A survey of GPU-based medical image computing techniques
Abstract: Medical imaging currently plays a crucial role throughout the entire clinical applications from medical scientific research to diagnostics and treatment planning. However, medical imaging procedures are often computationally demanding due to the large three-dimensional (3D) medical datasets to process in practical clinical applications. With the rapidly enhancing performances of graphics processors, improved programming support, and excellent price-to-performance ratio, the graphics processing unit (GPU) has emerged as a competitive parallel computing platform for computationally expensive and demanding tasks in a wide range of medical image applications. The major purpose of this survey is to provide a comprehensive reference source for the starters or researchers involved in GPU-based medical image processing. Within this survey, the continuous advancement of GPU computing is reviewed and the existing traditional applications in three areas of medical image processing, namely, segmentation, registration and visualization, are surveyed. The potential advantages and associated challenges of current GPU-based medical imaging are also discussed to inspire future applications in medicine.
Key words: Graphics processing unit (GPU); image segmentation; image registration; image visualization; highperformance computing
Introduction
Parallelism is the future of high-performance computation in medical applications. Over the past several years, GPU has been continuously developed as a higher-performance accelerator platform for data parallel computing, especially in medical image processing and analysis. Due to the rapidly increasing demands on high-performance computing for more sophisticated graphics and scientific applications, commercial graphics hardware has evolved significantly from a pipeline with fixed functionality into a programmable supercomputer (1). Meanwhile, the GPU has also quickly evolved into an efficient framework with excellent price/performance ratio for a wide range of computationally intensive tasks. Furthermore, the GPU is always designed for a particular class of applications with the following characteristics (2): (I) large computational requirements, (II) substantial parallelism, and (III) throughout is more important than latency.
In recent years, the computational speed of GPU has increased rapidly, thus the GPU can provide more significant acceleration for many computationally-heavy tasks compared to conventional CPU-based computing framework. Recently, GPU has emerged as a competitive platform for high-performance computing due to its massive processing capability. Nevertheless, it is not acceptable for general purpose or non-graphics computations. As a consequence, many efforts that implement general purpose computing by mapping general purpose applications onto graphics hardware are known as the general-purpose computing on graphics processing unit (GPGPU), which is introduced for non-graphics algorithms based on existing GPU hardware. The GPGPU computations are performed by using specialized graphics processing rather than vector and matrix operators (3). However, only the professional researchers and developers familiar with graphics APIs can fluently utilize the traditional GPU/GPGPU development platform, which brings the unfamiliar users more inconveniencies than advantages in practical applications (4). Fortunately, the appearance of Compute Unified Device Architecture (CUDA) technology can overcome these disadvantages which exist in current GPU/GPGPU versions to certain degrees. In late 2006, NVIDIA Corporation launched the CUDA development platform, which is a novel programming interface and environment for the general-purpose programming of its own GPU. For the convenience of general-purpose parallel programming on the NVIDIA GPU, the CUDA brings the C-like development environment to programmers and delivers (5). Figure 1 shows a schematic overview of the CUDA programming model.
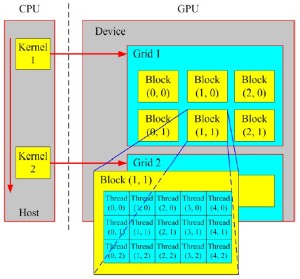
Generally speaking, each CUDA-enabled GPU is made of a collection of streaming multiprocessors and a global memory. As shown in Figure 1, kernels are basic building blocks of CUDA, which will be launched from the host (CPU) and executed on the graphics device (GPU). In the part of graphics device, each thread block is executed on a single stream multiprocessor, which is made up of a set of cores. Meanwhile, the threads are organized into blocks of threads within a grid of block (7). There also exist some other approaches for GPGPU computing, such as the Khronos Group’s OpenCL and Microsoft’s DirectCompute, all of them derive from the similar concepts for parallel computing (7). CUDA has been extensively studied and widely used in various application domains including medical physics, computer vision, computer graphics, and many more. Due to the convenient parallel computing for handling large data sets, OpenCL and DirectCompute will also be more mature as CUDA over the coming years. Meanwhile, compared to the previous GPU (GPGPU), CUDA has the following advantages (4,8,9):
General programming environment. CUDA is simple to understand as it brings the C-like development environment to programmers and delivers not acquainted with GPU. This C-like language makes programming better compatibility and portability.
More powerful parallel computing capacity. CUDA is a high-performance parallel computing platform, and is also well suited to make full use of the parallel capabilities of GPU acceleration.
Better development platform. It is a versatile development platform with well-documented references for CUDA, such as scientific libraries, open source compiler, debugger and profiler.
Shorter latency time. The data transfer rate and latency between the host (CPU) and the graphics device (GPU) have been improved obviously.
Although the CUDA has developed dramatically over the past few years, the manual development of CUDA codes remains very laborious and time-consuming for general-purpose multi-core systems. Consequently, how to automatically transform one favorite programming language into efficient parallel CUDA program is of considerable importance and interest, especially for the common CUDA users. A C-to-CUDA transformation system was proposed to generate two-level parallel CUDA code, which was optimized for efficient data access in (10). Figure 2 shows the sequence of steps in the implemented system.
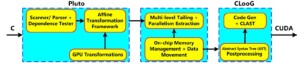
In Figure 2, CLooG is a powerful state-of-the-art code generator to generate transformed code without manual development of CUDA codes (10). This proposed source-to-source transformation framework can generate correspondingly efficient CUDA codes with handling arbitrary input C programming codes. Based on the competitive capacity of high-performance computing, GPU computing has been developed as an efficient research platform for a wide variety of applications in medical image processing and analysis, such as medical image reconstruction (11,12), real-time denosing (13,14), registration (15,16), deconvolution (17), segmentation (18,19) and visualization (20,21), due to the parallel computing power of the GPU designed to exploit the multi-thread capabilities of multi-core structures. A recent and detailed survey of GPU computing in medical physics can be found in (22), where Pratx and Xing surveyed existing applications in three areas of medical physics, namely image reconstruction, dose calculation and treatment plan optimization, as well as image processing. Medical image registration, the transformation of two or more images into a common frame of reference, has been also reviewed for physicians and researchers who are interested in using the multi-CPU and GPU applications (1,23). In addition to retaining the advantages of traditional GPU, for instance tremendous memory bandwidth and power efficiency in high-performance computing, newly developed GPU computing platform brings programmability and increased generality (24). In order to provide reference source for researchers who plan to develop modified GPGPU techniques for their research fields of interest, Owens et al. brought a detailed survey of general-purpose computation on graphics hardware and various general-purpose computing applications (25). Furthermore, many performance studies comparing GPGPU and traditional CPU were published. A paper by Bui and Brockman (26) made a performance analysis of accelerated image registration using GPGPU, and emphasized the demand to manage memory resources carefully to fully utilize the GPU and obtained maximum speedup. A parallel design for medical image reconstruction (MIR) was presented based on GPGPU implementation. Meanwhile, two approaches, Sobel edge detection and marching cube, in MIR framework were simultaneously implemented in CUDA on NVIDIA GPGPU (27). Moreover, a survey paper on recent trends in software and hardware for GPGPU computing was made by Neelima and Raghavendra (28). In this survey paper, we will review the latest GPU-based applications in three areas of medical imaging, namely segmentation, registration, and visualization, in the following sections.
Medical image segmentation
Image segmentation definition and approaches
Nowadays, medical image segmentation plays an important role in medical image analysis, for instance, computer-aided diagnosis (CAD), surgical planning and navigation (19). Its aim is to divide the target image into connected regions, which are meaningful for mathematical analysis and quantification of medical images. By classical definition, image segmentation represents dividing a target image into its non-overlapping structural regions according to some criterions such as color information, grayscale intensities or texture features (29). For the sake of image segmentation definition, the domain of an image I: Ω⊂R2 R is divided into a set of N class labels by a mapping S: Ω
R is divided into a set of N class labels by a mapping S: Ω  {1,2,...,N}, then the segmentation problem can be expressed as determining the sub-labels Sk ⊂ Ω whose union is the entire image domain Ω, while the sub-labels Sk must satisfy
{1,2,...,N}, then the segmentation problem can be expressed as determining the sub-labels Sk ⊂ Ω whose union is the entire image domain Ω, while the sub-labels Sk must satisfy
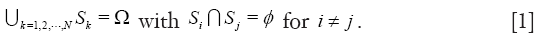
In the process of medical imaging, a segmentation approach should find those labels that correspond to distinct anatomical structures or regions of interest in the image (30). As a consequence, numerous segmentation algorithms have been extensively investigated for many years in a host of publications (31), but they remain open difficult tasks, due to the tremendous variability of object shapes and the variations in image quality. These segmentation methods can be broadly categorized into four groups: (I) pixel-based, (II) boundary-based, (III) region-based, and (IV) hybrid-based methods (32).
