickness and conventional CT scans’ resolution, it is impossible to fully appraise the ossicular lesions’ situation. However, with the widespread clinical use of HRCT, high-quality images of the human body can more easily and rapidly be obtained, enabling a much clearer display of the bone structures inside the temporal bone, particularly the ossicular chain (3).
HRCT is a form of CT imaging technology that uses high-resolution bone algorithms for reconstruction. It sacrifices some density resolution to highlight spatial resolution, providing the most optimal match between the two. Slice thicknesses <1 mm (the slice thickness used in the present study was 0.625 mm) results in a significant reduction in the partial volume effect due to structural overlap; therefore, the images obtained are more accurate, fine, and hierarchical. This is particularly the case for displaying bone changes, which is significantly superior to the images obtained through conventional CT scans.
When HRCT is applied to the temporal bone, the quality of imaging tests on the ear is greatly improved, making it one of the most crucial examination methods currently used for middle-ear disease and suspected ossicular chain abnormalities (9). We used this technique in the present study to obtain clearer and finer scan images with sharper edges. Such images are particularly suitable for displaying anatomical locations with multiple delicate bone structures. High-quality CT images can also be used in conjunction with high-speed computing workstations for reconstruction to obtain 2D and 3D images of the temporal bone and ossicles. This provides intuitive 3D image data, resulting in a clearer and more comprehensive display of the ossicles and surrounding structures.
In the present study, the highly integrated Mimics 16.0 medical 3D image generation and editing software was selected for converting data for subsequent CAD. This system can identify DICOM data and input CT, magnetic resonance imaging, ultrasound, and even positron emission tomography scan data. It can then use plane data accumulation to construct 3D models and segmentation tools, which are used to highlight the given target region rapidly. These are then output in an STL format. This system can provide images at the axial, coronal, and sagittal planes of the raw data in image visualization. After obtaining 3D images from the reconstruction of the target region, 3D image panning, zooming, and rotation can be carried out for multifaceted, multi-angle observation of the images. Besides restoring observation images, this system also offers measurement, analysis, surgery simulation, mirror imaging technology, and other functions. The system is currently widely used in orthopedics and cosmetic surgery, and there have even been reports on its relevant applications in otomicrosurgery. A possible reason for this is that the ossicles have a delicate structure, which current imaging methods cannot display.
In the present study, Mimics 16.0 software was first used to extract ossicles from temporal bone HRCT scans successfully. Different tissues or cells’ physical density and the characteristics of different grayscale shadows in the slice images were used in different algorithms, such as grayscale interpolation and contour interpolation (grayscale interpolation is a true sense of 3D interpolation). Region growing and region opening were used for the extraction and segmentation of ossicles. This was used to generate a digitized 3D model with triangles as its basic unit.
Although the data source for Mimics 16.0 3D conversion is the same as that of CT post-processing techniques (MPR and 3D VR), it was not possible to ascertain the consistency and accuracy of data after Mimics 16.0, MPR, and 3D VR processing due to differences in computing methods and users. The Mimics 16.0 restorations’ accuracy was verified via the identification of the delicate anatomical structures from individualized ossicles and comparison between images obtained by CT post-processing and normal ossicular structures. Statistical results also demonstrated these differences.
Ossicles are delicate and small, and the stapes is one of their smallest components. An adult human’s crus anterius stapedis and crus posterius stapedis are extremely delicate (10,11), with their smallest diameter being 0.3 mm. However, the HRCT slice thickness used for scanning in the present study was 0.625 mm, and its accuracy can only reach 0.4 mm after split-layer scanning. The display of the crus anterius stapedis and crus posterius stapedis was therefore poor (54.1% and 62.5%, respectively), and the display rate of the stapes footplate was even lower at only 25%. This is because the stapes are buried inside the vestibular window by the annular stapedial ligament as there is close contact between the annular stapedial ligament and the vestibular window. The density of the stapes footplate is only slightly lower than that of the incus and malleus and similar to that of the annular stapedial ligament; the grayscale shadows display very similarly in the image. These mixed shadows cannot be distinguished through technical methods, and intentional human distinction and segmentation would result in unavoidable assumptions and oversight, which would affect the accuracy and authenticity of the restoration.
Due to scanning accuracy and unique anatomical structure, it was not possible in the present study to reconstruct a satisfactory individualized model of the stapes (12,13). With further development of imaging technology, a decrease in the thickness of CT scanning slices, and an increase in the CT resolution rate, it would be possible to reconstruct accurate, individualized stapes.
Based on the present study, surgery simulations can be carried out using the CAD module by inputting the ossicles’ HRCT data with lesions or defects (14). Either mirror imaging or the use of the normal ossicular morphology as a standard can then be employed for the virtual realization of the anatomical repair of the defective malleus, incus, or even stapes (15-18). The application of suitable ossicular prosthesis materials in TDP rapid prototyping for the accurate repair of the ossicles is expected to change conventional ossicular reconstruction’s current status, making it less time-consuming and effort-intensive, and more accurate.
