
Detection of iron oxide nanoparticle (IONP)-labeled stem cells using quantitative ultrashort echo time imaging: a feasibility study
Introduction
Stem and immune cell therapies offer promising regenerative treatment solutions for many diseases such as Parkinson’s and multiple sclerosis (1). Non-invasive imaging of cellular therapeutics in patients during and after cell therapy is critical for assessing both the safety and efficacy of the treatment strategy. A variety of iron oxide nanoparticle (IONP)-based stem cell labeling agents have been utilized for magnetic resonance imaging (MRI)-based cell tracking (2-4). One such agent, ferumoxytol, is an FDA-approved intravenous iron supplement for chronic kidney patients that can be used off-label as a stem cell labeling agent. It is one of the very few available magnetically active IONP-based stem cell labeling agent and is visible with clinical MRI (5,6).
In MRI, the T1 and T2 values of an IONP-based labeling agent (e.g., ferumoxytol) decrease as the concentration of nanoparticles (5,7) does, creating a positive or negative contrast in the MR image depending on the respective T1 or T2 weighting (6,7). As a result, both conventional T1-weighted and T2-weighted MRI techniques can be used to localize the IONPs. Unfortunately, signal from highly accumulated IONPs may be difficult to detect with conventional MRI techniques because of the shortened T2 and T2* causing the signal to decay rapidly to near-background noise levels at an echo time (TE) on the order of milliseconds. Furthermore, clinical MRI techniques lack the ability to perform quantitative assessment of IONPs. An efficient quantitative MRI technique capable of both capturing the signal from IONPs and permitting quantitative evaluation would be highly advantageous to the field.
Ultrashort echo time (UTE) magnetic resonance imaging (MRI) technique has emerged as a promising technique able to image tissues with short T2 decays (8,9). UTE MRI achieves significantly shortened TEs (on the order of microseconds) that can capture rapidly decaying signal from tissues with short T2 relaxation times by utilizing a short radiofrequency (RF) pulse and rapid center-out non-Cartesian encoding schemes. UTE MRI has been evaluated in many preclinical applications and shown its efficacy in the imaging of short T2 tissues such as tendon (10), ligament (11), meniscus (12), bone (13), the osteochondral junction (14,15), and myelin lipid (16,17). Advanced, quantitative UTE techniques based on different MRI-based parameters have also been investigated to provide more sensitive assessment. Among them, UTE-T1 mapping (18), UTE-T2* mapping, and UTE quantitative susceptibility mapping (UTE-QSM) (19,20) have shown feasibility in assessing iron-laden tissues in human body. Detection of IONP with quantitative UTE-T1 mapping has proved to be a good strategy for imaging (21), which considers both negative and positive contrast mechanisms. Subtraction of later echo signal from early echo has shown to improve IONP specificity. UTE-QSM has been a popular technique of choice for quantification of iron (22) that evaluates echo dependence and transverse relaxation rate measurement. UTE-QSM is shown to be more sensitive towards TE selection than R2*. To improve QSM accuracy, continuous single point imaging has been explored in phantoms and ex vivo experiments suggesting novel ways of iron quantification (23). The proposed method can also be used for general QSM applications for tissues with longer T2*. UTE sequences have also been employed to image cartilage hemosiderin in patients affected by hemophilic joint disease (24). Using UTE techniques, studies have been conducted in detection of lung inflammation with respect to T1 weighted imaging (25) and for demonstrating quantitative contrast enhancement to measure ferumoxytol in vitro and in vivo (26). A potential limitation of using ferumoxytol can be its long intravascular half life that might influence MRI signal for days after administration. With prior knowledge, a radiologist can resolve this issue. With a polyglucose sorbitol carboxymethylether coating designed to minimize immunologic sensitivity and prolong intravascular half-life (27), ferumoxytol alone does not effectively label non-phagocytic cells such as stem cell and immune cells ex vivo. To overcome this challenge, a nanocomplex technology had been developed (5) and subsequently validated by multiple groups (28-30) to safely and effectively label cells with ferumoxytol while simultaneously providing a high enough iron concentration for adequate detection and long-term tracking in the clinical MR setting.
