Algorithms applied to spatially registered multi-parametric MRI for prostate tumor volume measurement
Introduction
Prostate cancer (PCa) in men is the most common non-cutaneous malignancy and the second leading cause of cancer-related death in the United States (1). Studies show that prostate tumor volume correlates with Gleason score (GS) grade, stage, disease progression and predicting biochemical-free survival after radical prostatectomy, probability of vascular invasion, involvement of seminal vesicles, and distant metastasis (2-6). For example, small tumors (<0.5 cm3) displaying no evidence of tumor aggressivity are considered to be clinically insignificant, and such tumors are often grouped within low risk or very low risk PCa that are appropriate for deferred therapy. However, metastasis is highly likely when tumor volume exceeds 12 cm3, but less likely for smaller tumor volumes (<4 cm3) (6). Others reported that tumor size is related to the risk of relapse following radical prostatectomy (7) and prostate-specific antigen (PSA) progression (8). Autonomous and accurate noninvasive measurement of prostate tumor volume could, therefore, substantially improve PCa disease management.
Exploiting tumor volume measurement for PCa management is currently problematic. Conventional assays such as preoperative PSA levels and findings from digital rectal examination and transrectal ultrasonography poorly predict tumor volume (9-11). In addition, the quality of MRI interpretations is highly dependent on the skill and experience of the reader and results in widely varying reported accuracy of tumor volume measurements using MRI (12-14). Therefore, current tumor volume measurements that only use MR imaging and other assays are not currently considered sufficiently reliable for clinical decision making (15-18).
Prostate tumor’s distinguishing physiology relative to normal prostate tissue is manifested in its appearance in MRI. MR imaging methods, such as MR spectroscopic imaging (19) and dynamic contrast material-enhanced MR imaging (20-24) have been investigated for their capability to improve prostate tumor volume measurement. Reports (17,25-29) suggested that apparent diffusion coefficient (ADC) maps may clinically aid PCa diagnosis.
Currently most conventional MRI and computer aided diagnosis (CAD) do not fully exploit the high spatial resolution of MRI (<1 mm in the transverse direction). Tumors are often considered to be single monolithic entities despite being heterogeneous with variable GS spread across the lesion. The common spatial processing of an image assesses the local roughness, entropy, etc. via feature extraction using co-occurrence matrices (greater than one dimension) to distinguish cancer and normal tissues. Texture-based imaging features in conjunction with machine learning-based classification have been applied for classifying malignant from noncancerous prostate tissues (30-34). The spatial processing requires analyzing an image over several voxels, thereby limiting analysis (such as GS) within a heterogeneous tumor.
The purpose of our study was to examine three novel assessments to measure tumor volume by analyzing spatially registered parametric MRI and comparing them to surgical pathologic examination as the reference standard. We analyzed a retrospective database composed of matched multi-parametric MRI (MP-MRI) and pathologic findings in patients treated with radical prostatectomy. In the present study, we used this database to evaluate MP-MR imaging accuracy in estimating PCa volume. This study tests and exploits the vector nature of spatially registered MP-MRI. Most clinical assessments of MP-MRI inefficiently examine each individual image to assess the presence of tumor. This study examines three approaches that fuse multiple (3 or 7) images into a single image. Target signatures were inserted into supervised target detection algorithms (STDA) (35) to non-invasively detect PCa at the voxel level and to measure the tumor volume. The tumor appearance was enhanced through colors (yellow) (36,37) visually identified by a reader and by also applying a version of the CIELAB process (38) to the multi-spectral hypercubes. The techniques described in the manuscript are general and apply for any MRI scanner that collect the requisite images. Comparing “blobs” between MRI and histology is technically difficult due to slice misregistration between the two modalities. The goal of this study was not direct detection of tumors, but to estimate tumor size. By computing a cumulative rather than an individual feature, errors due to misregistration will average out.
Methods
Overall approach
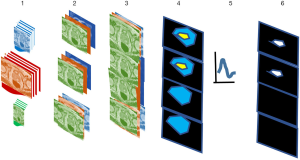
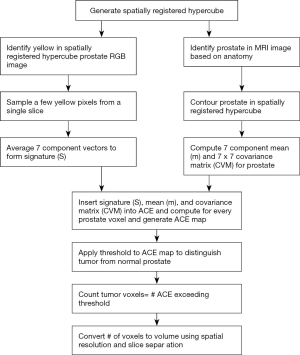
This study follows steps 1–6 (Figure 1) to analyze tumor (targets). This approach adapts procedures previously applied for Gleason scoring (35) and multispectral data and target detection for drone imagery (39,40). Each voxel is treated as a vector composed of MRI modality rather than a scalar value and can be exploited for color to highlight the tumor. Multispectral MRI data containing 7 components (35) [T1 (pre contrast), T1 (maximum contrast), T2, ADC, diffusion weighted image-high B (B=1,000 s/mm2), washout or kep from dynamic contrast enhanced (DCE)] (step 1), are registered by translating, scaling, and repositioning every MRI slice (step 2) into “cubes.” Further stitching spatially registered slices or cubes forms hypercubes (step 3), judiciously assigning red, green, blue (R, G, B) (step 4) to three modalities in the hypercube to enhance the tumor appearance as yellow after masking and displaying for the entire prostate. A reader and an algorithm identified the yellow color and assigned it to the tumor. The algorithms use multispectral tumor signatures (for tumor, GS) derived from training (steps 4, 5) in conjunction with the patient’s whole mount histology (ground truth) (35).
