
Automatic knowledge extraction from Chinese electronic medical records and rheumatoid arthritis knowledge graph construction
Introduction
Knowledge graphs, which are structural representations, have evolved as a critical research direction in intelligent cognitive systems. As a repository of knowledge, a knowledge graph elucidates various entities or concepts in the real world with their underlying relationships, effectively visualizing knowledge networks (1). Medicine is a key domain for the application of vertical knowledge graphs, and thus far, several medical knowledge graphs have been reported (2-5). Medical knowledge graphs were introduced earlier and developed more rapidly in countries other than China. There is not much research on Chinese medical knowledge graphs because the management of medical data is strict, medical data utilization is low, Chinese natural language processing tools and medical knowledge bases are relatively lacking, and Chinese is a much more complex language than English. Most current Chinese medical knowledge graphs have been obtained from medical literature, textbooks, network resources, clinical trials, and others (6,7). However, knowledge graphs based on electronic medical records (EMRs) are rarely reported. EMRs are the most valuable information in the medical domain. They are a repository of large amounts of medical knowledge associated with patient health status and a complete record of patient diagnosis and treatment. EMRs typically include the main complaints, disease course information, examination results, medical advice, and therapeutic regimens of patients, which are crucial references for the development of clinical medical research. The development and utilization of EMRs have always been active research topics in the medical field (8-10). In China, research on knowledge graph construction based on EMRs has only emerged recently (11,12). Generally, EMRs are primarily represented in three forms, i.e., tables, free text, and images. In China, free text entries are currently the predominant EMR mode. Much of the rich, expressive clinical data captured in EMRs are documented and stored within these unstructured free text (13).
Knowledge extraction is the primary task of knowledge graph construction and includes named entity recognition (NER) and relation extraction. English language automatic knowledge extraction models have achieved advanced performance, but they are not suitable for the knowledge extraction of Chinese EMRs (CEMRs) due to the different language characteristics of Chinese and English. Although existing neural network models have achieved strong performance in extracting Chinese knowledge in general fields, their performance in the medical domain is not ideal. This is mainly due to the characteristics of free text extraction in the medical domain versus free text extraction in general domains being markedly different. For example, the definitions of entity types between these two domains are different. The general domain focuses on general entities such as person names, place names, and organization names. The entity length is short, and the structure is relatively simple. Nevertheless, the entity types in the medical field, such as disease types, drug names, gene and protein names, and other professional terms, are highly domain specific and professional, and the entity structure is more complex and changeable, such as the identification of discontinuous entities. Most existing models in the Chinese medical domain usually require high-quality and large-scale manually labeled corpora for model training, which is challenging and expensive to obtain. Annotated CEMR data are even scarcer.
Here, we focus on CEMRs of rheumatoid arthritis (RA). RA is a systemic autoimmune disease primarily characterized by chronic, progressive, and invasive arthritis. RA affects up to 1% of the general population and is manifested by a high systemic inflammatory burden and an increased rate of disability (14,15). RA EMRs have unique characteristics, namely, more types, more patterns, larger quantities, high similarity, and low regional variability of data. Manual annotation is highly time-consuming and laborious. Therefore, how to extract knowledge from RA CEMRs with high efficiency and accuracy using a small number of manually labeled samples is a key scientific problem.
In this study, we detailed the process of RA knowledge graph construction based on CEMRs, conducted a preliminary assessment and presented an application. We focused on the challenges of automatic knowledge extraction based on a small number of manually annotated samples and proposed a pretrained language model combined with a deep neural network for knowledge extraction from CEMR tasks. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1158/rc).
Methods
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics committee of the Peking University People’s Hospital (No. 2020PHB356-01). Written informed consent was waived by the IRB of Peking University People’s Hospital because the medical records were analyzed anonymously. All methods were carried out in accordance with relevant guidelines and regulations.
In this section, the systematic procedure to construct a medical knowledge graph from CEMRs is described. This procedure involves 6 main steps: (I) data preparation; (II) NER; (III) relation extraction; (IV) entity linking; (V) knowledge graph storage and drawing; and (VI) knowledge graph quality evaluation and application. Steps (II) and (III) require a significant amount of human effort and experience. Therefore, this study aims to address this issue and proposes a machine learning model to automatically perform NER and entity relation extraction.
Labeling work for basic data
Data source
The data used in this study were obtained from Peking University People’s Hospital. A total of 1,986 medical CEMRs of RA from July 2015 to January 2021 were obtained. Each CEMR comprised the admission notes and discharge summary notes of a patient. The admission notes include the patients’ details, chief complaints, present illness history, past history, physical examination results, and preliminary diagnosis. Individual discharge records contain admission status, diagnosis and treatment processes, auxiliary examination results, discharge status, medical advice, and discharge diagnoses.
Definitions for RA entity and entity relations
Notably, complete and accurate definitions of disease-related entities in EMRs are vital in the construction of medical knowledge graphs. In this study, five experts (3 medical experts and 2 rheumatology experts) developed definitions for medical information related to RA CEMRs based on relevant research on entity recognition and entity relation classification as well as the practical and clinical characteristics of RA medical records. A total of 9 entity types and 11 relation categories were systematically defined, highlighting the symptoms, treatment, and diagnostic information of RA (Tables 1,2). In this paper, 11 entity relations were defined to summarize the potential relationships between entities in RA CEMR more comprehensively. As all labeled entities in this study except ‘diseases’ are related to RA, the first five relationship categories in Table 2 were established through rules in subsequent relation extraction tasks, and the last six relation categories were obtained through the proposed model based on deep learning in the subsequent relation extraction tasks.
