
Diagnostic performance of convolutional neural network-based Tanner-Whitehouse 3 bone age assessment system
Introduction
Bone age, more so than chronological age, reflects the actual growth and development status of a child. The theory of skeletal physiological maturity was first proposed by Franz Boas (1), and since then, bone age was used to describe different stages of skeletal development. Bone age assessment (BAA) plays a pivotal role in confirming the diagnosis of endocrine diseases, predicting the adult height, and evaluating the efficacy of the treatment. Nevertheless, the basis of these evaluations requires an accurate, consistent, and stable assessment approach.
Greulich and Pyle (GP) (2) and Tanner-Whitehouse 3 (TW3) (3) are the most prevalently used BAA techniques. GP method compares the patient’s radiographic information with the nearest standard radiograph in the atlas. Nevertheless, the degree of accuracy can only reach to half a year, and the doctor’s subjectivity may cause significant variation between reviewers (4). However, due to its convenience and speed, approximately 76% of doctors worldwide still prefer the GP method (5). On the other hand, the TW3 method is based on a scoring system enabling the bone age estimation accuracy to be within a month. Specifically, the reviewer will firstly identify 20 bones [13 radius, ulna and short bones (RUS) + 7 Carpal], each with a categorized stage. Then, each stage is replaced by a score. Finally, a total score is calculated and transformed into the bone age. This method requires at least 20 minutes to complete the bone age evaluation for manual assessment. Although the TW3 method is more precise compared with the GP method, it is more complex and time-consuming. And, even when adopting the computer-aided detection (CAD) system, the rating for each bone still relies on a human interpretation that also imposes unavoidable inter- and intra-reviewer variability. New and advanced artificial intelligence (AI) techniques are urgently needed to aid the radiologist and clinicians in BAA.
Deep learning is a type of machine learning. When properly trained with a vast number of training samples, the algorithm can make accurate predictions for new input (6). In deep learning, a convolutional neural network (CNN) is a kind of feedforward neural networks with a deep structure that includes convolution or related calculations. It is widely used in image and video recognition, recommender systems, image classification, natural language processing, and medical image analysis (7). A CNN usually consists of an input and an output layer, as well as multiple hidden layers. The activation function is commonly a rectified linear unit (RELU) layer and includes pooling layers and fully connected layers. By using CNN, local information can be effectively utilized without manually selecting features. Also, through the sharing of perceptual fields, we can learn large scale images with small scale parameters. However, CNN is not suitable for long-distance logical reasoning, nor is it good at dealing cases with the feature of large shift or rotation.
Furthermore, the physical information of the features extracted by the convolutional layer is ambiguous. Due to the availability of big data in medical fields and enhanced computing power with graphics processing units (GPU), deep learning has been widely applied in medical applications, including the identification of brain tumors (8) and diabetic retinopathy in retinal fundus (9), early warning of lymph node metastases in breast cancer (10), and classification of skin cancer (11). Many attempts to automating BAA, such as BoneXpert, a system developed by Harvard Medical School and Stanford University, have been proposed over the past few years (12-14). Recently, AI models based on the GP method have been proven to possess great potential in making accurate and time-saving predictions (15). The following study aimed to establish a new large-scale, fully automated CNN-based, TW3 BAA system, and to compare the accuracy, stability, and efficiency of the model with experienced endocrinologists and radiologists.
Methods
Data collection
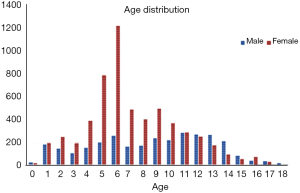
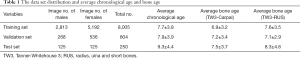
The institutional review boards of our hospital approved the study. A total of 9,059 left-hand radiographs were obtained from our hospital between January 2012 and December 2016. All images were drawn from the picture archives and communication systems (PACS). The radiology reports included the patient’s accession numbers, chronological age, sex, and bone age. Figure 1 shows the age distribution of the data sets, and Table 1 summarizes the average chronological age and the estimated bone age of each set.
Full table
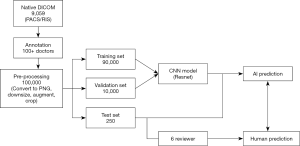
Among the radiographs, 8,005/9,059 (88%) were randomly selected for the training set, 804/9,059 (9%) were used for validation, and the remaining 250/9,059 (3%) were used for the test set. The training set was used to optimize the model parameters, and the validation set was used to tune hyper-parameters to optimize the model.