The supervised target algorithm uses the signature (step 5), to detect the patient’s tumor (step 6). The prostate tumor measurements are compared to the “gold standard”, i.e., a pathology evaluation of histology of slices of a patient’s resected whole mount prostate. Tumor volume measurements used a similar approach. No image registration for histology and MP-MRI is needed.
Supervised target detection, adaptive cosine estimator (ACE)
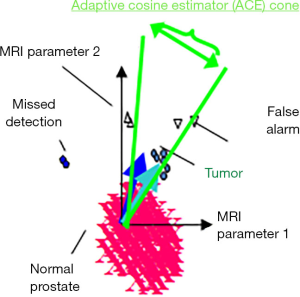
The multispectral STDA methods (35) were adapted for this medical application, shown in Figure 2 in two (of the seven) dimensions. Targets are characterized by their “target signature” (cyan vector in Figure 2) that is MRI modality dependent, extending from the background mean (normal tissue background displayed as magenta in Figure 2). After “training” or identifying the target signature within an image (Figure 2 cyan vector) taken from summing selected vectors from comparing MRI and histology assessments, (Figure 2 blue vector) and inserted into a STDA algorithm such as ACE {Eq. [1]} to classify each pixel (35,39). ACE uses the conical hyperspace decision surface to assess whether a voxel is background (large angle, small cosine) or target (small angle or large cosine). Mathematically, the ACE score at a given voxel i that has a seven-component vector is given by (35,39)
[1]
where ACE is the cosine between the test pixel i and the signature in the “whitened space” (35), m is the background (normal prostate) or mean value for each of the 7 modalities, S is the tumor signature from the mean of N target vector-voxels xk, summed over k target voxels, that are identified via MR/RP training data {Eq. [2]},
[2]
In Eq. [1], CVM is the covariance or clutter matrix (7×7), and the superscripts T and −1 denote a matrix transpose and inversion operation, respectively.
To generate the background statistics (m, CVM), the prostate image is manually outlined for all slices to generate an image mask and restrict computations to the prostate volume.
The identification of the pixel depends on the detection threshold set by the user based on previous examined data that optimizes a desired feature such as correlation with a standard or for hyperspectral analysts a maximum false alarm rate or minimum detection rate. A map of candidate targets can then be presented to the image analyst, radiologist or radiation oncologist.
Study design and population
The National Institutes of Health (NIH) prostate MRI and histology from wholemount prostatectomy were gathered from The Cancer Imaging Archive (TCIA) (41,42). This retrospectively designed, single institution study was approved by the local institutional review board, and was compliant with the Health Insurance Portability and Accountability Act of 1996. This subsequent retrospective analysis used the publicly available, anonymized data and did not require further review due to previous protections implemented by TCIA. A total of 26 consecutive patients were enrolled in the study between July 2008 and July 2009. Median patient age was 60 years (range, 49 to 75 years) and median PSA was 5.8 ng/mL (range, 2.3 to 23.7 ng/mL). All patients had biopsy proven adenocarcinoma of the prostate and median GS was 7 (range 6 to 9). In this study, 18 patients were selected based on tumor size >1 cc and evidence of contrast uptake. No restrictions were placed on tumor location within the prostate. A robotic assisted radical prostatectomy was performed with a median time of 60 days (maximum 180 days) of imaging without any intervening treatment. Additional exclusion criteria were contraindications to MRI or inability to have an endorectal coil placed.
Magnetic resonance imaging
The MRI collects DWI, DCE, and structural images in DICOM format. The pulse sequences were described in earlier studies (43-46). These studies (43-46) were performed using a combination of an endorectal coil (BPX-30, Medrad) tuned to 127.8 MHz and a 16-channel cardiac coil (SENSE, Philips Medical Systems) on a 3T magnet (Achieva, Philips Medical Systems) without the need for prior bowel preparation. Standard approaches were used to insert the endorectal coil. The MRI protocol included triplanar T2W turbo spin echo, DW MRI, 3DMR point resolved spectroscopy, and axial pre-contrast T1-weighted axial 3D fast field echo DCE MRI sequences, and their detailed sequence parameters were defined in a prior study (40). The mean interval between MRI and radical prostatectomy was 60 days (range, 3 to 180 days).
Whole mount prostatectomy and histology
3D models of each prostate (44,45) were generated by segmenting the prostate capsule on in vivo triplane T2W MRI, fusion of the binary objects, and surface extracting from high resolution 3D surfaces. Each mold was designed using commercially available 3D computer aided design software and the design incorporated the deformation of the endorectal coil. A 3D printer (Dimension Elite 3D printer, Stratasys, Inc.) deposited styrene to fabricate each mold. Following robotic radical prostatectomy, the specimen was fixed in formalin for 2 to 24 hours at room temperature, and the specimen was placed in the customized 3D mold and sliced in axial 6 mm sections. After robotic prostatectomy, the rostate were placed in room temperature formalin for 24–48 hours, sectioned, embedded in paraffin, fixed again, and stained with hematoxylin-eosin for histopathologic evaluation. Whole mount histopathology NIH patient specimens were sectioned in the customized mold for histopathology and mapped for individual tumor foci, dimensions and GS independently assessed by 2 experienced pathologists blinded to MRI. Sectioning of the gross specimen in the molds corresponded to the axial plane of the MRI sections.