Table 1
| Named entity type | Meaning | Example |
|---|---|---|
| Disease | The name of the disease | Hypertension |
| Symptom | Patient’s discomfort or abnormal feeling | Pain in knee joint |
| Treatment | Treatment procedures and drugs | Arthroscopic surgery |
| Physical examination | Results obtained from an examination | Swelling of knee joint |
| Imaging test | A method of examination imposed on a patient to detect or confirm symptoms/signs | MRI |
| Lab test | Laboratory process and items | ESR |
| Aggravating factor | Factors and behaviors aggravating the disease or symptoms | Catch a cold |
| Mitigation factor | Factors and behaviors relieving the disease or symptoms | Bed rest |
| Body part | Body part where the disease occurs | Knee |
MRI, magnetic resonance imaging; ESR, erythrocyte sedimentation rate.
Table 2
| Relation category | Semantic relation |
|---|---|
| Disease and symptom | The disease causes a symptom |
| Disease and physical examination | Assists in the diagnosis of diseases through a physical examination |
| Disease and lab test | Assists in the diagnosis of diseases through a lab test |
| Disease and imaging test | Assists in the diagnosis of diseases through an imaging test |
| Disease and treatment | Methods of disease treatment |
| Mitigation factor and symptom | Factors mitigate the symptoms |
| Aggravating factor and symptom | Factors aggravate the symptoms |
| Treatment did not improve the disease | Treatment did not improve the disease |
| Treatment improved the disease | Treatment improved the disease |
| Body part and physical examination | Body part for physical examination |
| Body part and symptom | Body part with symptoms |
Manual annotation
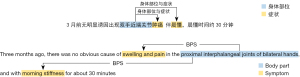
The data labeling work was carried out by three medical experts using the medical text processing platform of the Chinese Academy of Medical Sciences, Institute of Medical Information. The annotated data were then carefully inspected and corrected by another information expert to ensure the accuracy of the manual annotations. The annotation results (partial entities) are presented in Figure 1. Of these, “swelling and pain” and “morning stiffness” were the symptoms, and “proximal interphalangeal joints of bilateral hands” was the body part, with two relations between the three entities. A total of 367 RA medical records were manually annotated.
Data preprocessing
CEMRs were documented in text form. First, we split the CEMRs by periods, time points and line breaks. Unlike English, Chinese clinical text is written without spaces to indicate word boundaries, and there are many different combinations of characters. To avoid entity boundary recognition errors caused by word segmentation, we used characters as the input of the model.
We formulated the clinical NER task as a sequence-labeling task to determine the sequence of labels with the largest joint probability for the sequence of input tokens and a predefined set of labels. In this paper, we adopted the BIO (beginning, inside, and outside) tagging scheme to represent the position of the tokens within the entities. A token is labeled as ‘B-label’ if it is at the beginning of a named entity, ‘I-label’ if it is inside but not at the beginning of a named entity or ‘O-label’ otherwise. An example of BIO tagging can be seen in Figure 2, where ‘来氟米特’ (leflunomide) is a treatment entity and ‘来’ is the beginning of this entity.
In this paper, we formulated the entity relation extraction task as a classification task to extract relations between two entities of a certain type (e.g., disease, symptom, and treatment) and fall into a number of semantic categories (e.g., DCS: the disease causes a symptom, and TnID: the treatment did not improve the disease). Before entering the text into the relation extraction model, special tags were inserted before and after the target entity to identify the position of the two target entities. Specifically, for sentences with two target entities and , a special tag “$” was inserted at the start and the end positions of the first entity (i.e., the head entity), and a special tag “#” was inserted at the beginning and the end of the second entity (i.e., the tail entity), enabling the module to capture the location information of the two entities. Moreover, “[CLS]” was added at the beginning of each sentence. An example is shown in Figure 3.

The NER model
We utilized an improved deep learning model named bidirectional encoder representation from transformers (BERT)-bidirectional long short-term memory (BiLSTM)-conditional random field (CRF), which applies the medical pretrained language model, i.e., MC-BERT, to generate more accurate contextual word representations to enhance the basic BiLSTM-CRF model. A brief introduction of the network structure and working principle of the model are presented herein.
BERT
The process of learning word representations from a large amount of unannotated text has been a well-established method for a long time. Previous models [e.g., Word2Vec (16) and GloVe (17)] have focused on learning context-independent word representations, and recent works [e.g., ELMo (18) and BERT (19)] have focused on learning context-dependent word representations. BERT is one of the mainstream contextualized word representation models. It is based on bidirectional transformers (20) and pretrained using a masked language model. The network structure of BERT is presented in Figure 4. Compared with traditional unidirectional language models (e.g., left-to-right and right-to-left), BERT applies a masked language model that predicts randomly masked words in a sequence and hence can be used for learning bidirectional representations. In addition, BERT has achieved significant performance on most natural language processing (NLP) tasks while requiring minimal task-specific architectural modifications rather than unidirectional representations, which is crucial for representing words in natural language. It is critical to apply these bidirectional representations, rather than unidirectional representations, to represent biomedical terms existing in a biomedical corpus to mine their complex relationships. Due to space limitations, we refer readers to Devlin et al. (19) for a more detailed description of BERT.
As a general language representation model, BERT was pretrained using English Wikipedia and BookCorpus. However, biomedical domain texts contain a considerable number of domain-specific proper nouns and terms, which are understood mostly by biomedical researchers.
LSTM
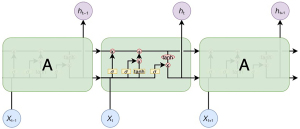
Recurrent neural networks (RNNs) are a family of neural networks that operate on sequential data. They use a sequence of vectors () as input and output another sequence () that represents some information about the sequence at every step in the input (Figure 5). Although RNNs can, in theory, learn long dependencies, in practice, they fail to do so and tend to be biased toward the most recent inputs in the sequence (20).