Data annotations
The radiograph annotation team included more than 100 professional radiologists and endocrinologists from Children’s Hospital in Fujian and Zhejiang Province. During the annotation process, each expert evaluated the same image at least 3 times, and the mode value was chosen as his/her final annotation result. Five different reviewers estimated the rank (from A to I) of each hand bone. The result was accepted as the estimated Ground Truth (eGT) only when the same result was obtained from at least 3 reviewers. Otherwise, a re-grade was needed.
Data pre-processing
Before training the model, each radiograph was first converted from Digital Imaging and Communications in Medicine (DICOM) to a portable network graphic (PNG) file format. The original images were further compressed to 256×256 pixels. The original training data set [8,005] was further expanded more robustly to train the model into more than 100,000 samples by rotating, shifting, and scaling the original images. The augmented parameters and selected value ranges are summarized in Table 2. The full implementation pipeline is shown in Figure 2.

Full table
Model implementation
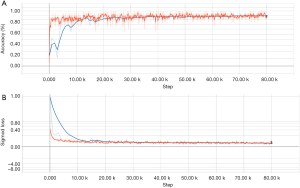
The primary components of the model included an alignment module and a later classification module. The two modules were built on the same backbone, known as a deep residual network (ResNet), which is a deep CNN with 50 layers and about 3.6×109 floating point operations (FLOPS). The model was built according to an open-source machine learning library (TensorFlow version 1.4.1; Google, Mountain View, CA, USA). Training of layers was performed by stochastic gradient descent in batches of 20 images per step, using an Adam Optimizer with a learning rate of 0.001. Training on all categories was run for 80,000 iterations since the training of the final layers for all classes had converged by then. After 80,000 iterations through the entire dataset, the training was stopped due to the absence of further improvement in both accuracy (Figure S1A) and sigmoid loss (Figure S1B).
Left-hand radiographs were used as input data, and the alignment module was trained to directly regress all the 20 ossification center regions of TW3. Figure S2 depicts a total of 13 ossification center regions and 1 carpal bone region, which were fitted by the regression algorithm.
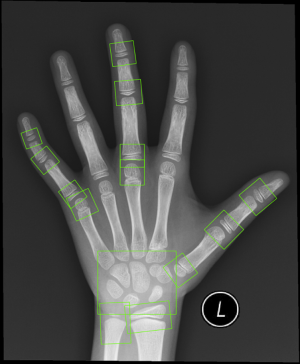
In the classification module, the relevant 20 bones (13 RUS, 7 Carpal) were labeled by the 59 localized points inferred from the alignment module, which were then cropped and impute into the same CNN. The relevant 20 ossification center regions (13 RUS, 7 Carpal) inferred from the alignment module were then passed through a classification network for the labeling of their ossification levels. The classification module used a softmax layer to output multi-classification ranks ranging from A to I for each of the bones. Finally, the ranks for all concerned bones were sent into a TW3-RUS/TW3-Carpal calculator, summed to get the respective final score and cross-referenced with the skeletal maturity table. The output of the classifier was an estimated bone age, according to TW3-RUS and TW3-Carpal method. The scores corresponding to different ranks of bone development are summarized in Table S1 (for males) and Table S2 (for females). The Python code (version 3.7.3) implementing the deep CNN and simulation algorithms can be found online at https://github.com/bmehighday/bone-age-algorIthm.

Full table
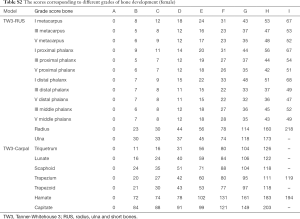
Full table
Statistical analysis
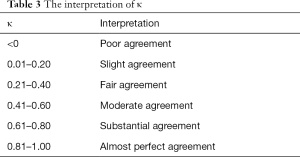
A mean paired inter-observer difference was calculated for each reviewer pair to compare the performance of the human reviewers to paired inter-observer. The overall performance of the model was assessed by comparing the root mean square (RMS) and mean values. RMS was calculated as the square root of the sum of the squares of the paired differences, and the mean was calculated as the average of the paired differences. To assess the overall agreement between reviewers along with the agreement between the model and each reviewer, 95% confidence limits of agreement were calculated. Bland-Altman (BA) plot was used to show the consistency between the model and reviewers. Individual bone agreements were performed by Fleiss’ kappa statistics (Table 3). Statistical significance was determined by using paired t-tests for comparing mean values and F-tests for comparing variances (i.e., RMS). A value with P value <0.05 was considered statistically significant. All the statistical analyses were conducted by R statistical software, version 3.3.2 (R Foundation).