Image processing, pre-analysis
This study exploits tumor physiology to help distinguish lesions from normal tissues by analyzing the DCE. DCE are time series images that follow the evolution of contrast material over several hundred seconds following its injection and uptake in the tissues. Prostate tumors are often highly vascularized. The vasculature is porous to material and the contrast material enters the small extravascular space (but not the cells). Therefore, prostate tumors can fill and empty MRI contrast material quickly relative to normal prostate organ. A simple two compartment model (47,48) describes the tracer concentration in the tissue that supplies and empties through the tumor vasculature and used to create the washout or kep image.
The MRI images were digitally resampled (35) to 1 mm resolution in the transverse direction and 6 mm in the superior-inferior direction based on patient’s table position, scaled, translated, resliced and spatially registered at the pixel level. Rigid registration is applied between the structural, diffusion, and DCE due to the short time intervals between scans (<20 minutes). The multiple axial cubes in three dimensions were “mosaicked” together by sequentially stitching them together into a narrow three-dimensional image. In this way, the four dimensions (three-dimensional body volume plus the fourth dimension composed of MRI modalities) are compressed into three dimensions using the mosaicked cubes.
Tumor volume measurements, coloring
Measuring the tumor volume is a critical component for assessing the patient’s condition and for helping to guide treatment decisions (2-8). Color displays (36) (left side of Figure 3) delineate the tumor. The images are normalized by setting minimum and maximum to the mean −3× standard deviation and mean +3× standard deviation, respectively. The color display can be generated by assigning red, green, and blue channels to the grey scale images of the washout or kep, DWI, and ADC images respectively. Tumors (denoted by arrows) can appear as yellow (or bright) due to exhibiting high vascularization (high kep, high red), low diffusion (high for high B-DWI, green, and low ADC, low blue). The number of pixels inside the brighter or yellow portion of RGB images or Region of Interest can be generated from standard image processing (see darkened or red area inside the contoured image). In addition, a matching tumor is delineated by a pathologist (see Figure 3 for before and after outlined histology images). A comparison of tumor areas in each slice can be made between the MRI and the histology images after accounting for different spatial resolutions for the two sets of images.
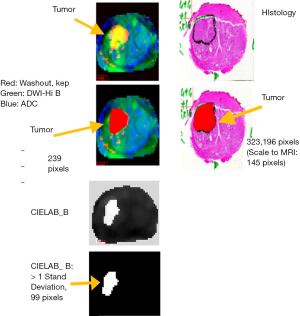
In Figure 3, the spatial resolution is 1 mm per pixel for the registered MRI image set and 47 pixels per mm or 1,200 dots per inch for the histology images (scaling of 47×47=2,209). Figure 3 shows tumor areas contain 239 and 323,196 pixels (scales to 145 pixels using 1/2,209) in the MRI and histology images, respectively. The displayed images in Figure 3 are scaled for the reader’s convenience. The histology images were stained with hematoxylin-eosin. To convert to tumor volume, both sets of images are matched to a slice thickness of 6 mm.
Histologic sectioning of wholemount prostatectomy shrinks the prostate and tumor volume (17,49,50). The tissues shrink by 15% (49) and the computed histology-based tumor volume is corrected accordingly.
CIELAB computation
The CIELab is a color space defined by the International Commission on Illumination (CIE) in 1976 (37,38). The unique goal of the L*a*b* model is to be ‘device-independent’. The colors should not depend on the device they are displayed. It expresses color as three values: L*, a*, and b*. The nonlinear relations between red, green, and blue components and L*, a*, and b* are intended to mimic the nonlinear response of the eye and derived from psychovisual studies. The b* axis is of particular interest in this study and represents the opposing blue-yellow component, with blue in the negative direction and yellow in the positive direction.
The calculation proceeds in two stages. All negative values are masked and the RGB are normalized after removing the lowest 2.5% and highest 2.5% values. The R, G, and B are normalized and assigned to X, Y, and Z, respectively, without admixing. It is observed that admixing (non-identity transform matrix) degrades (not shown) the performance for this particular application. The CIEXYZ tristimulus values for Xn, Yn, and Zn of the reference white point are 96.42, 100, and 82.52, respectively, based on the illuminant D50. The b* is given in Eq. [3] as (37,38)
[3]
where
[4]
where
[5]
In this study, “yellow” b* >0 and tumor is defined as b* >1 standard deviation to help filter out background or normal tissue.
Tumor volume measurements, supervised target detection
Figure 4 summarizes the procedure for estimating the tumor volume using the STDA. The voxels identified as seeds for the spectral signature are found by identifying the yellow in the RGB image. The 7-dimensional spectra from the 2 to 5 voxels are averaged to form a single 7-dimensional signature S {Eq. [2]}. The small number of pixels were chosen within the heterogeneous tumor for maximal contrast uptake and minimal diffusion and averaged to reduce temporal noise. The 7 components for ACE were inherited from a GS study (35) that used a relatively high number of components but also provided diverse information about the tumor and helped assess the tumor through high discrimination. The normal prostate is contoured using the spatially registered hypercube to help generate a mask for determining the mean (m) and covariance matrix (CVM) of the normal prostate. The ACE score {Eq. [1]} is computed for each voxel in the hypercube using the signature, mean, and covariance. A threshold is applied to the ACE map with tumor exceeding the threshold and normal tissue assigned to ACE scores residing below the threshold. ACE scores range from –1.0 to 1.0. The chosen thresholds in this study varied from 0.40 to 0.85 in 0.05 increments. The number of tumor voxels are converted to volume using the MRI spatial resolution (1 mm × 1 mm) and slice thickness (6 mm) to compute the single voxel volume (0.006 cm3). The MR-derived volume is matched against the histology determined volume (see above).