LSTMs have been designed to address this issue by incorporating a memory cell and have been shown to capture long-range dependencies well. They do so using several gates that control the proportion of the input to give to the memory cell and the proportion from the previous state to forget (21). The repeating module of LSTM contains four interaction layers. The network structure is presented in Figure 6, and its implementation is as follows:
where σ is the elementwise sigmoid function, which can map the output to a space from 0 to 1, and ⊙ is the elementwise product. xt is the input at time t, and W and b in these formulas are learnable parameters.
For a given sentence () containing n words, each represented as a d-dimensional vector, and an LSTM network computes a representation of the left context of the sentence at every word t. Naturally, generating a representation of the right context should add useful information. This can be achieved using a second LSTM network that reads the same sequence in reverse. We will refer to the former as the forward LSTM and the latter as the backward LSTM. These are two distinct networks with different parameters. This forward and backward LSTM pair is referred to as a bidirectional LSTM (22).
NER model: MC-BERT-BiLSTM-CRF + extra_feature model
The MC-BERT (23) model was applied to initialize the transformer parameters subjected to NER-labeled Chinese medical datasets in other domains, including cEHRNER and cMedQANER, for fine-tuning. First, contextual word vector representations were generated through MC-BERT, and the learned word vectors were input into the BiLSTM model. Then, high-dimensional word vectors were obtained by using the contextual information capturing ability of BiLSTM. Finally, the results were fed into the CRF model based on contextual information augmentation to obtain a predicted tag sequence. Extra_feature refers to the feature information related to sentence sequences obtained externally using ‘Jieba’ and other open-source tools and include pinyin, font, glyph, part-of-speech, and word segmentation tags of Chinese characters. As illustrated in Figure 7, the framework of the NER model contains three components: an embedding layer, an encoding layer, and a CRF layer.
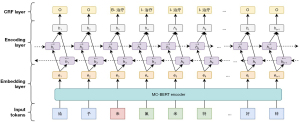
Embedding layer
We passed the sequence of input tokens to the MC-BERT model to obtain the contextual representations , where ei denotes the vector representation of the character xi calculated by MC-BERT, N is the length of the input sentence and d1 is the size of the BERT hidden states. The calculation process can be formulated as follows:
Next, the representations are given as the input to the encoding layer.
Encoding layer
To acquire high-dimensional representations, we utilized BiLSTM to further capture the context information of the input sentence.
In LSTM, the hidden states ht and the memory cell ctare a function of the previous ct-1 and ht-1 and the input vector et, or formally, as follows:
where the hidden state denotes the representation of position t while also encoding the preceding contexts of the position.
In this paper, we applied BiLSTM with two separate hidden states, i.e., a forward LSTM and a backward LSTM. For each token at position t, the new representation
is generated by concatenating the hidden states of the forward LSTM and backward LSTM. In this way, the forward and backward contexts are considered simultaneously, and we obtain the complete sequence of hidden states:
Finally, we utilize a linear layer to map the hidden state vector ht of BiLSTM from the d2-dimension to the k-dimension, where k is the number of labels defined in the tagging scheme. As a result, the sentence features are extracted and represented as a matrix
where the element Pi,j of the matrix is the score of the jth tag of the ith token in the sentence.
CRF layer
With respect to the sequence labeling task, there are strong dependency relationships between the tags of adjacent words. For example, if a token of a sentence is labeled as ‘B-label’ in the BIO tagging scheme, only the ‘I-label’ or ‘O-label’ can be considered to annotate the next token, and other tags cannot be used. Therefore, to make the most of the dependency relationships, we adopt a CRF layer to decode the best tag path in all possible tag paths.
In the CRF layer, the current input is predicted by the previous input and the state to which the input belongs. Thus, we defined the parameters of the CRF layer by a matrix , and Ti,j denotes the score by moving from the ith label to the jth label. Considering a sequence of predicted labels , the following formula is used to calculate the score of the label sequence:
The score of the whole sequence is equal to the sum of the scores of all words with the sentence, which is determined by the output matrix P of the BiLSTM layer and the transition matrix T of the CRF layer. Moreover, the softmax function can be used to obtain the normalized probability:
The maximum likelihood estimation is used as the loss function during the model training. The following equation gives the logarithmic likelihood of a training sample (X, Y):
The model is trained by maximizing the log likelihood function,
and the Viterbi algorithm is used to compute optimal tag sequences for inference by using dynamic programming.
The entity relation extraction model: MC-BERT + feedforward neural network (FFNN)
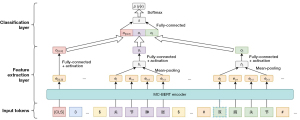
We exploited the MC-BERT pretrained language model to extract the features of the sentences and entities, and then the extracted sentence-level and entity-level features were fed into a classifier to predict the relation between entities. As illustrated in Figure 8, which contains two components, a feature extraction layer and a classification layer.
Feature extraction layer
First, we passed the processed sentence (in which the positions of two entities are marked, and the special token “[CLS]” is inserted at the beginning of the sentence, as shown in Figure 3) into MC-BERT to obtain a representation of individual words and sentences. Then, we considered the corresponding representations of the token [CLS] as the features of the whole sentence and applied a mean pooling operation on the word representations involved in each entity to obtain the entity-level features. In addition, a fully connected FFNN was applied to both the sentence-level and entity-level features. As shown in Figure 8, the representations of the head entity can be calculated as
where L+1 denotes the length of the head entity. Similarly, we obtained the representations of the tail entity as
where M+1 is the length of the tail entity, and the sentence representation is given as
Classification layer
The sentence representations were concatenated with two entity representations into a fully connected layer to obtain vectors with length C as the number of relation categories, which is denoted as:
Then, the softmax function is utilized to obtain the relation classification result.