Full table
Results
In total, 8,809 images were obtained to train and validate the model, and another independent 250 images were used to test it. The chronologic age distributions of the patients were 0–18 years old for males and 0–17 years old for females, and the average age was 7.8±3.8 years old (Figure 1). The data set, mean chronologic age, and bone age are shown in Table 1, and the male-to-female ratio of the training and validation set was 3,081/8,809:5,728/8,809 (35%:65%), and 125/250:125/250 (50%:50%) for the test set.
The efficiency of TW3-AI model
We compared the time consumption for BAA between the TW3-AI model and the endocrinologists in the test set. The average processing time for the TW3-AI model was 1.5±0.2 s, which was significantly shorter than the average time (525.6±55.5 s) needed for endocrinologists or radiologists to assess bone age according to the TW3 rule.
The diagnostic performance of the TW3-AI model
The accuracy of the diagnostic performance of the TW3-AI model was evaluated in the test set. Tables 4,5 shows the statistical difference of BAA by the TW3-AI model and reviewers. The average RMS of the model is 0.50 years, which is not significantly different from the average RMS of the 6 reviewers, which means that the performance of the model was not inferior to manual assessment. Meanwhile, the model’s RMS was significantly lower than reviewer 5 in both TW3-Carpal and TW3-RUS (P<0.05), but not lower than the other reviewers.

Full table
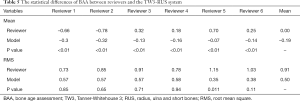
Full table
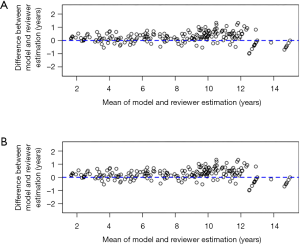
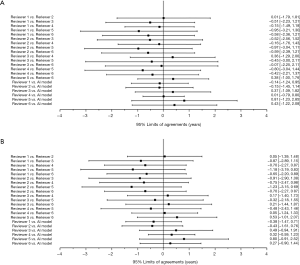
The BA plot shows the difference between the model and the mean of the 6 reviewers in TW3-Carpal (Figure 3A) and TW3-RUS (Figure 3B), which demonstrates a high consistency between the model and reviewers. However, when we compared the model with each reviewer individually, the BA plot showed a poor consistency between the model and reviewer 5 in TW3-Carpal (Figure S3A) and TW3-RUS (Figure S3B). The agreement between BAA made by the model and by the reviewers is shown in Figure 4A (TW3-Carpal) and Figure 4B (TW3-RUS). All assessments are within the 95% confidence limits of agreement between each other.

High variability of reviewer interpretation of individual bones
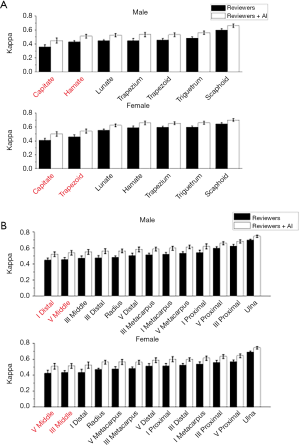
Kappa-test was used to evaluate the consistency both between the TW3-AI model and reviewers, and between reviewers. Table 3 shows the interpretation of κ. The overall consistency between the model and reviewers is better than the between reviewers. Further analysis revealed that for experienced endocrinologists and radiologists, their interpretations were most variable in the male capitate and hamate, the female capitate and trapezoid in TW3-Carpal (Figure 5A). The bones with the highest estimation variation in TW3-RUS were the male first distal and fifth middle phalanx, the female third phalanx, and the fifth middle phalanx (Figure 5B).
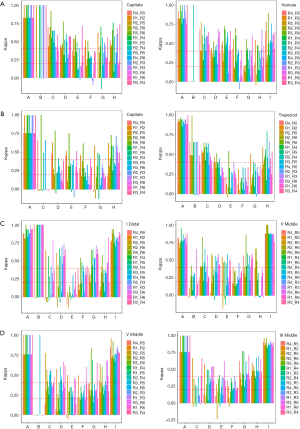
The variation between reviewers in the assessment of these bones was further investigated in this study. Firstly, in TW3-Carpal, different reviewer interpretations occurred in rank B, E, F, and G when assessing the male capitate, while for the male hamate, rank B, F, G, and H were easily misestimated (Figure S4A). Similarly, for female samples, reviewers mostly misinterpreted rank C, E, and G in the capitate and rank B, E, F, and G in the trapezoid (Figure S4B). Secondly, in TW3-RUS, reviewers mostly misinterpreted rank C, D, and E in the male first distal phalanx, and rank B, E, and H in the fifth middle phalanx (Figure S4C). For female samples, rank B, E, and F in the fifth phalanx, and rank C, E, and F in the third middle phalanx were the most misinterpreted ranks (Figure S4D).