Results
Tumor volumes were computed by applying color and STDA (following procedures described in the “Methods” section) to MP-MRI of 18 of the 26 TCIA archived patients. The analysis was confined to tumors that exceeded 1 cc in size as measured from the wholemount prostatectomy. Seven tumors were excluded due to size falling below the threshold. One tumor was excluded due to no apparent contrast material uptake as determined through DCE. No restrictions were applied to the analysis regarding location of the tumor in the prostate. Two of the tumors were very large (12 cc, 20 cc).
The analysis is summarized by intercomparing among the three different types of tumor volume measurements, i.e., color analysis vs. histology of wholemount prostatectomy and supervised target detection vs. histology of wholemount prostatectomy.
The comparison of tumor volume measurements that compared supervised target detection with color analysis and pathology of wholemount prostatectomy depends on the threshold for supervised target detection. As noted, this study analyzed the performance of supervised target detection that included 10 different thresholds (0.40 to 0.85 in 0.05 increments). Determination of how well a given measurement estimates the tumor volume provides a metric for assessing a given technique. A summary of the performance is displayed in Figures 5,6 and Table 1.

Full table
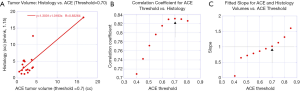
Figure 5A plots the computed linear correlation coefficient between the ACE supervised tumor volume measurements (with a fixed detection threshold =0.70) against the pathology assessment from wholemount prostatectomy. The linear correlation coefficient between ACE supervised target volume measurement for threshold =0.70 and wholemount prostatectomy is 0.83. The fitted slope is 1.02 and offset is −1.30 cc. Removing the largest and second largest tumors in this analysis results in a computed correlation coefficient of 0.415 and 0.912, respectively.
Figure 5B plots the correlation coefficients between the ACE supervised tumor volume measurement and tumor volume measurements and pathology assessment from wholemount prostatectomy versus ACE detection thresholds. The maximum correlation coefficient at threshold =0.70 is denoted by an arrow.
Figure 5C plots the fitted slopes between the ACE supervised tumor volume measurement and tumor volume measurements and pathology assessment from wholemount prostatectomy versus ACE detection thresholds. The fitted slope for threshold =0.70 is denoted by an arrow and intersects the unit slope.
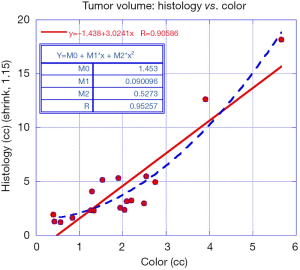
Figure 6 plots the data and a linear fit between the color-based tumor volume estimates against the pathology assessment from wholemount prostatectomy (red line). The correlation coefficient from a linear fit and wholemount prostatectomy is 0.91. The fitted slope is 3.02 and offset is −1.40 cc. A polynomial fit of order 2 was applied between color-based tumor volume estimates against the pathology assessment from wholemount prostatectomy (dashed blue line). The correlation coefficient from a polynomial fit tumor volume measurements using manual color assessments and wholemount prostatectomy is 0.95. Removing the largest and second largest tumors in this analysis results in a computed correlation coefficient of 0.795 and 0.892, respectively.
Figure 7A plots the data and a linear fit between tumor volume derived from the pathology assessment from wholemount prostatectomy against the b* component of the CIELAB formulation (red line) after applying Eq. [3] to the masked hypercubes. To avoid counting background voxels, voxels exceeding one standard deviation above the minimum yellow value (0) were counted as yellow. The correlation coefficient from a linear fit and wholemount prostatectomy is 0.71. The fitted slope is 1.99 and offset is −0.63 cc. Removing the largest and second largest tumors in this analysis results in a computed correlation coefficient of 0.606 and 0.920, respectively.
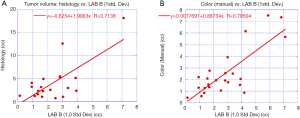
Figure 7B plots the data and a linear fit between tumor volume derived from the color assessment against the b* component of the CIELAB formulation (red line) after applying Eq. [3] to the masked hypercubes. Only voxels exceeding one standard deviation above the minimum yellow value (0) were counted as yellow. The correlation coefficient from a linear fit and wholemount prostatectomy is 0.80. The fitted slope is 0.89 and offset is 0.01 cc. Removing the largest and second largest tumors in this analysis results in a computed correlation coefficient of 0.687 and 0.887, respectively.
Table 1 summarizes the tumor volume measurements using supervised tumor volume measurements and color analysis. The table displays the slope and intercepts from linear fitting for the ACE supervised tumor measurement and color tumor volume analysis and correlation coefficients.