In this study, the efficacy of a series of 3D quantitative UTE techniques was investigated for the detection and quantification of IONP-labeled stem cells. UTE-T1, UTE-T2*, and UTE-QSM as well as clinical T2 mapping were evaluated in a phantom experiment with IONP-labeled human neural stem cells (NSCs) and in an ex vivo experiment with an NSC-injected mouse. In this work, for the first time we demonstrated that IONP-labeled stem cells could be detected by qUTE-MR techniques including UTE-QSM, UTE-T1, and UTE-T2* mapping. Especially, UTE-QSM based on cones trajectory has not been extensively studied for this application. We showed that UTE-QSM based on morphology enabled dipole inversion (MEDI) based algorithm yields reliable susceptibility mapping without major artifacts such as streaking.
Methods
Quantitative UTE (qUTE) MRI pulse sequence
UTE imaging is achieved by acquiring the free induction decay signal immediately after RF transmit/receive mode switching time (i.e., RF deadtime) based on the center-out readout scheme. Figure 1A shows the UTE MRI pulse sequence which is based on an efficient 3D cones trajectory and which was used in this study. A short hard pulse was used for RF excitation with minimum iso-delay (i.e., the duration between the peak and the end of the RF pulse), followed by 3D cones readout gradients placed after RF deadtime (32 µs), where data cannot be acquired due to transmit-receive mode switching. To achieve subsequent gradient recalled echoes, a fly-back encoding scheme was utilized where the same readout gradients were applied after rewinding (or cancelling) the dephasing induced by the preceding readout gradient. Figure 1B shows the 3D cones trajectory used to encode a 3D k-space.
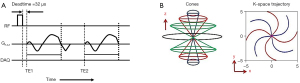
For UTE-T2* mapping and UTE-QSM, an interleaved dual echo acquisition technique (as shown in Figure 1A) was utilized in which scans were repeated multiple times with different readout gradients delays obtaining images at different echo times. For UTE-T1 mapping, variable flip angle (VFA) technique was used (31). Note that only the UTE echo was acquired without subsequent gradient recalled echo for T1 mapping, where the scans were repeated multiple time with different flip angles (FAs).
Stem cell labeling
NSCs were labeled using VMI-Trac Duo (Visicell Medical Inc., La Jolla, CA), a proprietary nanoparticle formulation that labels cells with ferumoxytol and a near-infrared (NIR) dye that, according to the manufacturer’s protocol, together enable cell tracking through optical imaging and MRI. Treated cells were washed to remove unincorporated nanoparticles, fixed with 4% paraformaldehyde, and examined by Prussian Blue staining and fluorescence microscopy.
Phantom design
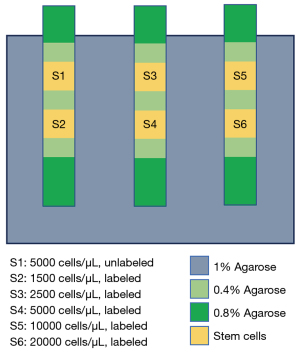
Figure 2 shows the phantom that was prepared for the validation of qUTE imaging. Specified numbers of unlabeled NSCs (5,000 cell/µL, S1) and iron-labeled NSCs (1,500, 2,500, 5,000, 10,000, and 20,000 cell/L, S2 to S6) were suspended in 0.4% low melt agarose and layered in 3-mL syringes. The three syringes were embedded in a secondary container filled with 1% agarose gel.
Mouse experiment
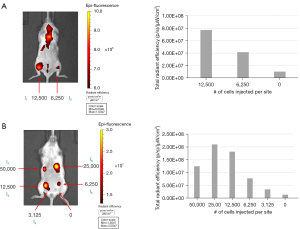
3,120, 6,250, 12,500, 25,000, or 50,000 cells/µL of labeled NSCs were injected into the mammary fat pads of a 14-week-old female NSG™ mouse. 6,250 and 12,500 cells/µL of labeled NSCs were also injected intramuscularly into the mouse’s gastrocnemius. Each injection was 50 µL and performed with a 29-gauge needle. Post-injection, optical imaging was performed using an IVIS Spectrum with excitation/emission filters set at 710/760 nm. Post optical imaging, the mouse was euthanized and underwent MR imaging. This study was approved by the Institutional Animal Care and Use Committee of Lumigenics LLC (Hercules, CA), in compliance of Lumigenics institutional guidelines for care and use of animals.