Where P(rk|X) represents the probability that sentence X belongs to the k-th relationship. Finally, we adopted the cross-entropy loss function to train the relation classification model as follows:
In this way, the relation type between the two entities was obtained in each sentence.
The parameter settings of the proposed model are detailed in Tables S1,S2.
Normalization-named entity linking (NEL)
There is usually a synonymy problem in the process of extracting knowledge from a single data source; that is, an entity has many different entity mentions, such as names, aliases, abbreviations, and even misrepresentations. NEL is an important method to solve the problem of entity ambiguity by mapping entity mentions to standard concepts in the knowledge base (24). Before carrying out NEL, we constructed 9 dictionaries corresponding to the entity type set in this study based on sources such as medical standard dictionaries (ICD-10, SNOMED CT, MedDRA 20.0, and Chinese Approved Drug Names) and the clinical medical knowledge base. In this way, a dictionary was constructed so that the set of entries in the dictionary consists of all strings that may denote a named entity. Then, each entry string in the dictionary is mapped to a set of entities that refer to the same semantic concept. As a result, a named entity e is included in the set if and only if the string is one of the redirect names or disambiguation names of e.
The NEL process can be divided into 3 steps. The first step is to remove duplicate entities and match them hierarchically between entities and categories. Specifically, the semantic type of an entity matches a category; then, the entity exactly matches the entities in this category. The second step is to use the constructed dictionaries to cover the unmatched entity mentions. Finally, the rank-based approach of cosine similarity is used to sort the unmatched entities and select the top-ranking entities as the NEL result after disambiguation. The vast majority of NEL was achieved by the above process, and the remaining unmatched entities were checked and matched manually. For example, “爱若华” is the trade name of leflunomide. “爱若华” belongs to the “treatment” class; it matches this class first and then matches the candidate entity ‘来氟米特’ (leflunomide). Errors in CEMRs are not uncommon if a doctor accidentally records “来氟米特” as “来福米特”. This type of entity is not well matched after the first and the second steps; however, in step 3, the cosine similarity indicated that this entity highly matched ‘来氟米特’, and “来氟米特” was selected as the NEL result.
RA knowledge graph storage and drawing
The RA knowledge graph was constructed using the Neo4j graph database. Previously identified entities and entity relations in the batch were imported into Neo4j from a CSV file. Neo4j applies Cypher to retrieve data and supports an ACID (atomicity, consistency, isolation, durability)-compliant transactional backend. The visual knowledge graph enables an improved understanding of the medical information content.
Evaluation of the model performance and knowledge graph quality
To evaluate the performance of the proposed automatic knowledge extraction model, we used precision, recall, and F1-score as the evaluation metrics (25).
To evaluate the reliability and practicability of the RA knowledge graph, the expert evaluation method was adopted to preliminarily evaluate the knowledge graph from 3 aspects, i.e., the data layer, schema layer, and application layer, covering 8 different quality dimensions. The evaluation indicators are listed as follows: the authority and value of the data score in the data layer; the rationality and scalability scores in the schema layer; and the data consistency, ease of use, readability of the results, and practicability score in the application layer. The quality evaluation process was divided into 3 steps: first, the main construction process of the RA knowledge graph based on CEMRs was introduced to the experts. Then, after the experts were assisted in employing the RA knowledge graph stored in Neo4j, they viewed the display of the knowledge graph and carried out knowledge retrieval. Finally, the evaluation indicators were scored with a secret ballot. A total of 4 experts (1 expert in computer science, 2 rheumatology experts, and 1 clinical expert) evaluated the quality of the knowledge graph using a 5-point scale, and the average values of the 4 experts were reported.
Results
A total of 367 CEMRs were manually annotated and divided into a training set, verification set, and test set at a ratio of 25:1:2. The training set comprises 327 medical records, the verification set contains 15 medical records, while the test set contains 25 medical records. The manually annotated RA data were used to train the model. Specifically, we first identified named entities and then further obtained the relationships between entities. After the completion of NER, the potential relation between entities was defined with more than 2 entities in a sentence. Therefore, a relation classification model for any two entities was established, and each sentence was labeled by different entity pairs to form a sample for the relation classification task. Note that we reported the results of all experiments in 5 runs with random seeds.
Results of NER
The entity type and number are presented in Table S3. The overall performance of the proposed model against other popular baselines for NER tasks with RA datasets is shown in Table 3. The proposed model outperformed all other popular models with the RA corpora. Specifically, the MC-BERT-BiLSTM-CRF model achieved an F1 score of 89.69%. When extra_features were added to the model, the F1 score was further improved, as an F1 score of 92.96% was obtained. Extra_features refers to the features related to sentence sequences obtained externally by using ‘Jieba’ and other open-source tools. The F1 score of the MC-BERT-BiLSTM-CRF + extra_feature model improved by +5.1% over that of the traditional method using a model pretrained with general domain knowledge. The main improvement of the proposed model comes from the fact that the pretrained model with rich knowledge from the medical field was utilized. The experimental results demonstrated the excellent ability of the proposed model to sequentially recognize named entities from the medical domain.
Table 3
| Model | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|
| BiLSTM-CRF | 83.45 | 82.12 | 82.78 |
| BERT-Chinese-wwm-ext-CRF | 86.32 | 89.46 | 87.86 |
| MC-BERT-sofmax | 88.15 | 88.97 | 88.56 |
| MC-BERT-CRF | 86.35 | 90.80 | 88.52 |
| MC-BERT-BiLSTM-CRF | 88.31 | 91.11 | 89.69 |
| MC-BERT-BiLSTM-CRF + extra_feature | 91.95* | 93.99* | 92.96* |
*, indicates that the result is statistically significantly better than that of the best result of all contrast models with t-test P<0.05. RA, rheumatoid arthritis; NER, named entity recognition; BiLSTM, bidirectional long short-term memory; CRF, conditional random field; BERT, bidirectional encoder representation from transformers; MC, medical.