Accurate segmentation of 2D, 3D, and even 4D medical images for isolation of anatomical structures for further medical analysis is necessary and important in almost any computer-aided diagnosis systems. In August 2006, Noble and Boukerroui presented a survey paper on ultrasound image segmentation by clinical applications, where the corresponding segmentation techniques were further classified in terms of use of prior information (33). Typically, achieving satisfactory segmentation performance always relies on quantitatively visual characterizations and image features, which help extract tissues/structures of interest from the image background. Due to the presence of sensor noise or low signal-to-noise ratio, segmentation techniques often fail to achieve the target objects. Consequently, in order to enhance segmentation performance, image features have been extensively and successfully utilized in medical image segmentation (34). Meanwhile, a statistical shape model-active shape model (ASM), where shape variations were described by using a point distribution model (PDM), was proposed to efficiently improve segmentation quality results (35). Based on wavelet transform, Davatzikos et al. further presented a hierarchical ASM approach in (36). Local and global threshold methods, based on target image intensity distribution, have also been widely used in image segmentation (37). Moreover, Osher and Sethian firstly presented the level set method in 1988, which was a powerful and flexible numerical technique for image segmentation (38). Currently, this level set method and its extensions have been widely used in denoising, registration, inpainting, and many more image processing applications.
The above-mentioned automatic segmentation techniques have generally struggled to achieve more accurate and robust segmentation results needed for clinical and practical applications (39). As a consequence, many other automated interactive mechanisms have recently become optimal selections in most real-life medical applications, for instance, interactive contour delineation (40) and seeded region growing (41,42). Ideally, the interaction process can occur in real-time to allow the users to receive immediate feedback on their actions and improve the accuracy of tissues/structures segmentation. Due to inequality with any semantic content, in-homogeneity, low contrast and additive noise, segmentation of medical images is still a challenging problem.
Related work on GPU-based segmentation
Although the aforementioned segmentation researches have become more and more active in recent decades, they are not suitable for real-time clinic applications because of their expensive computational time. Fortunately, the segmentation approaches could be implemented in a real-time, operational environment by the porting and adaptation of these approaches to the GPU architectures (19). Table 1 lists some GPU-based image segmentation approaches and their respective characteristics.
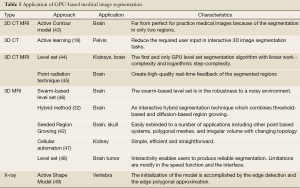
Full table
The first implementation of GPU-based medical image segmentation technique was achieved by formulating level set segmentation as a sequence of graphics operators of image blending (50). The introduced programmable shaders brought greater flexibility and enabled the segmentation of 3D images using curvature regularization to favor smooth isosurfaces (51). Then, a novel level set approach, one part of the integrated, interactive workflow for visualizing and segmenting neural processes, was implemented in CUDA (52). In general, segmentation methods can also be classified into broad two categories, namely, low-level and high-level approaches. Low-level approaches, which require no statistical information about the types of objects in the image, directly manipulate the pixel/voxel information to form connected objects/regions of interest (19). GPU-based implementations of watershed (53) and region growing methods (54) are typically low-level approaches. A more complex and robust statistical segmentation was implemented based on GPU platform using adaptive region growing process (55). Moreover, the Markov random fields (MRF) and graph cuts are two other types of low-level approaches found in literatures (56,57).
In high-level segmentation framework, geodesic active contours (58), a modification of traditional active contours (snakes) (59), has been efficiently implemented on GPU to segment interested structures according to the differences between foreground and background regions in 2D images (60). In the GPU computing framework, other methods for implementing active contours with gradient vector flow (GVF) external force have been introduced (61,62), but they were restricted to only 2D image segmentation. Then a parallel segmentation framework based on NVIDIA CUDA architecture was presented for segmentation of volumetric images using discrete deformable models (19). Different implementations of several medical image segmentation approaches via CUDA (63) and CUDA-enabled GPUs (64) have also been proposed, achieving high-performance in terms of speedup over the sequential version of the considered algorithms (65). Furthermore, medical images continue to increase in size and volume. For high-dimensional image segmentation in real-time clinical applications, it is crucial that image segmentation approaches could be implemented in a real-time for very large data sets on GPU/CUDA. Thus, the GPU/CUDA-based high-dimensional segmentation with low computational cost is still a challenge for future medical imaging.
Interactive seeded region growing
The seeded region growing approach was introduced to achieve satisfactory closed regions in the final segmentation results (41). However, computational costs for traditional CPU-based segmentation implementation are too high when the target image is rather large, which is always the negative case in biomedical applications. The GPU/CUDA can be utilized to accelerate and satisfy the requirements of clinical applications because of the parallelization potential of the seeded region growing approach.
In the interactive seeded region growing segmentation process, seed point selection is a crucial procedure. In the practical application, a fast volume segmentation framework, using programmable graphics hardware, was proposed based on the seeded region growing approach (54). In this framework, the users were allowed to interactively paint growing seeds by drawing on the sectional views of the volume. More recently, Schenke et al. have implemented a GPGPU-based seeded region growing method with fragment shaders and VTK (32). In seeded region growing segmentation framework, it is essential to draw as many seeded points as possible to make full use of the parallel performances of GPU/CUDA. In 2006, a sketch-based interface for the seeded region growing volume segmentation was proposed to prevent unexpected segmentation, where the user could freely sketched regions of interest (ROI) over the 3D volume (42). Meanwhile, a region growing approach with CUDA was presented for fast 3D organ segmentation, at a speed of about 10-20 times faster than the traditional segmentation methods on CPU (66). Kauffmann and Piche presented a seeded cellular automaton (CA) to perform an automated multi-label segmentation of organs for N-dimensional (ND) medical images, which was implemented on GPU with minimal user interaction for robust initialization (47). The interactive seeded region growing can be efficient and effective for 2D/3D medical image segmentation, but not on large amount of images that require online clinical analysis within a limited time.
Variational level set segmentation
The variational level set approach (LSA) has been widely used in medical image segmentation. Briefly, the main idea is to embed the initial position of the moving interface, at any time t, as the zero level-set of a higher-dimensional function (x,t), where the surface consists of all points C(x,t)={(x,t)∣I(x,t)=0} with I: Rn R. In the level-set framework, one can execute a wide variety of deformations by introducing an appropriate motion function
R. In the level-set framework, one can execute a wide variety of deformations by introducing an appropriate motion function 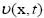 of the surface. For segmentation, the velocity often consist of a combination of two terms
of the surface. For segmentation, the velocity often consist of a combination of two terms
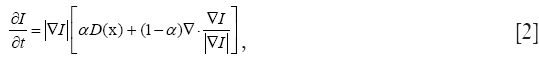
where D is a data term that forces the model toward desirable features in the input data, the term 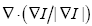 is the mean curvature of the surface, which forces the surface to have less area (and remain smooth), and α ∈[0,1] is a free parameter that controls the degree of smoothness in the solution. For 2D medical image segmentation n = 2 in I: Rn
is the mean curvature of the surface, which forces the surface to have less area (and remain smooth), and α ∈[0,1] is a free parameter that controls the degree of smoothness in the solution. For 2D medical image segmentation n = 2 in I: Rn R, this method represents an evolving segmentation boundary as the zero level set of a function on a two dimensional grid (67).
R, this method represents an evolving segmentation boundary as the zero level set of a function on a two dimensional grid (67).