MR imaging
MRI was performed on a 3T clinical MR scanner (MR 750, GE Healthcare, Milwaukee, WI, USA) using a 16-channel phase array receive-only RF coil (GEM Flex Medium, GE Healthcare) in the phantom experiment or a custom-made 3-inch surface coil in the mouse experiment. MR images were acquired using 3D UTE-QSM (19), VFA UTE-T1 (VFA-UTE-T1) mapping (31,32), and UTE-T2* mapping (33) techniques as well as a clinical Carr-Purcell Meiboom-Gill (CPMG) T2 mapping sequence. B1 mapping was additionally performed based on actual flip angle imaging (AFI) technique (32) utilizing two different steady-state signals acquired with an interleaved variable TR approach. The MRI parameters are shown in Table 1.
Table 1
| UTE-QSM and UTE-T2* mapping | VFA-UTE-T1 mapping | CPMG-T2 mapping | |
|---|---|---|---|
| Phantom (3T GE MR750 w/a 16-ch flexible coil) | - TE =0.032, 0.4, 0.8, 2.2, 4.4, 6.6, 8.8, 11, 15.4, 22 ms - TR =34 ms, FA =17 degrees - FOV =100×100×64 mm3 - Matrix =200×200×64 - Readout BW =125 kHz - Total scan time =50 min 8 s |
- TE =32 μs, TR =20 ms - FA=5, 10, 20, 30 degrees - FOV =100×100×64 mm3 - Matrix =200×200×32 - Readout BW =125 kHz - Total scan time =12 min 9 s - B1 mapping: 6min 41 s |
- TE =6.5, 13.1, 19.6, 26.1, 32.7, 39.2, 45.7, 52.3, 58.8, 65.3, 71.9, 78.4, 84.9, 91.4, 98.0, 104.5 ms - TR =600 ms, NEX =2 - Slice thickness =2 mm - # of slices =1 - FOV =100×100 mm2 - Matrix =256×256 - Readout BW =125 kHz - Total scan time =5 min 7 s |
| Ex vivo mouse (3T GE MR750 w/a 3-inch surface coil) | - TE =0.032, 0.4, 0.8, 2.2, 4.4, 6.6, 8.8, 11, 15.4, 22 ms - TR =34 ms, FA =17 degree – FOV =80×80×30 mm3 - Matrix =160×160×60 - Readout BW =125 kHz - Total scan time =40 min 42 s |
- TE =32 μs, TR =20 ms - FA =5, 10, 20, 30 degree - FOV =80×80×30 mm3 - Matrix =160×160×30 - Readout BW =125 kHz - Total scan time =9 min 54 s - B1 mapping: 5 min 24 s |
- TE =7.2, 14.5, 21.7, 29.0, 36.2, 43.5, 50.7, 58.0, 65.2, 72.4, 79.7, 86.9, 94.2, 101.4, 108.7, 115.9 ms - TR =500 ms, NEX =2 - Slice thickness =2 mm - # of slices =11 - FOV =80×80 mm2 - Matrix =256×256 - Readout BW =125 kHz - Total scan time =46 min 56 s |
MR, magnetic resonance; UTE-QSM, ultrashort echo time - quantitative susceptibility mapping; VFA, variable flip angle; CPMG, Carr-Purcell-Meiboom-Gill; TR, repetition time; FA, flip angle; BW, bandwidth; FOV, field of view.
Data processing
UTE MR images were reconstructed using an offline image reconstruction code written in Matlab (MathWorks, Natick, MA, USA) based on non-uniform fast Fourier transform (NuFFT) (34). The following gridding parameters were used: oversampling rate of 2 and kernel width of 5. The images reconstructed in the individual channels were combined to form a complex image. To process the data acquired from the phantom experiment, images were first individually reconstructed for each channel and then combined using coil sensitivity maps estimated by principal component analysis (PCA) based on singular value decomposition (SVD) (35).