As shown in Table 4, we analyzed the entity recognition performance of the MC-BERT-BiLSTM-CRF + extra_feature model for different entity types. The model achieved better performance for most types of entities, but it was worst for the “aggravating factor” entity. The limitation of the dataset is one possible reason for this result.
Table 4
| Named entity type | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|
| Disease | 93.10 | 96.43 | 94.74 |
| Symptom | 92.67 | 96.19 | 94.40 |
| Treatment | 92.93 | 94.30 | 93.61 |
| Physical examination | 82.46 | 80.56 | 81.50 |
| Imaging test | 83.33 | 91.46 | 87.21 |
| Lab test | 95.14 | 98.00 | 96.55 |
| Aggravating factor | 41.67 | 62.50 | 50.00 |
| Mitigation factor | 66.67 | 100.00 | 80.00 |
| Body parts | 92.99 | 93.40 | 93.19 |
| All | 91.95 | 93.99 | 92.96 |
NER, named entity recognition; MC, medical; BERT, bidirectional encoder representation from transformers; BiLSTM, bidirectional long short-term memory; CRF, conditional random field.
Results of entity relation classification
The entity relation type and number are presented in Table S4. As previously described, five relation types in this study were established through rules, and the other six relation types were extracted through models. This section presents the entity extraction results with the proposed deep learning model. Table 5 shows the seven relation types obtained by the proposed model. ‘Other’ refers to the entity relation extracted by the MC-BERT + FFNN model that does not belong to the categories defined in the study. Two sets of experiments were designed, i.e., one for relation prediction using the standard entity recognition results as input and the other using the model predicted entities as input. The results are reported in Table 5. The results showed that when using the model predicted entities as input, our model achieved an F1 score of 73.64%, while the F1 score reached 95.29% when standard entities were used. This suggests that the accuracy of the entity recognition task is critical to the accuracy of entity relation extraction. The results showed that the MC-BERT + FFNN model achieved an F1 score of over 90%, except for ‘Treatment did not improve the disease’ and ‘Treatment improved the disease’. The distances of the ‘treatment’ and ‘symptom’ entities were too large in most of the medical records, which decreased the accuracy of the relation extraction task to some extent.
Table 5
| Relationship category | Standard entity relation extraction | Model predicted entity relation extraction | |||||
|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | F1 score (%) | Precision (%) | Recall (%) | F1 score (%) | ||
| Treatment improved symptom | 87.12 | 76.37 | 81.59 | 64.31 | 70.03 | 66.70 | |
| Treatment did not improved symptom | 83.63 | 70.11 | 76.30 | 60.72 | 74.26 | 66.66 | |
| Body part and symptom | 95.90 | 98.73 | 97.30 | 76.65 | 81.68 | 79.09 | |
| Body part and physical examination | 97.65 | 90.22 | 93.79 | 66.49 | 68.68 | 67.57 | |
| Aggravating factor and symptom | 100 | 70.21 | 82.49 | 50.31 | 54.26 | 52.21 | |
| Mitigation factor and symptom | 100 | 84.53 | 91.61 | 53.80 | 57.75 | 55.58 | |
| Other | 91.73 | 94.94 | 93.31 | 72.52 | 75.28 | 73.87 | |
| All | 95.89 | 94.69 | 95.29 | 70.94 | 76.56 | 73.64 | |
The overall performance of the proposed model against other popular models for RA relation extraction tasks with RA datasets is shown in Table 6. Our proposed model outperformed all other popular models with the RA corpora. As a pretrained word vector model, BERT can extract word vector information well. Therefore, when the pretrained language model was used as the sentence feature extractor, the results of the BERT model were better than the results without the pretrained language model. Compared with the BERT base, the words extracted by MC-BERT led to a better performance in terms of the relation extraction task, which confirmed the effectiveness of the pretrained language model based on the medical field in the RA relation extraction task.
Table 6
| Model | Precision | Recall | F1 score |
|---|---|---|---|
| MC-BERT + FNN | 95.89* | 94.69* | 95.29* |
| PRNN | 74.63 | 80.22 | 77.41 |
| CNN + ATTENTION | 75.56 | 78.72 | 77.13 |
| BiLSTM + ATTENTION | 81.42 | 83.11 | 82.26 |
| BERT-base | 95.12 | 92.13 | 93.62 |
* indicates that a result is statistically significantly better than that of the best result of all contrast models with t-test P<0.05. RA, rheumatoid arthritis; MC, medical; BERT, bidirectional encoder representation from transformers; FNN, feedforward neural network; PRNN, pipelined recurrent neural network; CNN, convolutional neural network; Bi-LSTM, bidirectional long short-term memory.
Construction and visualization of the RA knowledge graph
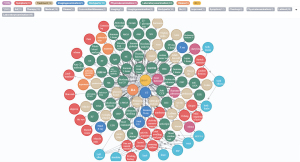
Eventually, based on the NER and entity relation models established, the remaining 1,619 CEMRs were automatically labeled. After entity alignment, entities and entity relations with a frequency of more than 5 were finally used to construct a knowledge graph with 6,465 entities and 15,268 entity relations. An RA knowledge graph was constructed by batch importing the previously identified medical entities and relations into the Neo4j graphic database. Figure 9 reveals part of the constructed RA knowledge graph (the English version was artificially translated, and the original version of the Chinese knowledge graph is presented in Figure S1), although the actual graph is much larger. It provides abundant information on different types of entities; the graphical display provides an overview of the underlying semantic relations.