Extensive researches on LSA have been completed to improve segmentation performance. The comprehensive reviews of this LSA and their associated numerical techniques were documented in the medical imaging literatures (29,30,33). The level set approaches handle well interfaces with sharp corners, cusps, topological changes, and 3D complications (68). Numerous extensions of LSA are good candidates for implementation on GPU because of the high degree of parallelism and high-speed computational requirements. In an early contribution, Lefohn and Whitaker demonstrated a full 3D level set solver using a graphic processor for MRI brain segmentation (69). This approach achieved the same performance attributed to a more highly optimized CPU-based implementation. Then Lefohn et al. devised an interactive LSA to segment target objects in real-time clinical applications based on GPU computing, with 10× to 15× speedup over the non-accelerated version (48). However, none of these methods took advantage of the sparse properties of level set partial differential equations (PDEs), and therefore the computational performance is modified marginally compared with existing highly-optimized CPU implementations (70). In (71), an efficient GPU-based segmentation approach obtained high segmentation performance through packing the level set isosurface data into a dynamic, sparse texture format. By relying on graphics hardware, this level set segmentation approach could operate at interactive rates in real-time clinical applications.
Pervious level set segmentation approaches always suffer from the computationally expensive problem even when running on the GPU. In a work-efficient parallel algorithm framework, Roberts et al. presented a novel GPU-based level set segmentation algorithm which was both work-efficient and step-efficient. This algorithm reduced the number of processed level set field elements by 16× and converged 14× faster than previous GPU-based algorithms without segmentation accuracy reduction (44). This segmentation approach was the first GPU-based level set segmentation algorithm with linear work-complexity and logarithmic step-complexity. Meanwhile, another novel CUDA accelerated level set segmentation approach was presented with significantly improved performance over most previous approaches, for instance, 16× reduction in the size of the computational domain as well as 9× speedup compared to previous GPU approaches and no reduction in segmentation accuracy (72). Extending a 2D level set segmentation algorithm to 3D is still a relatively straightforward but difficult task. For instance, computing for the level set update required many more derivatives. Moreover, the storage and computational complexity of 3D medical image segmentation must also be appreciated (73).
Medical image registration
Image registration definition and approaches
As one of the most important procedures in medical image processing, image registration aims to obtain integrated analysis of information gathered from multiple sources. In general, the medical image registration should establish correspondence measure between a reference image, Ir, and a target image, It, using a parameter transformation, Tt(•), of image geometry in line with a similarity function, ρ(•), to specify the registration performance. When two images have different dimensions, projection operators, Pr and Pt, may be incorporated to project a higher-dimensional image domain into a lower-dimensional image domain. Then, the image registration problem can be expressed via maximizing the following similarity measure function:
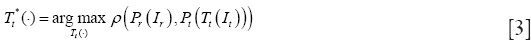
The optimization in Eq. [3] is mostly numerical to determine the optimal transformation Tt(•). Starting from an initial guess, Tt(•) converges to the optimum in a series of iterative steps depending on the corresponding objective function, image transformations and optimization technique (31,74). As shown in (75,76), the image registration procedure generally consists of the following four steps:
Feature detection. In this step, salient and distinctive features/structures are manually or preferably automatically extracted. The region-like, line and point features are the considered important objects, which are appropriate for registration task. Meanwhile, this feature detection process should not be sensitive to the additive noise or missing data in the degraded images.
Feature matching. In this step, the feature correspondence between two sets of features in the target and reference images is established. The corresponding mapping algorithms should also be robust and efficient.
Transform model estimation. After the feature correspondence and mapping function are established and constructed, respectively. It should transform the target image according to the reference image using the constructed mapping function to overlay the two images.
Image resampling and transformation. In the last step, the transformation process can be realized in a forward or backward manner. Meanwhile, appropriate interpolation techniques should be proposed to calculate the intensity values at the non-integer coordinates of the target image.
In practical registration applications, the implementation of each registration step has its typical problems. Consequently, users have to decide what kind of features is appropriate for the given medical images to improve registration accuracy progressively. Moreover, current techniques used for image registration can be divided into two main categories: namely, feature-based and pixel-based methods (77). These categories are also known as geometric registration and iconic registration in medical imaging, respectively.
If both the reference and target images contain obviously distinctive and easily cognizable objects/regions, the feature-based registration methods should be selected. However, if the simple and salient features are difficult to extract from both images, the pixel-based methods may be more effective and efficient. In feature-based methods, the reference and target images are extracted using a finite set of features. The basic principle of feature-based methods is to minimize the total error between the warped points of the reference image and their corresponding points in the target image using a robust M-estimator method (78). Pilet et al. proposed a more advanced method wherein the registration can be subjected to very large non-affine deformations (79). The main advantages of feature-based registration algorithm are that it can handle large deformations and it is efficient in terms of computational complexity. Pixel-based registration is the other useful approach for image registration, in which warp parameters are estimated by minimizing the pixel-wise dissimilarities between the reference image and the warped target image. The main advantage of this approach is that the data used for the parameter estimation is denser than that in the feature-based approach. As in the feature-based approach, it is not possible to estimate the hyperparameters with the direct approach (77).
Similarity measure, geometric transformation, and optimization procedure
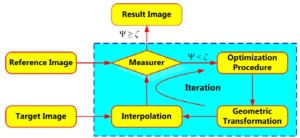
The goal of image registration technique is to maximize similarities and minimize matching errors between two images: the reference image (also called the static image) and the target image (moving image). Figure 3 shows the process of medical image registration which can be represented by four components: similarity measure (measurer), geometric transformation, optimization procedure and interpolation. As shown in Figure 3, the parameters Ψ and ζ represent the matching degree between two temporal images and the preselected threshold, respectively.
GPU-based similarity measure
Similarity measure is a method of measuring the similarity of images, which is required for automatic image registration. This measure can evaluates how closely the images are aligned. Ideally, the similarity measure ρ attains its maximum, where the images are perfectly aligned. The similarity measures can be divided into two main classes: (I) feature-based, and (II) intensity-based measures. Pixel intensity-based similarity measures utilize a large portion of the image data and therefore usually achieve more accurate registration results than those in feature-based approaches (80). Thus we mainly survey these intensity-based similarity measures in this paper:
(I) Sum of squared differences (SSD)
The SSD has been implemented on GPU, such as in (81), which is the simplest voxel similarity measure. Mathematically, SSD is defined as follows:

where Ir,t is the overlap of the two images Ir and It, and ir and it are the intensities of the two images Ir and It, respectively.
(II) Normalized cross correlation (NCC)
Grabner et al. investigated NCC as a similarity measure, which verified the existence of an affine relationship between the intensities in the images (82). NCC is given by
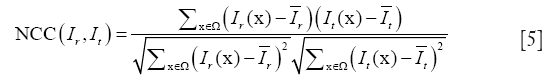
(III) Correlation ratio (CR)
The CR generalizes the correlation coefficient, which is a symmetrical measure of linear dependent between two images (83):
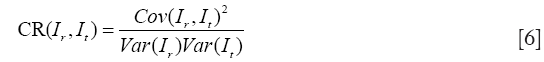
(IV) Mutual information (MI)
MI has been utilized with success for a large variety of combinations including MR, CT, PET, and SPECT. Given the reference image Ir and target image It, we define their joint probability density function, P(i,j), by simple normalization of their 2D-histogram. Let Pr(i) and Pr(j) denote the corresponding marginal probability density functions (83,84). MI between Ir and It is defined as follows:
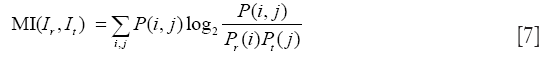
MI might be the most currently popular multimodal measure, and the corresponding MI-based image registration technique has received much attention in the literature (85). In (86), a proposed approximate histogram computation method speeded up MI computation and registration on GPU using NVIDIA CUDA, but at the expense of reduced accuracy. Then the same first author further presented an efficient method for parallel computation of MI similarity measure, which could achieve high speed performance (less than 1 s) for 3D medical image registration using a commodity GPU (87). Other researchers have also realized the potential capacity of GPU computing for computing similarity measure, and presented much efficient similarity measures. Ruiz et al. proposed a landmark-based similarity measure on the GPU for registering microscopic images non-rigidly (88). In (89), a new similarity measure was introduced by combining eight similarity measures between digitally reconstructed radiographs (DRRs) and X-ray image to compute the similarity measure more precisely and robustly. To generate similarity measure in a low-dimensional space, Khamene et al. presented another novel approach utilizing projection and performed a comparative study on various similarity measures on GPU (90). According to an empirical study (81), gradient correlation (GC), another example of a specific similarity measure implemented on the GPU, could contribute to improve the robustness of registration performance.