For the qUTE parameter mappings, codes written in MATLAB were utilized. For T1 and T2* parameter mapping, the Levenberg-Marquardt method was used for nonlinear least-squares curve fitting (13). In UTE-QSM, MEDI based QSM reconstruction was utilized (36) after performing Laplacian phase unwrapping and background field removal based on projection onto dipole field (PDF) technique (37). A Lagrange multiplier (i.e., regularization parameter) of 1000 was used for MEDI which was empirically tuned to achieve a susceptibility map with suppressed streaking artifacts. The T2 map was calculated using the toolkit on the scanner along with images acquired with the CPMG sequence. The CPMG based T2 map was generated online using the vendor provided software in the host PC (GE software version DV26R01).
In the phantom experiment, the estimated parameters including susceptibility, R1 (i.e., 1/T1), R2* (i.e., 1/T2*), and R2 (i.e., 1/T2) were fitted with a linear equation with respect to the known density of the labeled NSCs (i.e., 1,500, 2,500, 5,000, 10,000, and 20,000 cells/µL) including unlabeled NSCs as zero density. Goodness of fit (R2) as well as slope and y-intercept were evaluated.
Results
Stem cell labeling
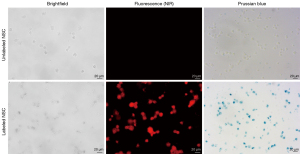
Figure 3 shows that 100% of the labeled NSCs stained positive for Prussian blue and were visualized by NIR fluorescence microscopy, illustrating efficient loading of both NIR dye and ferumoxytol into stem cells by VMI-Trac Duo.
NSC phantom
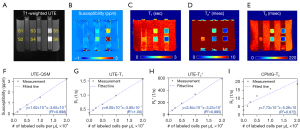
Figure 4 shows parameter maps from UTE-QSM, UTE-T1, UTE-T2*, and CPMG-T2 mapping techniques (Figure 4B-4E) and the corresponding mean values of susceptibility, R1, R2*, and R2 (Figure 4F-4I) in each stem cell compartments (S1-S6). All parameters showed monotonic increase with the number of labeled stem cells. The estimated susceptibilities for the cell layers S1 to S6 were 0.03±0.02, 0.16±0.03, 0.42±0.07, 0.82±0.19, 1.50±0.28, and 3.23±1.52 ppm, respectively. The estimated T1 values for the cell layers S1 to S6 were 2.7±0.1, 2.2±0.1, 1.8±0.2, 1.5±0.3, 1.0±0.3, and 0.6±0.2 s, respectively. The estimated T2* values for the cell layers S1 to S6 were 95.3±12.0, 13.9±1.1, 8.9±1.8, 5.4±1.2, 3.0±0.9, and 1.7±0.4 ms, respectively. The estimated T2 values for the cell layers S1 to S6 were 345.2±14.8, 187.6±20.5, 128.5±19.9, 87.4±19.6, 62.8±29.9, and 53.1±34.7 ms, respectively.
All parameters except for CPMG-based T2 showed highly linear relationships with the number of labeled stem cells. The estimated line equations can be given below:
where the independent parameter x indicates density of iron-labeled NSC, and dependent parameter y indicates MRI-based parameters (i.e., susceptibility, R1, R2*, and R2). UTE-QSM, UTE-T1, and UTE-T2* showed high linearity (R2>0.99), while CPMG-T2 exhibited nonlinearity in the curve (R2=0.87) especially for the data points with high densities of labeled stem cells.
In vivo IVIS imaging with a NSC-injected mouse
To demonstrate the feasibility of multi-modality cell tracking, NSCs labeled with VMI-Trac Duo were injected into the gastrocnemius muscle (Figure 5A) or mammary fat pads (Figure 5B) of a 14-week-old NSG™ mouse, and immediately imaged on a IVIS Spectrum. As few as 3,125 injected labeled cells could be detected by NIR fluorescence imaging, and there was a positive correlation between the number of injected labeled cells and the measured fluorescence intensity. These results demonstrated that VMI-Trac Duo-labeled cells can be tracked by optical imaging on an IVIS scanner.