Preliminary application of the RA knowledge graph
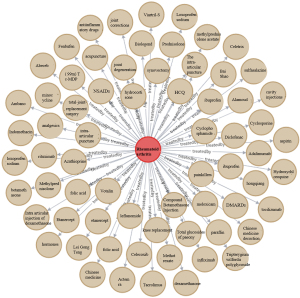
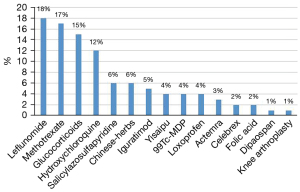
Notably, information searching and reasoning from a knowledge graph are achieved through the Neo4j graph database. In this database, the commonly used treatment information of RA can be searched using MATCH (n: entities)-[r: treatment]-> RETURN n, c LIMIT 200, and the database will return the results. Figure 10 shows the subgraph (the English version was artificially translated, and the original version of the Chinese knowledge graph is presented in Figure S2). RA treatment emphasizes the importance of standardized and overall treatments, i.e., the use of disease-modifying antirheumatic drugs (DMARDs) in the early stage of the disease, combined with NSAIDs and other therapies to relieve symptoms, achieve optimal treatment effects, and minimize other adverse effects. Intelligent analysis based on the RA knowledge graph showed that leflunomide (18%) and methotrexate (17%) are the most popular and widely used DMARDs for RA patients. The top 15 treatments are listed (Figure 11), and these data may guide clinicians. The medical specialists indicated that these treatment results were consistent with actual clinical scenarios.
Preliminary evaluation of the RA knowledge graph
Four experts evaluated the RA knowledge graph, and the results showed that the experts thought that the RA knowledge graph was generally good, with a mean score of 4.31. Comprehensively, the authority and value of the data scores in the data layer were 4.75 and 3.69, respectively. The data were derived from CEMRs from a 3A-class hospital in China, which had a high degree of authority and professionalism. The experts thought that the number of CEMRs was relatively small; thus, the data score was low. The rationality and scalability scores in the schema layer were 4.73 and 4.57, respectively. The experts thought the RA knowledge graph had good performance and a reasonable structure and was easy to expand. The data consistency, ease of use, readability of the results, and practicability scores in the application layer were 4.45, 3.71, 4.52, and 4.06, respectively. In all the evaluation metrics, the “ease of use” of the knowledge graph was the lowest, and although the Cypher query language is easy to learn, it is still difficult for users who do not understand programming, so the experts gave it a very low score. There is still a long way to go before this RA knowledge graph is ready for clinical application, so experts gave “practicability” a relatively low score.
Discussion
There are three main knowledge extraction methods: dictionary-and-rule-based, machine-learning-based, and neural-network-based methods. Dictionary-and-rule-based approaches require manual customization of many characteristic rules, which are usually specific to a particular document. Methods based on machine learning require much feature engineering. The above methods have poor generalizability, which is not conducive to the generalization and transfer learning of the results. The methods based on neural networks use text as a vector input, automatically discovering entity, relation, and attribute features. These methods are suitable for dealing with large-scale knowledge and have become the mainstream method of knowledge extraction. The advanced methods for knowledge extraction are mainly based on convolutional neural networks (CNNs), RNNs, and their variants.
Various variants of RNNs, such as LSTM and BERT, are more commonly used and have shown extraordinary competitiveness in recent years (26,27). Cheng et al. (28) applied a BiLSTM-CRF model in a clinical NER task and achieved an F1-score of 87.00%. Gao et al. (29) proposed a medical entity recognition model based on a character and word attention-enhanced neural network for Chinese resident admission notes and achieved an F1-score of 94.44%. Yin et al. (30) applied CNNs to extract radical-level semantic information, and then a self-attention mechanism was used to directly capture the dependencies between characters. The performance of the model based on the radical-level features and a self-attention mechanism outperformed the BiLSTM-CRF model in clinical NER tasks, and the F1 score improved by at least 0.55%. The introduction of the BERT model has made entity recognition technology more efficient, and the superiority of BERT in representational word vectors has been proven. Li et al. (31) trained a pretrained BERT model on Chinese clinical records and added it to bidirectional LSTM and CRF layers and additional dictionary and radical features. The proposed model attained superior performance against the existing models. Wu et al. (32) embedded a pretrained BERT model in a BiLSTM-CRF model and achieved an F1 score of 96.2% in the NER task from Chinese adverse drug reaction reports.
Relation extraction is also an important NLP task. Liu et al. (33) combined Bi-LSTM and an attention mechanism as a joint model to process the text features of sentences and classify the relation between two entities. The experimental results demonstrated that the model can effectively deal with data noise and achieve better relation classification performance at the sentence level. Recently, a pretrained BERT model achieved very successful results in many NLP classification/sequence labeling tasks (34). Zhang et al. (35) embedded a pretrained BERT model into a BiLSTM-CRF model and achieved 93.52% and 96.73% F1 scores in the NER and relation extraction tasks, respectively, indicating superior performance compared to those of previous state-of-the-art models. Chen et al. (36) combined BERT with a one-dimensional convolutional neural network (1D-CNN) to fine-tune a pretrained model for relation extraction. Extensive experiments on three datasets, namely, the BioCreative V chemical disease relation corpus, traditional Chinese medicine literature corpus, and i2b2 2012 temporal relation challenge corpus, showed that the proposed approach achieved state-of-the-art results. The BERT-CNN model demonstrated improved performance for efficiently extracting medical relations.