The present study had several limitations. First, this trial was not a randomized controlled trial. Second, this was only a single-center trial, and the sample size was small. Finally, both methods used were based on the same original CT data.
Conclusions
The accuracy of the visualization of the malleus and incus after restoration via Mimics 16.0 software, based on temporal bone HRCT data, was high, and the degree of restoration was good. However, the accuracy and degree of restoration of the stapes footplate require further improvement.
Acknowledgments
We are grateful to all the people who assisted with our article.
Funding: This research was supported by the Innovation and Cultivate Fund of Navy General Hospital (No. CXPY201405).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-894). The authors have no conflicts of interest to declare.
Ethical Statement: The study was conducted following the Declaration of Helsinki and was approved by the Ethics Committee of the Sixth Medical Center of People’s Liberation Army General Hospital. All participants signed informed consent forms before participation.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhao D, Sun J. The research situation and application prospect that 3 d printing technology in the field of otolaryngology department. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2015;50:83-5. [PubMed]
- Peltola SM, Melchels FP, Grijpma DW, Kellomäki M. A review of rapid prototyping techniques for tissue engineering purposes. Ann Med 2008;40:268-80. [Crossref] [PubMed]
- Guo Y, Lu QH, Liu Y, Wang QJ, Ren AJ. Value of CT Three-dimensional Volume Rendering in Assessment of the Oossicular Chain in Chronic Suppurative Otitis Media. Chinese Journal of Medical Imaging 2013;21:508-11.
- Venema HW, Phoa SS, Mirck PG, Hulsmans FJ, Majoie CB, Verbeeten B Jr. Petrosal bone: coronal reconstructions from axial spiral CT data obtained with 0.5-mm collimation can replace direct coronal sequential CT scans. Radiology 1999;213:375-82. [Crossref] [PubMed]
- Huang YH, Liang BL. A study of normalization for CT muttiplaner reconstruction of temporal bone. Chinese Journal of CT and MRI 2009;7:14-7.
- Chuang MT, Chiang IC, Liu GC, Lin WC. Multidetector row CT demonstration of inner and middle ear structures. Clin Anat 2006;19:337-44. [Crossref] [PubMed]
- Sun JJ, Liu Y, Guo Y, Lu QH. The diagnostic value of 2D and 3D computed tomography (CT) in the evaluation of the pathologic conditions of ossicular chain. Chinese Journal of Otology 2011;9:117-23.
- Gardner EK, Jackson CG, Kaylie DM. Results with titanium ossicular reconstruction prostheses. Laryngoscope 2004;114:65-70. [Crossref] [PubMed]
- Liu ZH, Xian JF, Wang ZC, Chen QH, Zhang H. Investigation of HRCT in chronic supprative otitis media. Journal of Medical Imaging 2007;17:123-7.
- Dass R, Grewal BS, Thapar SP. Human stapes and its variations. 3. Influence of age and sex. J Laryngol Otol 1966;80:1023-37. [Crossref] [PubMed]
- Anson BJ. Stapedial, capsular and labyrinthine anatomy in relation to otologic surgery. Ann Otol Rhinol Laryngol 1961;70:607-31. [Crossref] [PubMed]
- Ghesu FC, Georgescu B, Zheng Y, Grbic S, Maier A, Hornegger J, Comaniciu D. Multi-Scale Deep Reinforcement Learning for Real-Time 3D-Landmark Detection in CT Scans. IEEE Trans Pattern Anal Mach Intell 2019;41:176-89. [Crossref] [PubMed]
- Dahiya N, Yezzi A, Piccinelli M, Garcia E. Integrated 3D Anatomical Model for Automatic Myocardial Segmentation in Cardiac CT Imagery. Comput Methods Biomech Biomed Eng Imaging Vis 2019;7:690-706. [Crossref] [PubMed]
- Hosseini MP. Designing a new CAD system for pulmonary nodule detection in High Resolution Computed Tomography (HRCT) images. Tehran Univ Med J 2012;70:250-6.
- Alvi SA, Jones JW, Lin J. Bilateral Ossicular Head Dehiscence Into the Middle Cranial Fossa. Ann Otol Rhinol Laryngol 2018;127:209-12. [Crossref] [PubMed]
- Ricci JL, Clark EA, Murriky A, Smay JE. Three-dimensional printing of bone repair and replacement materials: impact on craniofacial surgery. J Craniofac Surg 2012;23:304-8. [Crossref] [PubMed]
- Chen C, Chen Y, Li Q, Kau CH. The Esthetic Difference of Chinese Beauty Evaluated by Two Different Human Races Based on Three-Dimensional Average Face Analysis. J Craniofac Surg 2019;30:1435-40. [Crossref] [PubMed]
- Aydin HE, Kaya I, Aydin N, Kizmazoglu C, Karakoc F, Yurt H, Hüsemoglu RB. Importance of Three-Dimensional Modeling in Cranioplasty. J Craniofac Surg 2019;30:713-5. [Crossref] [PubMed]