Discussion
This study tested tumor volume estimators, specifically a semi-autonomous ACE supervised target detection and highlighting and quantitatively analyzing the appearance of tumors using a color. Both methods used spatially registered MP-MRI. This study applied color (3-component) and supervised target detection (7-component) to estimate prostate tumor volume. This study compared the MP-MRI derived tumor volume measurements to tumor volume measurements derived from pathology evaluation of wholemount prostatectomy. This study found that MP-MRI derived techniques correlate with the pathology analysis of wholemount prostatectomy (R>0.7). This study examined tumors that exceeded 1 cc in size, displayed contrast material uptake, but no restrictions were placed on tumor location within the prostate.
Pathology analysis of histologic samples derived from wholemount prostatectomy is designated as the “gold standard” for determining prostate tumor volume in this study. Generating the freely suspended histology slides requires resecting, slicing, staining etc. the prostate. Such manipulations may deform, shrink, distort, and perturb the prostate’s geometry and result in tissue loss relative to an MR imaged prostate. The in vivo MR imaged prostate is pressed by other organs and soft tissue and supported against a gravitational field. As noted in the “Methods” section, did not distort, rotate, twist, and register etc. the MRI to the histology slides. This study, like earlier work (43-46,49), assumed that the distortion in the histology slides did not affect the tumor volume except for the shrinkage determined from other studies (49). Measures of agreement such as DICE separability (51,52), receiver operator curves (ROC) (53), and Hausdorff distances (54) require voxel level registration between the “gold standard” and test samples to match the positions and locations of tumors. Therefore, these metrics are not suitable nor applicable in the present study. Instead, correlation coefficient compares tumor volumes (not tumor edges, positions) and do not require voxel-level registration.
These MP-MRI based measurements used the dynamic contrast enhancement images to detect contrast flow and uptake within a tumor. Tumors are heterogeneous with regions that are highly vascularized and that are detected with DCE. The highly vascularized portion of the tumor may occupy a portion of the tumor. Most of the tumor volume is composed of non-vascularized tumor such as hypoxic and necrotic regions. Therefore, the detected tumor volumes using the techniques described in this study should yield estimates that are lower than the tumor volumes derived from wholemount prostatectomy as confirmed in the negative bias of the Bland Altman plot for uncorrected ACE (not shown) and the color-based polynomial fit (Figure 6). The linear fitted slope (Figure 6) of roughly 3 for tumor volume estimation from color analysis relative to wholemount prostatectomy suggests that, on average, the color analysis yields estimates roughly one-third of the total tumor volume. The factor of 3 agrees with an earlier (55) determination of tumor volume using MRI. The better fits from polynomial functions also implies that the disparity in vascularized regions relative to the total tumor volumes increases due to the tumor outgrowing its nutritionally supporting system resulting in increasing hypoxia and necrosis. The green areas attached to the yellow volumes are regions of low vascularity and low diffusion and are associated with the tumor. Similarly, thresholding of the b* part of CIELAB was correlated with the yellow display as well as pathology assessment of wholemount prostatectomy (Figure 7A,B). The linear fitted slope (Figure 7A) of roughly 2 for tumor volume estimation from CIELAB analysis relative to wholemount prostatectomy suggests that, on average, the color analysis yields estimates roughly half of the total tumor volume.
ACE yielded lower correlation coefficients relative to the color and CIELAB approaches for predicting tumor volume. The highly selective target signature was chosen in to be regions of high contrast uptake and low diffusion. ACE uses more components (7) than color approaches (3) and further elevates the discrimination between highly vascularized and non-vascularized and possibly necrotic regions within the tumor. The tumor volume includes highly heterogenous regions and some that may not be counted in the ACE tumor volume estimate.
Most of the studied tumor volumes were small (<3 cc) resulting in possible spurious high correlation coefficients due to distortion by the larger tumors. A limitation of this study is the dominance of small tumors. Excluding larger tumors in the analysis reduces the correlation coefficient between the tumor volume measurements, although the reprocessed correlation coefficient remains significant. Including the large tumor shows that the approach can be possibly extended to especially aggressive tumors.
The optimal ACE detection threshold value of 0.70 was chosen due to the highest observed correlation coefficient (Figure 5B) and the closeness of fitted slope to 1 (Figure 5C). The peak in correlation coefficients (Figure 5B) is large and flat for higher ACE thresholds. Therefore, ACE thresholds greater than 0.60 maybe more useful and deserve examination in future studies.
Other studies (20-25) relied on experienced radiologists to delineate putative tumors in the MRI. Visually discriminating the border between tumor from normal tissue using MRI is often difficult and subjective. Tumor volume determination by different radiologists often lead to significant differences. Digital tumor volume assessment and elevating the appearance of the tumors may lead to more consistent and objective staging and thus more reliably inform treatment options and clinical and treatment decisions for patients. Employing and thresholding the b* component of CIELAB may further guide the radiologist in objectively finding the yellow (in this scheme) or putative tumor.
This study focused on tumors that display uptake of contrast material. Many clinics do not inject contrast in their patients in order to simplify the scanning process and because not all tumors uptake contrast. Future research efforts can examine the efficacy of MP-MRI tumor volume estimators that do not include DCE. Adding other modalities, such as textures, may also be incorporated into the MP-MRI hypercube for tumor display and tumor volume estimation. Currently, this study expended effort in algorithm development for spatially registering the MRI and generating hypercubes. Future developments, automation, and improvements should reduce the construction time. Actual computations such as ACE, CIELAB, etc., are quick using standard, readily available computer technology.