GPU-based geometric transformation
Due to the six degrees of freedom in geometric transformation, image registration methods can be divided into three classes: rigid, affine, and non-rigid (parametric or non-parametric) approaches (80). Meanwhile, the rigid and affine geometric transformations only depend on a few global parameters (such as image size, position and orientation), because they do not need nonlinear displacements of pixels (1). These two transformation methods are suitable for rigid tissues, such as pelvis, femur, and brain motion which is constrained by skull. Comparatively, non-rigid registration is utilized when the body parts undergo non-rigidly motions or deform during medical image acquisitions, which is suitable for soft tissues, such as breast and liver (91). To date, only a small number of non-rigid applications have been published, compared to the rigid 2D/3D registration publications.
Currently, GPU-based rigid geometric transformation approaches have been reviewed in (1,23). To the best of our knowledge, Strzodka et al. firstly proposed a fast 2D deformable image registration on DX9 graphics hardware in 2003 (92). In the regularized gradient flow (RGF) approach of (92), gradients are regularized by Jacobi iterations during a multigrid-cycle. Furthermore, an extension of the RGF registration algorithm effectively implemented 2D and 3D deformable image registration via GPU acceleration in (93). A popular method in image registration is the Demon’s algorithm, which is an optical flow variant (94). In (95), Sharp et al. implemented the Demon’s algorithm using the Brook programming environment. Moreover, the accelerated Demon’s algorithm has been further implemented on GPU utilizing CUDA, and high quality and excellent performance were achieved (96). Meanwhile, Rezk-Salama et al. proposed an appropriate mathematical model and illustrated how the deformation of volumes can be accelerated by data-parallel processing using graphics hardware (97). Meanwhile, the geometric transformation approach spends the majority of its total computing time performing interpolations. These interpolation methods, such as linear, quadratic, cubic, cubic B-spline, and Gaussian interpolation, have been commonly used for geometric transformation (98).
GPU-based optimization procedure
Image registration aims to find an optimal geometric transformation that pulls one image into the best possible spatial correspondence with other image by optimizing the similarity measure function in Eq. 3. In image registration, the optimization procedures can be broadly categorized as gradient-based or gradient-free, global or local, and serial or parallelizable (23). Gradient-based methods require computation of the partial derivatives of a cost function. Thus, gradient-based methods are more involved than gradient-free methods from an implementation perspective. A large number of algorithms have been implemented for medical image registration (91,99,100), but only the small parts can be directly utilized on the GPU parallel-computing platform to meet the requirements of practical clinical applications. In 2007, Vetter et al. presented a gradient-based registration approach, including a GPU-friendly computation of 2D histograms using vertex texture fetches, as well as an implementation of recursive Gaussian filtering on the GPU (101). Meanwhile, another fast GPU implementation was also proposed in (102), which employed the new hardware features of the DX10-compatible GPU and a series of optimization strategies for fast non-rigid multi-modal volume registration. The computation of the similarity measure and geometric transformation is the computational bottleneck of registration. Thus, researchers should further pay more attention on developing more effective parallelization techniques for these components.
Medical image visualization
The general approaches and challenges in medical image visualization
In general, image visualization has become an increasingly important tool for visual analysis, for instance, in scientific, engineering, and medical disciplines. Especially in medical imaging applications, visualization is essential for medical diagnosis and surgical planning to mine the important information included in 2D/3D imaging datasets. To gain a further understanding and insight into the data behind the generated images, visualization technique is a proper choose, which could explore and view the medical datasets as visual images for convenience (103). For instance, the collected medical data, which originates from numerical simulations of sensor measurements such as CT and MRI, always trends to be very large. Consequently, the medical data visualization is indispensable for understanding and making full use of this medical data (104,105).
According to the different methods, image visualization approaches can be categorized into two distinctive groups (103): surface rendering, and volume rendering. The surface rendering has traditionally been implemented via extracting the corresponding isosurface as polygonal mesh from a 3D scalar field, usually by using some variants of Marching Cubes (MC) algorithm (106,107). The first step in surface rendering is to construct a mathematical model of the object surface. The surfacing rendering techniques are utilized to reconstruct the continuous surface, and then to compute the texture coordinates and norm vectors on the final surface. Furthermore, the pixel values in rendered image are directly proportional to the amount of light that is reflected towards the observer from all visible surface regions (108). However, the surface rendering has several drawbacks and problems. First, it is difficult to implement the more complex lighting models that can be useful for better visual perception of shapes (107). Second, the piecewise linear approximation of volume using polygonal mesh can be topologically different from the actual isosurface in the volume data. Such undesirable behavior can be unacceptable in some scientific applications (109,110). Third, although the surface rendering has a satisfactory performance on rendering time, it could only display the surface characteristics of the medical object.
Unlike with surface rendering methodology, volume rendering (also called direct volume rendering) is a technique used to visualize 3D discretely sampled data set by computing 2D projections of a colored semitransparent volume, which can show the whole information of the 3D scalar fields (111). In general, the volume is usually regarded as a distribution of gaseous particles in volume rendering. To further understand the principle of volume rendering, the incident light along a viewing ray that passes the volume and then reaches the observer is modeled in Figure 4.
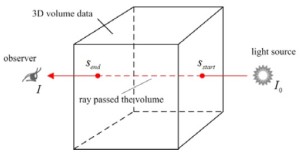
Mathematically, the differential change of the light intensity I at a position s along a ray is defined by the following differential equation:

where τ(s) is the extinction coefficient, which attenuates the light intensity I(s) at position s. q(s) is the source term that gives the amount of light emitted at position s. The finally intensity I starting with the initial intensity I0 has to be approximated numerically in differential Eq. [8].
Volume rendering approaches can be classified into two categories: space-domain and transform-domain methods. The space-domain methods could be further divided into object-order and image-order approaches in Table 2 (103). In recent years, researches have proposed some hybrid approaches (120,121), but their fundaments are still based on the two main categories. Because the projections of image pixel are discrete, the object-order volume rendering approaches are simple and fast, but often suffer from unwanted rendering artifacts (122). Fortunately, the proposed splatting (or footprint) method can overcome this disadvantage (113,114). Due to the view-dependent resampling in the splatting approach, shear-warp volume rendering method, considered to be the fastest type of volume rendering, was proposed to overcome the drawbacks of resampling for arbitrary perspective views (115). Nevertheless, this method has not been preferable in modern medical and clinical applications, due to its unsatisfactory rendering quality and required complex preprocessing (123). Comparatively, the ray casting algorithm, one image-order volume rendering method, achieves higher rendering performance than shear-warp volume rendering method (124). However, the image-order volume rendering approaches are contrary to the way GPU generates images. Other object-order 3D texture-based volume rendering algorithms have also been widely applied to the shading volume rendering, because they could be accelerated greatly by data-parallel processing via GPU acceleration (116).
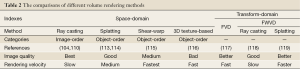
Full table
By using Fourier slice theorem, volume rendering can also be implemented in the frequency domain. After the volume data is transformed into frequency domain, the inverse Fast Fourier or Fast Hartley Transform are used to generate final image by transforming the extracted slice back into the spatial domain (117,118). To the best of our knowledge, the transform-domain rendering approaches include: Fourier volume rendering (FVR) and Fourier-wavelet volume rendering (FWVR). The FWVR makes full use of the advantages of ray casting rendering and wavelet splatting in wavelet space (125,126). By using the wavelets as reconstruction threshold filters, the wavelet splatting can modify the standard splatting approach in practical applications (119). Furthermore, the FWVR and standard FVR have the same time complexity, i.e. ![]() , whereas wavelet splatting has complexity
, whereas wavelet splatting has complexity ![]() . Nevertheless, the FWVR often suffers from a disadvantage that the slice needs resampling in Fourier space at full resolution (127).