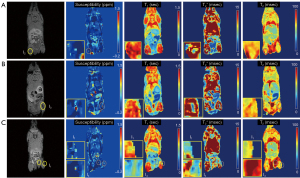
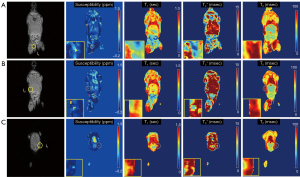
Ex vivo MRI with an NSC-injected mouse
In the ex vivo experiment with an NSC-injected mouse, qUTE techniques detected all seven injected regions (I1–I7). Figures 6,7 show all parameter maps in the corresponding slices. UTE-QSM showed elevated susceptibility in the regions injected with the labeled stem cells more clearly than other techniques (>1.5 ppm). UTE-T2* mapping exhibited decreased relaxation times in the most injected regions but was obscured by surrounding tissues with high inhomogeneity. UTE-T1 (ranges between 0–0.75 s) and CPMG-T2 (around 100 ms) showed no significant changes in relaxation times of the injected regions.
Discussion
In this study, we demonstrated that IONP-labeled stem cells could be detected by qUTE-MR techniques including UTE-QSM, UTE-T1, and UTE-T2* mapping. The efficacy of UTE imaging over non-UTE imaging in the detection of iron nanoparticles has been shown in the previous study (22) which showed the improved quantification of nanoparticles with UTE based T2* and QSM (R2>0.99). In the phantom experiment, qUTE techniques showed feasibility in the quantification and localization of IONP-labeled stem cells, where a highly linear relationship was found between the density of labeled stem cells and the measured parameters. R2 parameter from CPMG-T2 showed non-linearity especially for the high densities (e.g., 10,000 and 20,000 cells/µL) which implies impaired sensitivity for highly accumulated iron contents. This can be due to the T2 relaxation being shortened by highly concentrated IONPs, resulting in a lower SNR in later echo times where the measured signal tends to be overestimated relative to its actual intensity by Rician noise being added to the small signal. Consequently, this may produce an overestimated T2 parameter in signal fitting. Another hypothesis is that the R2 or the labeled stem cells naturally does not linearly correlate with iron contents, which needs further investigation. In either case, CPMG- T2 is not likely the best option for quantification of stem cells due to the reduced sensitivity in high density.
In the ex vivo mouse experiment, only UTE-QSM and UTE-T2* demonstrated noticeable quantitative changes in the injected regions of the ex vivo mouse, with UTE-QSM showing the best performance. As most soft tissues are slightly diamagnetic (negative susceptibility) in a living system, it is expected that paramagnetic IONPs (i.e., a substance with high positive susceptibility) can provide high contrast in UTE-QSM. UTE-T2* is also an effective approach in this regard since T2* depends directly on a substance’s magnetic susceptibility. However, T2* only reflects the magnitude of susceptibility (T2’), which cannot separate substances between positive and negative susceptibilities. For example, both bone (i.e., including calcium with strong diamagnetism) and hemosiderin (i.e., including iron with paramagnetism) have ultrashort T2* relaxation times, which can create strong negative contrast in T2*-weighted MRI despite their opposite polarities with regard to susceptibility. Another confounding factor in T2* mapping is the natural inhomogeneity of the surrounding tissues. Likewise, T1 in a living system is highly inhomogeneous, in much the same way that T2* is. This explains why T1 was not effective in the ex vivo mouse imaging. T1 with labeled stem cells ranged from 0.6 to 2.7 s in the phantom experiment. Because most tissues have T1 relaxation times in this range, it is possible for the injected IONP-labeled stem cells to be obscured by the inhomogeneous surrounding tissues. However, depending on the targeted organ, T1, T2, and T2* can also be effective in detecting IONP-labeled stem cells. For example, in the brain and liver, which both have relatively homogenous structures, T1, T2 and T2* may provide good detection of the labeled cells, creating positive or negative contrast to the surrounding tissues. With the given technical complexity of QSM which requires multiple steps including phase unwrapping (38), background field removal (37), and dipole inversion with iterative reconstruction (36), those conventional MRI parameters could be considered reasonable alternatives.