In addition, a large tumor (>5 cc) can occupy a relatively large portion of prostate organ and perturb the image statistics that characterize the background (normal prostate) for ACE calculations. Such cases may require masking for a smaller section of the normal prostate. There are a significant number of pixels (much more than the number of components) in the normal prostate subsection to achieve accurate statistics and avoid rank degeneracy for the covariance matrix.
Conclusions
Supervised target detection derived from security and defense applications and color display and analysis that are applied to registered MP-MRI, can non-invasively estimate prostate tumor volumes. Color (and CIELAB) analysis generates tumor volumes that are 3 (and 2) times lower than the tumor volume derived from histology of wholemount prostatectomy, in accord with earlier MRI results that uses different methodology. Non-invasive tumor volume measurements may improve clinical decision making, elevate patient care and reduce discomfort and side effects.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-137a). Dr. RM has a patent null pending and has submitted a non-provisional patent application related to this study and is waiting for judgement but reports no other conflicts of interest. Dr. CBS received an honorarium from Varian Medical Systems but reports no other conflicts of interest, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This retrospective study of data that is publicly available, accessible and anonymized, collected and previously collected and anonymized did not require institutional board approval. TCIA had earlier obtained informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- McNeal JE. Cancer volume and site of origin of adenocarcinoma in the prostate: relationship to local and distant spread. Hum Pathol 1992;23:258-66. [Crossref] [PubMed]
- Villers AA, McNeal JE, Redwine EA, Freiha FS, Stamey TA. Pathogenesis and biological significance of seminal vesicle invasion in prostatic adenocarcinoma. J Urol 1990;143:1183-7. [Crossref] [PubMed]
- Friedersdorff F, Groß B, Maxeiner A, Jung K, Miller K, Stephan C, Busch J, Kilic E. Does the Prostate Health Index Depend on Tumor Volume?—A Study on 196 Patients after Radical Prostatectomy. Int J Mol Sci 2017;18:488. [Crossref] [PubMed]
- May M, Siegsmund M, Hammermann F, Loy V, Gunia S. Visual estimation of the tumor volume in prostate cancer: a useful means for predicting biochemical-free survival after radical prostatectomy? Prostate Cancer Prostatic Dis 2007;10:66-71. [Crossref] [PubMed]
- McNeal JE, Villers AA, Redwine EA, Freiha FS, Stamey TA. Histologic differentiation, cancer volume, and pelvic lymph node metastasis in adenocarcinoma of the prostate. Cancer 1990;66:1225-33. [Crossref] [PubMed]
- Epstein JI, Carmichael M, Partin AW, Walsh PC. Is tumor volume an independent predictor of progression following radical prostatectomy? a multivariate analysis of 185 clinical stage B adenocarcinomas of the prostate with 5 years of followup. J Urol 1993;149:1478-81. [Crossref] [PubMed]
- Kikuchi E, Scardino PT, Wheeler TM, Slawin KM, Ohori M. Is tumor volume an independent prognostic factor in clinically localized prostate cancer? J Urol 2004;172:508-11. [Crossref] [PubMed]
- Bostwick DG, Graham SD Jr, Napalkov P, Abrahamsson PA, di Sant'agnese PA, Algaba F, Hoisaeter PA, Lee F, Littrup P, Mostofi FK. Staging of early prostate cancer: a proposed tumor volume-based prognostic index. Urology 1993;41:403-11. [Crossref] [PubMed]
- Noldus J, Stamey TA. Limitations of serum prostate specific antigen in predicting peripheral and transition zone cancer volumes as measured by correlation coefficients. J Urol 1996;155:232-7. [Crossref] [PubMed]
- D’Amico AV, Chang H, Holupka E, Renshaw A, Desjarden A, Chen M, Loughlin KR, Richie JP. Calculated prostate cancer volume: the optimal predictor of actual cancer volume and pathologic stage. Urology 1997;49:385-91. [Crossref] [PubMed]
- Palken M, Cobb OE, Warren BH, Hoak DC. Prostate cancer: correlation of digital rectal examination, transrectal ultrasound and prostate specific antigen levels with tumor volumes in radical prostatectomy specimens. J Urol 1990;143:1155-62. [Crossref] [PubMed]
- Kahn T, Burrig K, Schmitz-Drager B, Lewin JS, Furst G, Modder U. Prostatic carcinoma and benign prostatic hyperplasia: MR imaging with histopathologic correlation. Radiology 1989;173:847-51. [Crossref] [PubMed]
- Sanchez-Chapado M, Angulo JC, Ibarburen C, Aguado F, Ruiz A, Viaño J, García-Segura JM, Gonzalez-Esteban J, Rodriquez-Vallejo JM. Comparison of digital rectal examination, transrectal ultrasonography, and multicoil magnetic resonance imaging for preoperative evaluation of prostate cancer. Eur Urol 1997;32:140-9. [Crossref] [PubMed]
- Ponchietti R, Di Loro F, Fanfani A, Amorosi A. Estimation of prostate cancer volume by endorectal coil magnetic resonance imaging vs pathologic volume. Eur Urol 1999;35:32-5. [Crossref] [PubMed]