. Nevertheless, the FWVR often suffers from a disadvantage that the slice needs resampling in Fourier space at full resolution (127).
The advantages of GPU-based medical image visualization approaches
Due to the rapidly increasing computational complexities of medical imaging, the processing time is now limiting the development of advanced technologies in medical and clinical applications. Originally designed for data-parallel accelerating in the process of computer graphics, the GPU has positioned itself as a versatile platform for running parallel computation to deal with the medical data sets (22). Especially nowadays in medical diagnostics, it highly depends on volumetric imaging methods that must be visualized in real-time. All of the aforementioned volume rendering approaches can be partially or entirely implemented in GPU for acceleration. Surface rendering (SR) is a common method of displaying 3D images, which can be divided into two categories: direct surface rendering (DSR) and indirect surface rendering (ISR) (128,129). DSR can be regarded as a special case of direct volume rendering (DVR), while ISR is considered as object surface modeling. In recent medical applications, by using the advantages of GPU acceleration and geometry shaders, the ISR rendering technique has been accelerated greatly (130). For DSR, the surface rendering can be achieved without the intermediate geometric primitive representations.
Surface rendering is often implemented for contrast-enhanced CT data to display skeleton and vasculature structures. However, it is sometimes difficult for researchers and physicians to justify the accuracy and reliability of the images generated with shaded-surface rendering (131). On the other hand, DVR is a major technique for 3D medical data display, implementing entire images for the data sets without explicitly extracting surfaces corresponding to the interest features. The ray casting is probably the most explored rendering approach of DVR, and is well suited for parallel implementation based on GPU since the rays are processed independently. The original ray casting approach can be divided into three parts: initialization and ray setup, ray traversal, and writing the rendering results (132). Simultaneously, the GPU-based ray casting approaches were first proposed by Krüger and Westermann (133) and by Röttger et al. (134) in 2003. Through distributing some of data management work to the CPU, Kim implemented a bricked ray casting volume rendering on CUDA, which was focused on streaming volume data not fitting in GPU memory (135). To accelerate volume rendering, a slab-based ray casting technique was presented based on experience gained from comparing fragment shader implementation of original ray casting to implementations directly translated to CUDA kernels (132).
Recently, the technique of real-time 4D cardiac data acquisition has become a reality via multi-dimensional medical imaging modalities in actual clinical environment. To efficiently handle the intractable problem of massive medical volumetric data in 4D volume rendering, an accelerated 4D medical image visualization and manipulation framework was constructed for the display of cardiac data sets in (136), which was effectively implemented using the parallel computing power of modern GPU. The novel schematic descriptions of the programmable pipeline for graphics hardware and the GPU-based ray casting volume rendering approach were also described in detail (136). This GPU-based ray casting volume rendering approach was directly implemented on the programmable vertex and fragment processors in the graphics hardware. In order to further enhance the 3D rendering performance, an accelerated ray casting algorithm was effectively implemented based on a novel space leaping acceleration technique (137). However, rendering a single homogenous volume is not sufficient for advanced clinical applications in modern medicine. Simultaneous rendering of multiple volumes is both necessary and important when multiple datasets have been acquired to examine patients. To improve the volume rendering performance for multiple data sets, a new GPU-based rendering system for ray casting of multiple volume data sets was presented in (138). This presented rendering technique was realized based on the fact that rasterization of the proxy geometry was implemented by CUDA rather than traditional graphics pipeline. Meanwhile, multi-frame rate volume rendering is also especially challenging because of the need for transparent volumetric objects (139). Traditional multi-frame rate systems reconstructed the whole image scene as a single surface, where the motion parallax perception was destroyed. To improve the rendering quality, Hauswiesner et al. presented a multi-frame rate volume rendering system with superior capacities of high quality reconstruction and fast transparent volume rendering (140). Other volume rendering techniques, such as pixel ray images (141) and light field representations (142), showed richer descriptions of the target volume, but demanded considerable analytic preprocessing and were less suitable for frequent bus transfer because of their large size.
Discussion and conclusion
With the rapidly increasing development of high-performance computing and recent programmability for graphics hardware, the graphics hardware has evolved into a compelling platform for a wide range of computationally demanding tasks, such as medical image processing (143), dose calculation and treatment plan optimization (144,145), computer vision (146), and many more. Nowadays, the GPU is one of the standard tools in high-performance computing, and is being widely adopted throughout industry and academia (22). Many researchers and developers have become interested in utilizing the power of GPU for general-purpose computing. The crucial advantages of GPU-based medical imaging benefit from high throughput computing, high memory bandwidth, supporting 32-bit floating-point arithmetic, excellent price-to-performance ratio and specialized hardware for interpolation (1).
We have presented a comprehensive survey of GPU-based medical image computing techniques in this article. A broad categorization of all such medical imaging is proposed on the basis of three major medical image processing components: segmentation, registration and visualization. Traditional CPU-based implementations are too slow to be suitable for practical clinical applications. In order to overcome this limitation, current medical image computing is always implemented in modern GPU. Especially in the clinical practice of medical imaging, GPU plays an important role in medical diagnosis and analysis. Despite much work has been done in the domain of GPU-based medical imaging, there still remain many long-standing unsolved problems. We would like to conclude this survey by pointing out a number of possible issues and directions for future research. To summarize, these directions are:
(I) Unifying framework for partial volume tissue segmentation.
(II) Medical image registration with partial or missing data.
(III) Comprehensive volume rendering framework.
Image segmentation plays an important role in practical clinical applications and acts as a preliminary stage in various diagnosis techniques, the corresponding high segmentation performance of brain MRI images is essential and crucial to provide correct diagnosis reference information for researchers and physicians (147). For brain images, usually three tissue categories are considered: gray matter (GM), white matter (WM), and cerebro-spinal fluid (CSF). Each of the partial volume (PV) voxels in an image is modeled as belonging to one of these three categories (148). In nature, the PV voxels in brain contain a mixture of two or more tissue types, thus it is difficult to accurately segment these three tissue structures simultaneously using only one special approach. Most existing segmentation approaches [interactive seeded region growing, (geodesic) active contours, and level set, et al.] are unlikely to correctly extract the objects/regions of interest, which only contain the homogenous voxels. Statistical pattern recognition might be the efficient and robust technique as a unifying framework for PV tissue segmentation, according to the statistical features of voxel intensities in brain MRI images. In addition, Markov random field (MRF) (149), Expectation-Maximization (EM) algorithm (150), and Hidden Markov Chains (HMC) (151) have been used to improve image segmentation performance. To speed up the process of statistical segmentation using GPU-acceleration, we should further rethink these approaches and adapt them for a massively parallel processing environment in real-time practical clinical applications.
In medical and clinical applications, medical images from similar or different modalities often need to be aligned with the reference image as a preprocessing scheme for many further procedures, for instance, atlas-based segmentation, automatic tissue identification and visualization tasks. Over the past few years, medical image registration has been extensively studied in the medical imaging domain. However, the former class of registration algorithms is mainly presented based on the assumption that each region in the reference image has a correspondence match in the target image. In practical applications for medical registration, there may not be a one-to-one correspondence between these two images, which are degenerated by partial or missing data sets (152). In this case, without high-quality segmentation performance or accurate localization of these missing regions, most traditional registration approaches are unlikely to correctly register these images (153). In a survey paper (154), Liu and Ribeiro presented a promising direction by combining the variational method with statistical model (155), which could improve the robustness of variational method and reduce the training requirement of statistical model. Nevertheless, this comprehensive framework will still suffer from the disadvantage of high computational cost in real-time applications. A more promising direction might be to develop a GPU-based general-purpose registration framework by combining the aforementioned variational method and statistical method. Moreover, this comprehensive framework can be further modified for image segmentation task. As a consequence, this comprehensive framework might has acceleration capacity of simultaneously segmenting and registering images in the presence of partial or missing data sets.