Discussion
Our group successfully established a CNN-based TW3-AI BAA system, which was developed from a training set size of 8,005 and a validation set size of 804 clinical hand radiographs. The BAA of the model was nearly real-time. After continuous optimization, the model reached an accuracy within 95% of the confidence limits of agreement compared with that of the experts’ assessment. In a head-to-head comparison, the consistency between the TW3-AI model and reviewers is better than that between reviewers. We concluded that our TW3-AI model performed similarly to experienced endocrinologists and radiologists in terms of the accuracy of BAA, with better stability than manual interpretations.
BAA is a crucial tool in pediatric clinics, which can be used to evaluate the current status of children's growth and development, to tell the future growth potential and predicted adult height, and to inform the efficacy of the treatment for the diseases and includes characteristics like short stature, congenital adrenal hyperplasia, and precocious puberty. Consequently, an accurate, consistent, and stable BAA is the prerequisite for clinical endocrine work. However, in the endocrinology department of our hospital, there are more than 47,000 outpatients annually which challenges our endocrinologists to consistently, accurately, and rapidly assess bone age. Thus, the development of a machine learning model would solve many of the problems that pediatricians face every day.
To the best of our knowledge, our model is the first AI model based on the TW3 rule for BAA. Developed on a TW3 scoring system, which rates for both carpal and RUS bones from A to I, our model is different from most of the previous works based on the extraction of morphological features just from carpal or RUS bones (16-18). In this study, the age distribution of the included patients covered the infancy to late adolescence stages, and both carpal and RUS bones were used to train and validate the model. As a result, our model had high accuracy and stability in BAA and was applicable not only to young children but also to older teenagers.
Further analysis revealed that the bones that were most variably interpreted by reviewers were the capitate, hamate, the first distal and fifth middle phalanx of male patients; and the capitate, trapezoid, and the third and fifth middle phalanx of female patients. The underlying reason for the variation was that the grading and scoring of these bones are subjective, so inter- and intra-reviewer variation is inevitable. By strengthening the learning of these individual bones and ranks scoring, we could improve the accuracy and consistency of BAA in clinicians, which in turn can be used as a reference for future model optimization for clinicians in the evaluation of bone age.
Much work has been done on refining an automatic system to evaluate bone age. HANDX and CASAS systems were the earliest attempts for automatic BAA (19,20). Both systems were based on feature extraction of hand bones, and they showed better consistency than manual assessment. Nevertheless, they are more time-consuming than the manual evaluation of bone age (16). Recently, Harvard Medical School and Stanford University School of Medicine individually developed an automated deep learning system for BAA; both models are based on feature extraction (13,14). Our model was based on the TW3 rule, which is recognized as the most objective method used to evaluate bone age. It also demonstrated superior stability in BAA, as the RMS was 0.50 years in both TW3-Carpal and TW3-RUS, which was smaller compared to the RMS of 0.67 years in the model developed by Stanford University.
There are several limitations to our model. Firstly, similar to the previous works in BAA, there is no gold standard for bone age evaluation, because the inter- and intra-reviewer variations are inevitable (21,22). Previous studies have reported a standard error of the inter-reviewer variation from 0.45 to 0.83 years (standard deviation of 0.64 to 1.17 years) (12,23). In this study, the RMS of inter-reviewer variation was 0.72 years, which is comparable with previous research, while the RMS between the model and reviewer was 0.50 years, which showed superior stability compared to the manual assessment. Secondly, all the images were obtained from our hospital, suggesting more images should be collected from other medical centers to reduce the bias. Thirdly, our model cannot detect certain diseases that human specialists might identify when analyzing the images, such as rickets, hypochondroplasia, and other congenital syndromes (22). Nonetheless, we believe that with the development of medically oriented machine learning techniques, the advantages of AI-model in BAA will become increasingly apparent.
In summary, we developed an automated CNN-based TW3-AI model that can estimate bone age with similar accuracy and superior stability compared to manual assessment. The highly accurate and efficient TW3-AI model will spare clinicians from the tedious clinical viewing process, and thoroughly improve the level of diagnosis and treatment for children’s endocrine diseases.