- Kirkham AP, Emberton M, Allen C. How good is MRI at detecting and characterizing cancer within the prostate? Eur Urol 2006;50:1163-74. [Crossref] [PubMed]
- Jager GJ, Ruijter ET, van de Kaa CA, de la Rosette JJ, Oosterhof GO, Thornbury JR, Barentsz JO. Local staging of prostate cancer with endorectal MR imaging: correlation with histopathology. AJR Am J Roentgenol 1996;166:845-52. [Crossref] [PubMed]
- desouza NM, Reinsberg SA, Scurr ED, Brewster JM, Payne GS. Magnetic resonance imaging in prostate cancer: the value of apparent diffusion coefficients for identifying malignant nodules. Br J Radiol 2007;80:90-5. [Crossref] [PubMed]
- Sommer FG, Nghiem HV, Herfkens R, McNeal J, Low RN. Determining the volume of prostatic carcinoma: value of MR imaging with an external-array coil. AJR Am J Roentgenol 1993;161:81-6. [Crossref] [PubMed]
- Coakley FV, Kurhanewicz J, Lu Y, Jones KD, Swanson MG, Chang SD, Carroll PR, Hricak H. Prostate cancer tumor volume: measurement with endorectal MR and MR spectroscopic imaging. Radiology 2002;223:91-7. [Crossref] [PubMed]
- Bratan F, Melodelima C, Souchon R, Dinh AH, Mège-Lechevallier F, Crouzet S, Colombel M, Gelet A, Rouvière O. How Accurate Is Multiparametric MR Imaging in Evaluation of Prostate Cancer Volume? Radiology 2015;275:144:54.
- Nakashima J, Tanimoto A, Imai Y, Mukai M, Horiguchi Y, Nakagawa K, Oya M, Ohigashi T, Marumo K, Murai M. Endorectal MRI for prediction of tumor site, tumor size, and local extension of prostate cancer. Urology 2004;64:101-5. [Crossref] [PubMed]
- Mazaheri Y, Hricak H, Fine SW, Akin O, Shukla-Dave A, Ishill NM, Moskowitz CS, Grater JE, Reuter VE, Zakian KL, Touijer KA, Koutcher JA. Prostate tumor volume measurement with combined T2-weighted imaging and diffusion weighted MR: correlation with pathologic tumor volume. Radiology 2009;252:449-57. [Crossref] [PubMed]
- Knoedler JJ, Karnes RJ, Thompson RH, Rangel LJ, Bergstralh EJ, Boorjian SA. The association of tumor volume with mortality following radical prostatectomy. Prostate Cancer Prostatic Dis 2014;17:144-8. [Crossref] [PubMed]
- Villers A, Puech P, Mouton D, Leroy X, Ballereau C, Lemaitre L. Dynamic contrast enhanced, pelvic phased array magnetic resonance imaging of localized prostate cancer for predicting tumor volume: correlation with radical prostatectomy findings. J Urol 2006;176:2432-7. [Crossref] [PubMed]
- Yoshimitsu K, Kiyoshima K, Irie H, Tajima T, Asayama Y, Hirakawa M, Ishigami K, Naito S, Honda H. Usefulness of apparent diffusion coefficient map in diagnosing prostate carcinoma: correlation with stepwise histopathology. J Magn Reson Imaging 2008;27:132-9. [Crossref] [PubMed]
- Haider MA, van der Kwast TH, Tanguay J, Evans AJ, Hashmi AT, Lockwood G, Trachtenberg J. Combined T2-weighted and diffusion weighted MRI for localization of prostate cancer. AJR Am J Roentgenol 2007;189:323-8. [Crossref] [PubMed]
- Sato C, Naganawa S, Nakamura T, Kumada H, Miura S, Takizawa O, Ishigaki T. Differentiation of noncancerous tissue and cancer lesions by apparent diffusion coefficient values in transition and peripheral zones of the prostate. J Magn Reson Imaging 2005;21:258-62. [Crossref] [PubMed]
- Shimofusa R, Fujimoto H, Akamata H, Motoori K, Yamamoto S, Ueda T, Ito H. Diffusion-weighted imaging of prostate cancer. J Comput Assist Tomogr 2005;29:149-53. [Crossref] [PubMed]
- Viswanath SE, Bloch NB, Chappelow JC, Toth R, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A. Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery. J Magn Reson Imaging 2012;36:213-24. [Crossref] [PubMed]
- Tiwari P, Viswanath S, Kurhanewicz J, Sridhar A, Madabhushi A. Multimodal wavelet embedding representation for data combination: integrating magnetic resonance imaging and spectroscopy for prostate cancer detection. NMR Biomed 2012;25:607-19. [Crossref] [PubMed]
- Fehr D, Veeraraghavana H, Wibmerb A, Gondo T, Matsumoto K, Vargas HA, Sala E, Hricak H, Deasy JO. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A 2015;112:E6265-73. [Crossref] [PubMed]
- Vos PC, Barentsz JO, Karssemeijer N, Huisman HJ. Automatic computer-aided detection of prostate cancer based on multiparametric magnetic resonance image analysis. Phys Med Biol 2012;57:1527-42. [Crossref] [PubMed]
- Vos PC, Hambrock PT, Barenstz JO, Huisman HJ. Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI. Phys Med Biol 2010;55:1719-34. [Crossref] [PubMed]
- Mayer R, Simone CB 2nd, Skinner W, Turkbey B, Choyke P. Pilot study for supervised target detection applied to spatially registered multiparametric MRI in order to non-invasively score prostate cancer. Comput Biol Med 2018;94:65-73. [Crossref] [PubMed]
- Schuler JM, Bassett WE, Howard JG, Klein RB, Kruer MR, Mayer R, Satyshur MP, Scribner DA, Warren PG, Waterman JR. Robust color fusion of diurnal multi-spectral imagery: Empirical Color Constancy. 2002 Proceedings of MSS specialty group on Passive Sensors, April 2002.