Finally, a number of techniques have been presented for medical volume rendering. They can be broadly classified as space-domain (splatting, ray casting, shear-warp and 3D texture-based) and transform-domain techniques. A detailed comparison between these popular volume rendering approaches can be found in (111,156), the aim is to provide both researchers and developer with guidelines on which approach is most suited in which scenario. For instance, the splatting and ray casting present the highest quality images on the expense of rendering speed, whereas shear-warp and 3D texture-based methods are able to maximize an interactive frame rate at the cost of image quality (111). Meanwhile, the frequency domain methods perform fast rendering, but are restricted to parallel projections and X-ray type rendering (157). In order to improve rendering quality and accelerate rendering speed, a promising comprehensive framework could be constructed via combining the space-domain and transform-domain methods, or combining the different space-domain methods. Thus, the comprehensive method might have excellent capabilities of creating high-quality rendering and reducing computational time, which could be utilized on different types of rendering, and be implemented efficiently in parallel on GPU for real-time medical applications.
Acknowledgements
The work described in this paper was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (Project No.: CUHK 475711), a direct grant from The Chinese University of Hong Kong (Project No. 2041698), a grant from the National Natural Science Foundation of China (Project No. 81101111), and a grant from the Science, Industry, Trade and Information Commission of Shenzhen Municipality (Project No. JC201005250030A).
Disclosure: The authors declare no potential conflict of interest.
References
- Fluck O, Vetter C, Wein W, et al. A survey of medical image registration on graphics hardware. Comput Methods Programs Biomed 2011;104:e45-e57.
- Owens JD, Houston M, Luebke D, et al. GPU computing. Proceedings of the IEEE 2008;96:879-99.
- Baskaran MM, Bondhugula U, Krishnamoorthy S, et al. A compiler framework for optimization of affine loop nests for GPGPUs. Proceedings of ICS 2008:225-34.
- Yang ZY, Zhu YT, Pu Y. Parallel image processing based on CUDA. Proceedings of CSSE 2008:198-201.
- Collins R, Li CH, Carloni LP, et al. An experimental analysis of general purpose computing with commodity data-parallel multicore processors. Technical Report RC25070, IBM TJ Watson Research Center 2010.
- Salvo RD, Pino C. Image and video processing on CUDA: State of the art and future directions. Proceedings of MACMESE 2011:60-6.
- Forsberg D, Eklund A, Andersson M, et al. Phase-based non-rigid 3D image registration: from minutes to seconds using CUDA. Lect Notes Comput Sc 2011;6688:414-32.
- Castaño-Díez D, Moser D, Schoenegger A, et al. Performance evaluation of image processing algorithms on the GPU. J Struct Biol 2008;164:153-60.
- Munawar A, Wahib M, Munetomo M, et al. Hybrid of genetic algorithm and local search to solve max-sat problem using nVidia CUDA framework. Genet Program Evol M 2009;10:391-415.
- Baskaran MM, Ramanujam J, Sadayappan P. Automatic C-to-CUDA code generation for affine program. Lect Notes Comput Sc 2010;6011:244-63.
- Jia X, Dong B, Lou Y, et al. GPU-based iterative cone-beam CT reconstruction using tight frame regulation. Phys Med Biol 2011;56:3787-807.
- Schellmann M, Gorlatch S, Meilander D, et al. Parallel medical image reconstruction: from graphics processing units (GPU) to grids. J Supercomput 2011;57:151-60.
- Fontes F, Barroso G, Coupe P, et al. Real time ultrasound image denoising. J Real-Time Image Pr 2010;6:15-22.
- Eklund A, Andersson M, Knutsson H. True 4D image denoising on the GPU. Int J Biomed Imaging 2011;2011:952819-16.
- Huang TY, Tang YW, Ju SY. Accelerating image registration of MRI by GPU-based parallel computation. Magn Reson Imaging 2011;29:712-6.
- Mousazadeh H, Marami B, Sirouspour S, et al. GPU implementation of a deformable 3D image registration algorithm. Proceedings of EMBS 2011:4897-900.
- Mazanec T, Hermanek A, Kamenicky J. Blind image deconvolution algorithm on NVIDIA CUDA platform. Proceedings of DDECS 2010;125-6.
- Top A, Hamarneh G, Abugharbieh R. Active learning for interactive 3D image segmentation. Med Image Comput Comput Assist Interv 2011;14:603-10.
- Schmid J, Guitian JAI, Gobbetti E, et al. A GPU framework for parallel segmentation of volumetric images using discrete deformable models. Visual Comput 2011;27:85-95.
- Hachaj T, Ogiela MR. Visualization of perfusion abnormalities with GPU-based volume rendering. Comput Graph 2012;36:163-9.
- Kutter O, Shams R, Navab N. Visualization and GPU-accelerated simulation of medical ultrasound from CT images. Comput Methods Programs Biomed 2009;94:250-66.
- Pratx G, Xing L. GPU computing in medical physics: a review. Med Phys 2011;38:2685-97.
- Shams R, Sadeghi P, Kennedy RA, et al. A survey of medical image registration on multicore and the GPU. IEEE Signal Proc Mag 2010;27:50-60.
- Sen A, Aksanli B, Bozkurt M. Speeding up cycle logic simulation using graphics processing units. Int J Parallel Prog 2011;39:639-61.
- Owens JD, Luebke D, Govindaraju N, et al. A survey of general-purpose computation on graphics hardware. Comput Graph Forum 2007;26:80-113.
- Bui P, Brockman J. Performance analysis of accelerated image registration using GPGPU. Proceedings of GPGPU 2009:38-45.
- Archirapatkave V, Sumilo H, See SCW, et al. GPGPU acceleration algorithm for medical image reconstruction. Proceedings of ISPA 2011:41-6.
- Neelima B, Raghavendra PS. Recent trends in software and hardware for GPGPU computing: a comprehensive survey. Proceedings of ICIIS 2010:319-24.
- Hu YC, Grossberg MD, Mageras GS. Survey of recent volumetric medical image segmentation techniques. Biomed Eng 2009:321-46.
- Pham DL, Xu CY, Prince JL. A survey of current method in medical image segmentation. Annu Rev Biomed Eng 2000;2:315-37.
- El-Baz A, Acharya R, Laine AF, et al. eds. Multi modality state-of-the-art medical image segmentation and registration methodologies. New York: Springer, 2011.
- Schenke S, Wuensche B, Denzler J. GPU-based volume segmentation. Proceedings of IVCNZ 2005:171-6.
- Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imaging 2006;25:987-1010.
- Yan P, Xu S, Turkbey B, et al. Discrete deformable model guided by partial active shape model for TRUS image segmentation. IEEE Trans Biomed Eng 2010;57:1158-66.
- Cootes TF, Taylor CJ, Cooper DH, et al. Active shape models-their training and application. Comput Vis Image Und 1995;61:38-59.
- Davatzikos C, Tao X, Shen D. Hierarchical active shape models, using the wavelet transform. IEEE Trans Med Imaging 2003;22:414-23.
- Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging 2004;13:146-68.
- Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: algorithms based on Hamilton-Jacobi formulation. J Comput Phys 1988;79:12-49.
- Top A, Hamarnech G, Abugharbieh R. Active learning for interactive 3D image segmentation. Med Image Comput Comput Assist Interv 2011;6893:603-10.
- Mortensen EN, Barrett WA. Intelligent scissors for image composition. Proceedings of SIGGRAPH 1995:191-8.
- Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell 1994;16:641-7.
- Chen HLJ, Samavati FF, Sousa MC, et al. Sketch-based volumetric seeded region growing. Proceedings of Eurographics 2006:123-9.
- Bojsen-Hansen M. Active contours without edges on the GPU. Project Paper For The Course In Parallel Computing For Medical Imaging And Simulation 2010:1-8.
- Roberts M, Packer J, Sousa MC, et al. A work-efficient GPU algorithm for level set segmentation. Proceedings of HPG 2010:123-32.
- Chen HLJ, Samavati FF, Sousa MC. GPU-based Point Radiation for Interactive Volume sculpting and segmentation. Visual Comput 2008;24:689-98.
- Feltell D, Bai L. Level set brain segmentation with agent clustering for initialization. Proceedings of BIOSIGNALS 2008:1-8.
- Kauffmann C, Piche N. Seeded ND medical image segmentation by cellular automaton on GPU. Int J Comput Assist Radiol Surg 2010;5:251-62.