This study introduced the whole process of RA knowledge graph construction in detail, focusing on the automatic knowledge extraction progress. In total, 9 entity types and 11 relation categories involved in the RA knowledge graph were systematically defined with the supervision of experts. For the challenge of having only a few labeled training samples, a pretrained model-based approach was used to learn text features from unlabeled text in the medical field. In this paper, we used the MC-BERT transformer model for text feature extraction, integrated a pretrained MC-BERT model into a neural network, and proposed an MC-BERT-BiLSTM-CRF model for clinical NER and MC-BERT-FFNN model for entity extraction of RA CEMRs. The proposed model achieved a 92.96% F1 score on the NER task and a 95.29% F1 score on the relation extraction task, reflecting superior results against those of state-of-the-art models. Notably, BERT adopts a neural network architecture based on an attention mechanism that provides a self-supervised method to learn the sentence features in a large-scale dataset, indicating strong generalizability. In addition, as an advanced language feature extractor, the BERT model can learn high-quality word vectors to further enhance the generalizability. The MC-BERT medical pretrained language model was used to extract word representations. Unlike general pretrained language models, MC-BERT involves large-scale text from the medical field as pretraining data to capture medical domain-specific features. Previous work revealed that training for relevant domain-specific text can effectively improve the task of a specific field (23). Furthermore, BiLSTM-CRF and fine-tuning of the pretraining language model with the RA CEMR data were modified to fit into the task. The proposed model achieved superior performance for entity identification and entity relation extraction tasks. In future work, we may consider using the proposed model to perform other NLP tasks with CEMRs.
In the automatic text analysis and knowledge extraction of CEMRs, the most important aspect for a deep learning model is to learn and extract relevant features. In the present study, several countermeasures were taken in the model for samples with only a small number of annotations. First, we used a pretrained language model as a feature extractor that was trained with many unlabeled data in an unsupervised learning manner. Second, we conducted an upsampling method during the model training, that is, in each training batch, the probability of small sample data being learned by the model was increased to ensure the balance of various samples in the training. Third, we used a data enhancement method to expand this kind of small sample by using manual rules. Through processing, the proposed automatic knowledge extraction model based on a small number of manually labeled samples achieved good performance.
We conducted a preliminary application with the knowledge graph. As a result, the RA knowledge graph allowed information retrieval, where a user could directly extract the required concepts, including ‘treatments’, ‘imaging examination’, and ‘symptoms’, and their semantic relationships to construct a schema. Since the application of knowledge graphs is a relatively new direction, we only conducted preliminary intelligent applications. Future research efforts will focus on the application-level development of the RA knowledge graph, including providing more intelligent retrieval services and auxiliary health care decision-making systems. Furthermore, we constructed an RA knowledge graph based on the identified entities and extracted entity relations by the proposed model from 1,986 CEMRs. Moreover, the reliability of the constructed RA knowledge graph was visualized before evaluating its practicability from different dimensions. Experts confirmed the effectiveness of the constructed RA knowledge graph (with a mean score of 4.31). Based on these results, it can be concluded that the RA knowledge graph has a reasonable structure and higher accuracy and can easily expand. Nevertheless, because the number of CEMRs was insufficient and the cyber query language was poorly used, there remains considerable room for practicability and ease of use of the knowledge graph.
The major contributions and strengths of this paper are summarized as follows:
- By combining the practical needs of clinical experts and relevant research on NER and relation extraction, we systematically defined 9 entity types and 11 relation categories involved in an RA knowledge graph. Then, 367 datasets were manually labeled to systemically and comprehensively summarize the main contents of the CEMRs. Future efforts will be made to share these manually labeled data via an open source platform.
- Two automatic knowledge extraction models (a pretrained language model combined with a deep neural network) were proposed to extract knowledge from unstructured CEMR data, which addressed the challenge of limited manually annotated labeled data.
- Furthermore, a systematic procedure was introduced to construct a medical knowledge graph from CEMRs, rather than only focusing on specific steps in previous works. To our knowledge, this is the first RA-related knowledge graph based on CEMRs in the real world. This work could provide a preliminary step for advanced intelligent applications and might provide guidance for knowledge graph construction in other medical fields based on CEMRs.
While the present study uncovered some insightful findings, it had a number of limitations. First, the entities and relations were relatively small, and further enriching the properties of entities and relations is considered to be a necessary next step. Second, the evaluation of the knowledge graph was relatively simple and lacked a comparative evaluation of different graphs generated by the different methods. Third, the application of knowledge graphs was preliminary, and the breadth and depth of RA knowledge graph applications can be further improved.