- Kather JN, Weidner A, Attenberger U, Bukschat Y, Weis CA, Weis M, Schad LR, Zöllner FG. Color-coded visualization of magnetic resonance imaging multiparametric map. Sci Rep 2017;7:41107. [PubMed]
- Jain AK. Fundamentals of Digital Image Processing. Upper Saddle River, NJ: Prentice Hall; 1989.
- Manolakis D, Shaw G. Detection algorithms for hyperspectral imaging applications. IEEE Sign. Process Mag 2002;19:29-43. [Crossref]
- Richards JA, Jia X. Remote Sensing Digital Image Analysis, New York: Springer-Verlag, 1999.
- Choyke P, Turkbey B, Pinto P, Merino M, Wood B. Data From PROSTATE-MRI. The Cancer Imaging Archive. Available online: http://doi.org/, 2016. [Crossref]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J Digit Imaging 2013;26:1045-57. [Crossref] [PubMed]
- Turkbey B, Pinto PA, Mani H, Bernardo M, Pang Y, McKinney YL, Khurana K, Ravizzini GC, Albert PS, Merino MJ, Choyke PL. Prostate Cancer: Value of Multi-parametric MR Imaging at 3 T for Detection-Histopathologic Correlation. Radiology 2010;255:89-99. [Crossref] [PubMed]
- Shah V, Pohida TJ, Turkbey B, Bernardo M. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum 2009;80:104301. [Crossref] [PubMed]
- Turkbey B, Mani H, Shah V, Rastinehad AR, Bernardo M, Pohida T, Pang Y, Daar D, Benjamin C, McKinney YL, Trivedi H, Chua C, Bratslavsky G, Shih JH, Linehan WM, Merino MJ, Choyke PL, Pinto PA. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol 2011;186:1818-24. [Crossref] [PubMed]
- Metzger GJ, Kalavagunta C, Spilseth CB, Bolan PJ, Li X, Hutter D, Bolan PJ, Li X, Hutter D, Nam JW, Johnson AD, Henriksen JC, Moench L, Konety B, Warlick CA, Schechel SC, Koopmeiners JS. Detection of Prostate Cancer: Multiparametric MR Imaging Models Developed by Using Registered Correlative Histopathologic Results. Radiology 2016;279:805-16. [Crossref] [PubMed]
- Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, Port RE, Taylor J, Weisskoff RM. Estimating Kinetic Parameters from Dynamic Contrast-EnhancedT1-Weighted MRI of a Diffusable Tracer: Standardized Quantities and Symbols. J Magn Reson Imaging 1999;10:223-32. [Crossref] [PubMed]
- Toft PS. T1-weighted DCE Imaging Concepts: Modelling, Acquisition and Analysis. Magnetom Flash 2010;3:31-9.
- Turkbey B, Mani H, Aras O, Rastinehad AR, Shah V, Bernardo M, Pohida T, Daar D, Benjamin C, McKinney YL, Linehan WM, Wood BJ, Merino MJ, Choyke PL, Pinto PA. Correlation of Magnetic Resonance Imaging Tumor Volume with Histopathology. J Urol 2012;188:1157-63. [Crossref] [PubMed]
- Jonmarker S, Valdman A, Lindberg A. Tissue shrinkage after fixation with formalin injection of prostatectomy specimens. Virchows Arch 2006;449:297. [Crossref] [PubMed]
- Chen W, Lin M, Gibson E, Bastian-Jordan M, Cool DW, Kassam Z, Liang H, Feng G, Ward AD, Chiu B. A self-tuned graph-based framework for localization and grading prostate cancer lesions: An initial evaluation based on multiparametric magnetic resonance imaging. Comput Biol Med 2018;96:252-65. [Crossref] [PubMed]
- Comelli A, Stefano A, Russo G, Sabini MG, Ippolito M, Bignardi S, Petrucci G, Yezzi A. A smart and operator independent system to delineate tumours in Positron Emission Tomography scans. Comput Biol Med 2018;102:1-15. [Crossref] [PubMed]
- Fusco R, Sansone M, Granata V, Setola SV, Petrillo A. A systematic review on multiparametric MR imaging in prostate cancer detection. Infect Agent Cancer 2017;12:57. [Crossref] [PubMed]
- Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15:29. [Crossref] [PubMed]
- Priester A, Natarajan S, Khoshnoodi P, Margolis DJ, Raman SS, Reiter RE, Huang J, Grundfest W, Marks LS. Magnetic Resonance Imaging Underestimation of Prostate Cancer Geometry: Use of Patient Specific Molds to Correlate Images with Whole Mount Pathology. J Urol 2017;197:320-6. [Crossref] [PubMed]