- Lefohn AE, Cates JE, Whitaker R. Interactive, GPU-based level sets for 3D segmentation. Lect Notes Comput Sc 2003;2878:564-72.
- Mahmoudi SA, Lecron F, Manneback P, et al. GPU-based segmentation of cervical vertebra in X-ray images. Proceedings of Cluster Workshops 2010:1-8.
- Rumpf M, Strzodka R. Level set segmentation in graphics hardware. Proceedings of ICIP 2001;3:1103-6.
- Cates JE, Lefohn AE, Whitaker RT. GIST: an interactive GPU-based level set segmentation tool for 3D medical images. Med Image Anal 2004;8:217-31.
- Jeong WK, Beyer J, Hadwiger M, et al. Scalable and interactive segmentation and visualization of neural processes in EM datasets. IEEE Trans Vis Comput Graph 2009;15:1505-14.
- Stoev S, Straßer W. Extracting regions of interest applying a local watershed transformation. Proceedings of the Conference on Visualization 2000:21-8.
- Sherbondy A, Houston M, Napel S. Fast volume segmentation with simultaneous visualization using programmable graphics hardware. Proceedings of the IEEE Visualization 2003:171-6.
- Narayanaswamy A, Dwarakapuram S, Bjornsson C, et al. Robust adaptive 3-D segmentation of vessel laminae from fluorescence confocal microscope images and parallel GPU implementation. IEEE Trans Med Imaging 2010;29:583-97.
- Walters J, Balu V, Kompalli S, et al. Evaluating the use of GPUs in liver image segmentation and HMMER database searches. Proceedings of IPDPS 2009:1-12.
- Pan W. Improving interactive image segmentation via appearance propagation. Proceedings of Eurographics 2009:93-6.
- Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int J Comput Vision 1997;22:61-79.
- Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vision 1988;1:321-31.
- Santner J, Unger M, Pock T, et al. Interactive texture segmentation using random forests and total variation. Proceedings of BMVC 2009.
- Kienel E, Brunnett G. Tile-based image forces for active contours on GPU. Proceedings of Eurographics 2009:89-92.
- He Z, Kuester F. GPU-based active contour segmentation using gradient vector flow. Lect Notes Comput Sc 2006;4291:191-201.
- Pan L, Gu LX, Xu JR. Implementation of medical image segmentation in CUDA. Proceedings of ITAB 2008:82-5.
- Ruiz A, Kong J, Ujaldon M, et al. Pathological image segmentation for neuroblastoma using the GPU. Proceedings of ISBI 2008:296-9.
- Salvo RD, Pino C. Image and video processing on CUDA: state of the art and future directions. Proceedings of WSEAS 2011:60-6.
- Yang F, Zhai WM, Wang H. Fast organ segmentation in CT images with CUDA. Proceedings of SPIE 2009;7497:749729-749729-5.
- Aubert G, Kornprobst P, eds. Mathematical problems in image processing: partial differential equations and the calculus of variations. New York: Springer, 2009.
- Sethian JA. Curvature and the evolution of fronts. Commum Math Phy 1985;101:487-99.
- Lefohn AE, Whitaker R. A GPU-based, three-dimensional level set solver with curvature flow. Technical Report, University of Utah 2002.
- Lefohn AE, Kniss JM, Hansen CD. Interactive deformation and visualization of level set surfaces using graphics hardware. Proceedings of VIS 2003:75-82.
- Lefohn AE, Kniss JM, Hansen CD, et al. A streaming narrow-band algorithm: interactive computation and visualization of level sets. IEEE Trans Vis Comput Graph 2004;10:422-33.
- Roberts M, Packer J, Mitchell JR, et al. CUDA accelerated sparse field level set segmentation of large medical data sets. Poster at the Nvidia GPU Technology Conference Research Summit 2009.
- Mostofi H. Fast level set segmentation of biomedical images using graphics processing units. Final Year Project, Keble College 2009.
- Maintz JBA, Viergever MA. A survey of medical image registration. Med Image Anal 1998;2:1-36.
- Zitova B, Flusser J. Image registration methods: a survey. Image Vision Comput 2003;21:977-1000.
- Kumuda MN, Kiran GC, Rajesh KS, et al. Corrective information based registration. Int J Comput Sci Inf Tech Secur 2012;2:40-6.
- Brunet F, Bartoli A, Navab N, et al. Pixel-based hyperparameter selection for feature-based image registration. Proceedings of VMV 2010:33-40.
- Belongie S. Shape matching and object recognition using shape contexts. IEEE Trans Pattern Anal Mach Intell 2002;24:509-22.
- Pilet J, Lepetit V, Fua P. Fast non-grid surface detection, registration and realistic augmentation. Int J Comput Vision 2007;76:109-22.
- Mousazadeh MH. Fast 3D deformable image registration on a GPU computing platform. Diploma Thesis, McMaster University 2011.
- Penney GP, Weese J, Little JA, et al. A comparison of similarity measures for use in 2-D-3-D medical image registration. IEEE Trans Med Imaging 1998;17:586-95.
- Grabner M, Pock T, Gross T, et al. Automatic differentiation for GPU-accelerated 2D/3D registration. Lect Notes Comput Sci Eng 2008;64:259-69.
- Roche A, Malandain G, Pennec X, et al. The correlation ratio as a new similarity measure for multimodal image registration. Lect Notes Comput Sc 1998;1496:1115-24.
- Viola P, Wells III WM. Alignment by Maximization of Mutual Information. Int J Comput Vision 1997;24:137-54.
- Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging 2003;22:986-1004.
- Shams R, Barnes N. Speeding up mutual information computation using NVIDIA CUDA hardware. Proceedings of DICTA 2007:555-60.
- Shams R, Sadeghi P, Kennedy R, et al. Parallel computation of mutual information on the GPU with application to real-time registration of 3D medical images. Comput Methods Programs Biomed 2010;99:133-46.
- Antonio R, Manuel U, Lee C, et al. Non-rgrid registration for large set of microscopic images on graphics processors. J Signal Process Sys 2009;55:229-50.
- Kubias A, Deinzer F, Feldmann T, et al. 2D/3D image registration on the GPU. Pattern Recogn Image Anal 2008;18:381-9.
- Khamene A, Bloch P, Wein W, et al. Automatic registration of portal images and volumetric CT for patient positioning in radiation therapy. Med Image Anal 2006;10:96-112.
- Markelj P, Tomazevic D, Likar B, et al. A review of 3D/2D registration methods for image-guided interventions. Med Image Anal 2012;16:642-61.
- Strzodka R, Droske M, Rumpf M. Fast image registration in DX9 graphics hardware. J Med Inf Tech 2003;6:43-9.
- Köhn A, Drexl J, Ritter F, et al. GPU accelerated image registration in two and three dimensions. Bildverarbeitung für die Medizin 2006:261-5.
- Horn BK, Schunck BG. Determing optical flow. Tech Rep, Massachusetts Institute of Technology 1980.
- Sharp GC, Kandasamy N, Singh H, et al. GPU-based streaming architectures for fast cone-beam ct image reconstruction and demons deformable registration. Phys Med Biol 2007;52:5771-83.
- Samant SS, Xia J, Muyan-Özçelik, et al. High performance computing for deformable image registration: towards a new paradigm in adaptive radiotherapy. Med Phys 2008;35:3546-53.
- Rezk-Salama C, Scheuering M, Soza G, et al. Fast volumetric deformation on general purpose hardware. Proceedings of SIGGRAPH 2001:17-24.
- Lehmann TM, Gönner C, Spitzer K. Survey: Interpolation methods in medical image processing. IEEE Trans Med Imaging 1999;18:1049-75.
- Turgeon GA, Lehmann G, Guiraudon G, et al. 2D-3D registration of coronary angiograms for cardiac procedure planning and guidance. Med Phys 2005;32:3737-49.
- Munbodh R, Tagare HD, Chen Z, et al. 2D-3D registration for prostate radiation therapy based on a statistical model of transmission images. Med Phys 2009;36:4555-68.
- Vetter C, Guetter C, Xu C, et al. Non-rigid multi-modal registration on the GPU. Proceedings of SPIE Medical Imaging 2007;6512:651228.