In conclusion, we established an RA knowledge graph based on CEMRs and detailed the processes of data annotation, automatic knowledge extraction, and knowledge graph construction, and presented a preliminary assessment and an application. At the same time, the effectiveness of a pretrained language model combined with a deep neural network to address knowledge extraction tasks based on a small number of manually annotated samples was also proven.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 82071930 to J Zhu).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-1158/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1158/coif). ML is an employee of Beijing Lanzhou Technology Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work and ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics committee of Peking University People’s Hospital (No. 2020PHB356-01). Written informed consent was waived by the IRB of Peking University People’s Hospital because the medical records were analyzed anonymously. All methods were carried out in accordance with relevant guidelines and regulations.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rotmensch M, Halpern Y, Tlimat A, Horng S, Sontag D. Learning a Health Knowledge Graph from Electronic Medical Records. Sci Rep 2017;7:5994. [Crossref] [PubMed]
- Mohamed SK, Nováček V, Nounu A. Discovering protein drug targets using knowledge graph embeddings. Bioinformatics 2020;36:603-10. [Crossref] [PubMed]
- Li L, Wang P, Yan J, Wang Y, Li S, Jiang J, Sun Z, Tang B, Chang TH, Wang S, Liu Y. Real-world data medical knowledge graph: construction and applications. Artif Intell Med 2020;103:101817. [Crossref] [PubMed]
- Nováček V, Mohamed SK. Predicting Polypharmacy Side-effects Using Knowledge Graph Embeddings. AMIA Jt Summits Transl Sci Proc 2020;2020:449-58. [PubMed]
- Hasan SMS, Rivera D, Wu XC, Durbin EB, Christian JB, Tourassi G. Knowledge Graph-Enabled Cancer Data Analytics. IEEE J Biomed Health Inform 2020;24:1952-67. [Crossref] [PubMed]
- Cheng B, Zhang J, Liu H, Cai M, Wang Y. Research on Medical Knowledge Graph for Stroke. J Healthc Eng 2021;2021:5531327. [Crossref] [PubMed]
- Huang Z, Hu Q, Liao M, Miao C, Wang C, Liu G. Knowledge Graphs of Kawasaki Disease. Health Inf Sci Syst 2021;9:11. [Crossref] [PubMed]
- Melzer G, Maiwald T, Prokosch HU, Ganslandt T. Leveraging Real-World Data for the Selection of Relevant Eligibility Criteria for the Implementation of Electronic Recruitment Support in Clinical Trials. Appl Clin Inform 2021;12:17-26. [Crossref] [PubMed]
- Abe T, Sato H, Nakamura K. Extracting Safety-II Factors From an Incident Reporting System by Text Analysis. Cureus 2022;14:e21528. [Crossref] [PubMed]
- Wang L, Xie H, Han W, Yang X, Shi L, Dong J, Jiang K, Wu H. Construction of a knowledge graph for diabetes complications from expert-reviewed clinical evidences. Comput Assist Surg (Abingdon) 2020;25:29-35. [Crossref] [PubMed]
- Xiu X, Qian Q, Wu S. Construction of a Digestive System Tumor Knowledge Graph Based on Chinese Electronic Medical Records: Development and Usability Study. JMIR Med Inform 2020;8:e18287. [Crossref] [PubMed]
- Han S, Zhang RF, Shi L, Richie R, Liu H, Tseng A, Quan W, Ryan N, Brent D, Tsui FR. Classifying social determinants of health from unstructured electronic health records using deep learning-based natural language processing. J Biomed Inform 2022;127:103984. [Crossref] [PubMed]
- Ross MK, Wei W, Ohno-Machado L. "Big data" and the electronic health record. Yearb Med Inform 2014;9:97-104. [Crossref] [PubMed]
- van der Woude D, van der Helm-van Mil AHM. Update on the epidemiology, risk factors, and disease outcomes of rheumatoid arthritis. Best Pract Res Clin Rheumatol 2018;32:174-87. [Crossref] [PubMed]
- McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med 2011;365:2205-19. [Crossref] [PubMed]
- Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013. arXiv:1301.3781.
- Pennington J, Socher R, Manning C. GloVe: Global Vectors for Word Representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha: ACL, 2014:1532-43.
- Sarzynska-Wawer J, Wawer A, Pawlak A, Szymanowska J, Stefaniak I, Jarkiewicz M, Okruszek L. Detecting formal thought disorder by deep contextualized word representations. Psychiatry Res 2021;304:114135. [Crossref] [PubMed]
- Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018. arXiv:1810.04805.
- Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw 1994;5:157-66. [Crossref] [PubMed]
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. [Crossref] [PubMed]
- Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw 2005;18:602-10. [Crossref] [PubMed]
- Zhang N, Jia Q, Yin K, Dong L, Gao F, Hua N. Conceptualized Representation Learning for Chinese Biomedical Text Mining. In: WSSM ’20. Houston, TX, USA: WSSM, 2020.
- Zhu G, Iglesias CA. Exploiting semantic similarity for named entity disambiguation in knowledge graphs. Expert Syst Appl 2018;101:8-24. [Crossref]
- Liu Y, Yin B. Sensors (Basel) 2020;20:4995. [Crossref] [PubMed]
- Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020;36:1234-40. [Crossref] [PubMed]
- Guan H, Devarakonda M. Leveraging Contextual Information in Extracting Long Distance Relations from Clinical Notes. AMIA Annu Symp Proc 2020;2019:1051-60. [PubMed]
- Cheng M, Li L, Ren Y, Lou Y, Gao J. A Hybrid Method to Extract Clinical Information from Chinese Electronic Medical Records. IEEE Access 2019;7:70624-33.
- Gao Y, Wang Y, Wang P, Gu L. Medical Named Entity Extraction from Chinese Resident Admit Notes Using Character and Word Attention-Enhanced Neural Network. Int J Environ Res Public Health 2020;17:1614. [Crossref] [PubMed]
- Yin M, Mou C, Xiong K, Ren J. Chinese clinical named entity recognition with radical-level feature and self-attention mechanism. J Biomed Inform 2019;98:103289. [Crossref] [PubMed]
- Li X, Zhang H, Zhou XH. Chinese clinical named entity recognition with variant neural structures based on BERT methods. J Biomed Inform 2020;107:103422. [Crossref] [PubMed]
- Wu H, Ji J, Tian H, Chen Y, Ge W, Zhang H, Yu F, Zou J, Nakamura M, Liao J. Chinese-Named Entity Recognition From Adverse Drug Event Records: Radical Embedding-Combined Dynamic Embedding-Based BERT in a Bidirectional Long Short-term Conditional Random Field (Bi-LSTM-CRF) Model. JMIR Med Inform 2021;9:e26407. [Crossref] [PubMed]
- Liu Z, Di X, Song W, Ren W. A Sentence-Level Joint Relation Classification Model Based on Reinforcement Learning. Comput Intell Neurosci 2021;2021:5557184. [Crossref] [PubMed]
- Wu SC, He YF. Enriching Pre-trained Language Model with Entity Information for Relation Classification. In: 28th ACM International Conference on Information and Knowledge Management (CIKM). Beijing: ACM, 2019:2361-4.
- Zhang X, Zhang Y, Zhang Q, Ren Y, Qiu T, Ma J, Sun Q. Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform 2019;132:103985. [Crossref] [PubMed]
- Chen T, Wu M, Li H. A general approach for improving deep learning-based medical relation extraction using a pre-trained model and fine-tuning. Database (Oxford) 2019;2019:baz116. [Crossref] [PubMed]