Acknowledgments
Funding: We are incredibly grateful to all the patients who took part in this study, along with the whole team, including the technicians, engineers, clerical workers, research scientists, and nurses. This study was supported by the National Key Research and Development Program of China (No. 2016YFC1305301), the National Natural Science Foundation of China (No. 81570759 and 81270938), the Fundamental Research Funds for the Central Universities (2017XZZX001-01), the Research Fund of Zhejiang Major Medical and Health Science and Technology & National Ministry of Health (WKJ-ZJ-1804), the Public Welfare Technology Application Research Program of Zhejiang Provincial Science and Technology Project (2016C33130), and the Zhejiang Province Natural Sciences Foundation Zhejiang Society for Mathematical Medicine (LSZ19H070001).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The institutional review boards of our hospital approved the study (No. 2016-IRB-018). All patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tanner JM. A history of the study of human growth. London: Cambridge University Press, 1981.
- Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. Stanford: Stanford University Press, 1959:238-393.
- Tanner JM, Healy MJR, Cameron N, Goldstein H. Assessment of Skeletal Maturity and Prediction of Adult Height (TW3 Method). London: W.B. Saunders, 2001.
- Roche AF, Rohmann CG, French NY, Dávila GH. Effect of training on replicability of assessments of skeletal maturity (Greulich-Pyle). Am J Roentgenol Radium Ther Nucl Med 1970;108:511-5. [Crossref] [PubMed]
- De Sanctis V, Soliman AT, Di Maio S, Bedair S. Are the new automated methods for bone age estimation advantageous over the manual approaches? Pediatr Endocrinol Rev 2014;12:200-5. [PubMed]
- Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, Kim N. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18:570-84. [Crossref] [PubMed]
- Wang S, Wang R, Zhang S, Li R, Fu Y, Sun X, Li Y, Sun X, Jiang X, Guo X, Zhou X, Chang J, Peng W. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤3 cm using HRCT. Quant Imaging Med Surg 2018;8:491-9. [Crossref] [PubMed]
- Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018;95:43-54. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak JAWM. CAMELYON16 Consortium. Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Öner MÜ, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The BoneXpert method for automated determination of skeletal maturity. IEEE Trans Med Imaging 2009;28:52-66. [Crossref] [PubMed]
- Lee H, Tajmir S, Lee J, Zissen M, Yeshiwas BA, Alkasab TK, Choy G, Do S. Fully automated deep learning system for bone age assessment. J Digit Imaging 2017;30:427-41. [Crossref] [PubMed]
- Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018;287:313-22. [Crossref] [PubMed]
- Halabi SS, Prevedello LM, Kalpathy-Cramer J, Mamonov AB, Bilbily A, Cicero M, Pan I, Pereira LA, Sousa RT, Abdala N, Kitamura FC, Thodberg HH, Chen L, Shih G, Andriole K, Kohli MD, Erickson BJ, Flanders AE. The RSNA pediatric bone age machine learning challenge. Radiology 2019;290:498-503. [Crossref] [PubMed]
- Seok J, Hyun B, Kasa-Vubu J, Girard A. Automated classification system for bone age X-ray images. Seoul: 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC), 2012:208-13.
- Zhang A, Gertych A, Liu BJ. Automatic bone age assessment for young children from newborn to 7-year-old using carpal bones. Comput Med Imaging Graph 2007;31:299-310. [Crossref] [PubMed]
- Somkantha K, Theera-Umpon N, Auephanwiriyakul S. Bone age assessment in young children using automatic carpal bone feature extraction and support vector regression. J Digit Imaging 2011;24:1044-58. [Crossref] [PubMed]
- Michael DJ, Nelson AC. HANDX: a model-based system for automatic segmentation of bones from digital hand radiographs. IEEE Trans Med Imaging 1989;8:64-9. [Crossref] [PubMed]
- Pietka E, McNitt-Gray MF, Kuo ML, Huang HK. Computer-assisted phalangeal analysis in skeletal age assessment. IEEE Trans Med Imaging 1991;10:616-20. [Crossref] [PubMed]
- Kim JR, Lee YS, Yu J. Assessment of bone age in prepubertal healthy Korean children: comparison among the Korean standard bone age chart, Greulich-Pyle method, and Tanner-Whitehouse method. Korean J Radiol 2015;16:201-5. [Crossref] [PubMed]
- van Rijn RR, Thodberg HH. Bone age assessment: automated techniques coming of age? Acta Radiol 2013;54:1024-9. [Crossref] [PubMed]
- Bull RK, Edwards PD, Kemp PM, Fry S, Hughes IA. Bone age assessment: a large scale comparison of the Greulich and Pyle, and Tanner and Whitehouse (TW2) methods. Arch Dis Child 1999;81:172-3. [Crossref] [PubMed]