- Fan Z, Vetter C, Guetter C, et al. Optimized GPU implementation of learning-based non-rigid multi-modal registration. Proceedings of SPIE Medical Imaging 2008;6914:69142Y-1-169142Y-10.
- Liu JH, Ma WN, Liu F, et al. Study and application of medical image visualization technology. Lect Notes Comput Sc 2007;4561:668-77.
- Tatarchuk N, Shopf J, DeCoro C. Advanced interactive medical visualization on the GPU. J Parallel Distrib Comput 2008;68:1319-28.
- Zhao Y, Cui XY, Cheng Y. High-performance and real-time volume rendering in CUDA. Proceedings of BMEI 2009:1-4.
- William EL, Harvey EC. Marching cubes: A high resolution 3d surface construction algorithm. SIGGRAPH Comput Graph 1987;21:163-9.
- Wald I, Friedrich H, Marmitt G, et al. Faster isosurface ray tracing using implicit kd-tress. IEEE Trans Vis Comput Gr 2005;11:562-72.
- Hsieh J, eds. Computed tomography: principles, design, artifacts, and recent advances. Bellingham: SPIE Publications, 2003.
- Knoll A, Wald I, Parker S, et al. Interactive isosurface ray tracing of large octree volumes. Proceedings of IEEE Symposium on Interactive Ray Tracing 2006;115-24.
- Pavlík I. Advanced volume ray casting on GPU. Diploma Thesis, Charles University in Prague 2009.
- Shen R, Boulanger P. Hardware-accelerated volume rendering for real-time medical data visualization. Lect Notes Comput Sc 2007;4842:801-10.
- Rößler FA. Bridging the gap between volume visualization and medical applications. Diploma Thesis, Universität Stuttgart 2009.
- Mueller K, Shareef N, Huang J, et al. High-quality splatting on rectilinear grids with efficient culling of occluded voxels. IEEE Trans Vis Comput Gr 1999;5:116-34.
- Westover P. Footprint evaluation for volume rendering. SIGGRAPH Comput Graph 1991;24:367-76.
- Lacroute P, Levoy M. Fast volume rendering using a shear-wrap factorization of the viewing transformation. Proceedings of SIGGRAPH 1994:451-8.
- Engel K, Hadwiger M, Kniss J, et al. Real-time volume graphics. CRC Press, Boca Raton 2006.
- Xiao DG, Liu Y, Yang L, et al. Fourier volume rendering on GPGPU. Lect Notes Comput Sc 2009;5553:648-56.
- Viola I, Kanitsar A. GPU-based frequency domain volume rendering. Proceedings of SCCG 2004;55-64.
- Lippert L, Gross MH. Fast wavelet based volume rendering by accumulation of transparent texture maps. Comput Graph Forum 1995;14:431-43.
- Hadwiger M, Berger C, Hauser H. High-quality two-level volume rendering of segmented data sets on consumer graphics hardware. Proceedings of VIS 2003:301-8.
- Mora B, Jessel JP, Caubet R. A new object-order ray-casting algorithm. Proceedings of VIS 2002:203-10.
- Upson C, Keeler M. V-buffer: visible volume rendering. Comput Graph 1990;22:59-64.
- Muraki S, Kita Y. A survey of medical applications of 3D image analysis and computer graphics. Syst Comput JPN 2006;37:13-46.
- Schlegel P, Makhinya M, Pajarola R. Extinction-based shading and illumination in GPU volume ray-casting. IEEE Trans Vis Comput Graph 2011;17:1795-802.
- Westenberg MA, Roerdink JBTM. Frequency domain volume rendering by the wavelet X-ray transform. IEEE Trans Image Process 2000;9:1249-61.
- Nagy Z, Miiller G, Klein, R. Classification for Fourier volume rendering. Proceedings of PG 2004:51-8.
- Westenberg MA, Roerdink JBTM. An extension of Fourier-wavelet volume rendering by view interpolation. J Math Imaging Vis 2001;14:103-15.
- Dougherty G. eds. Medical image processing: techniques and applications. New York: Springer, 2011.
- Zhang Q, Eagleson R, Peters TM. Volume visualization: a technical overview with a focus on medical applications. J Digit Imaging 2011;24:640-64.
- Petrik S, Skala V. Technical section: space and time efficient isosurface extraction. Comput Graph 2008;32:704-10.
- Park SH, Choi EK, Lee SS, et al. Linear polyp measurement at CT colonography: 3D endoluminal measurement with optimized surface-rendering threshold value and automated measurement. Radiology 2008;246:157-67.
- Mensmann J, Ropinski T, Hinrichs K. An advanced volume raycasting technique using GPU stream processing. Proceedings of GRAPP 2010:190-8.
- Krüger J, Westermann R. Acceleration techniques for GPU-based volume rendering. Proceedings of VIS 2003:287-92.
- Röttger S, Guthe S, Weiskopf D, et al. Smart hardware-accelerated volume rendering. Proceedings of VISSYM 2003:231-8.
- Kim J. Efficient rendering of large 3-D and 4-D scalar fields. PhD thesis, University of Maryland, College Park 2008.
- Zhang Q, Eagleson R, Peters TM. Dynamic real-time 4D cardiac MDCT image display using GPU-accelerated volume rendering. Comput Med Imaging Graph 2009;33:461-76.
- HuY, Xu XH. Accelerate ray casting algorithm based on ray coherence. J Image Graph 2004;9:234-40.
- Kainz B, Grabner M, Bornik A, et al. Ray casting of multiple volumetric datasets with polyhedral boundaries on manycore GPUs. ACM T Graphic 2009;28:1-9.
- Springer JP, Beck S, Weiszig F, et al. Multi-frame rate rendering and display. Proceedings of VR 2007:195-202.
- Hauswiesner S, Kalkofen D, Schmalstieg D. Multi-frame rate volume rendering. Proceedings of EGPGV 2010;1-8.
- Shareef N, Lee TY, Shen HW, et al. An image-based modeling approach to GPU-based rendering of unstructured grids. Proceedings of the Eurographics 2006:31-8.
- Rezk-Salama C, Todt S, Kolb A. Raycasting of light field galleries from volumetric data. Comput Graph Forum 2008;27:839-46.
- Owens JD, Luebke D, Govindaraju N, et al. A survey of general-purpose computation on graphics hardware. Comput Graph Forum 2007;26:80-113.
- de Greef M, Crezee J, Eijk JC, et al. Accelerated ray tracing for radiotherapy dose calculation. Med Phys 2009;36:4095-102.
- Lo WCY, Han TD, Rose J, et al. GPU-accelerated Monte Carlo simulation for photodynamic therapy treatment planning. Proceedings of SPIE 2009;7373:737313-737313-12.
- Fung J, Mann S. Computer vision signal processing on graphics processing units. Proceedings of ICASSP 2004:V-93-6.
- Balafar MA, Ramli AR, Saripan MI, et al. Review of brain MRI image segmentation methods. Artif Intell Rev 2010;33:261-74.
- Balafar MA. Gaussian mixture model based segmentation methods for brain MRI images. Artif Intell Rev 2012;35:1-11.
- Held K, Kops ER, Krause BJ, et al. Markov random field segmentation of brain MR images. IEEE Trans Med Imaging 1997;16:878-86.
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random filed model and the expectation-maximization algorithm. IEEE Trans Med Imaging 2001;20:45-57.
- Ibrahim M, John N, Kabuka M, et al. Hidden Markov models-based 3D MRI brain segmentation. Image Vision Comput 2006;24:1065-79.
- Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage 2010;53:1181-96.
- Periaswamy S, Farid H. Medical image registration with partial data. Med Image Anal 2006;10:452-64.
- Liu W, Ribeiro E. A survey on image-based continuum-body motion estimation. Image Vision Comput 2011;29:509-23.
- Rueckert D, Frangi A, Schnabel J. Automatic construction of 3D statistical deformation models using non-rigid registration. Lect Notes Comput Sc 2001;2208:77-84.
- Meißner M, Huang J, Bartz D, et al. A practical evaluation of popular volume rendering algorithms. Proceedings of VV 2000:81-90.
- Entezari A, Scoggins R, Möller T, et al. Shading for Fourier volume rendering. Proceedings of VVS 2002:131-8.