Although UTE-QSM showed more clear detection of the injected stem cells, the susceptibility values should be understood in the context of the anatomical structure. For example, any tissues with air can show elevated values as the air contains paramagnetic oxygen which is a strong source of susceptibility. Therefore, the regions with inherently high susceptibility (e.g., intestine and lung) can be easily excluded when interpreting the UTE-QSM results from whole-body imaging. In addition, connective tissues such as tendons and ligaments have paramagnetism, while cortical bone has strong diamagnetism, which should be also considered in the interpretation of susceptibility values. The best approach is to image the subject before and after the injection of the stem cells so that reference parameter maps are obtained. Certainly, there are a lot of scopes for optimization and improvement which were not covered in this pilot study. We plan to perform more extensive experiments to investigate this in our future studies.
To better comprehend the efficacy and safety of cell-based therapies, researchers are increasingly using medical imaging to track adoptively transferred cells in vivo. Although single photon emission computed tomography (SPECT) and positron emission tomography (PET) have been used to track genetically engineered immune cells in vivo in both preclinical and clinical settings by direct labeling with 111In-Oxine, 89Zr-Oxine, 89Zr-DFO, or indirect labeling with 18F-TMP and 18F-FHBG, the use of ionizing radiation poses a particular concern with cell therapy products because it can cause DNA damage and thereby impair the viability or therapeutic efficacy of the cells (39,40). In contrast, the multimodality iron-based imaging contrast agent VMI-Trac Duo used in this study provides non-invasive and radiation-free molecular imaging with high soft tissue contrast via MRI. Moreover, the inclusion of an NIR dye with fluorescence emission in the near-infrared spectral range (700–900 nm) enables in vivo detection of the labeled cell by optical imaging owing to deep penetration of the fluorescence signal while minimizing interference from tissue autofluorescence. This unique combination allows cell therapy developers and clinical researchers to leverage different imaging modalities at different stages of preclinical research and translation while utilizing a single nanoparticle formulation. We envision the quantitative UTE MR techniques can provide accurate, sensitive, and quantitative assessment on the biodistribution of administered therapeutic cell products, track their presence at the intended target sites, and provide better understanding of their in vivo behaviors for a more-informed treatment strategy, with further improvement and validation. Considering the mechanism of action for adoptive immune therapy and the positive correlations observed in animal cancer models between early tumor targeting by CAR T-cells and therapeutic response, the imaging insights from qUTE could be employed as an imaging biomarker for anti-tumor treatment efficacy in the future. This quantitative imaging-based approach would enable oncologists to not only better understand and potentially predict therapeutic efficacy, but also to stratify patients for adoptive cell therapy in a more personalized medicine approach.
In vivo imaging will demonstrate the feasibility of qUTE imaging in clinical applications. Further studies are planned to investigate our findings by performing in vivo MR imaging pre- and post-injection with mice in two groups (e.g., a control group injected with unlabeled NSCs and an experimental group injected with labeled NSCs). There are several challenges anticipated in the in vivo experiment such as lower SNR, motion, and scan time. To allow enough SNR and shorten the scan time, small surface coil will be effective rather than the volume coils. Motion is a major confounding factor that can impact the qUTE results. UTE imaging with center-our radial or cones trajectory is known to be robust to motion due to the oversampled k-space center (41). In addition, prospective gating will be readily done without navigation data in the UTE sequence. Additional retrospective gating will be also beneficial to minimize effect of intra-scan motion. Inter-scan bulk motion can be simple compensated using image registration methods (42).
This feasibility study has several limitations. First, only one mouse was included. Inclusion of more mice is likely to reconfirm our findings. Second, histology, which could have provided a ground truth for the ex vivo mouse experiment, was not performed. Third, only ex vivo imaging with a post-mortem mouse was performed, while injection was done in vivo. In vivo imaging will demonstrate the feasibility of qUTE imaging in clinical applications. Further studies are planned to investigate our findings by performing in vivo MR imaging pre- and post-injection with mice in two groups (e.g., a control group injected with unlabeled NSCs and an experimental group injected with labeled NSCs). Another drawback of this study could be its lengthy scan times which can potentially be improved using acceleration techniques like compressed sensing or deep learning based reconstruction methods (43,44).
Conclusions
In this study, we evaluated a series of qUTE imaging techniques (i.e., UTE-T1, UTE-T2*, and UTE-QSM) as well as conventional T2 mapping for the detection of IONP-labeled stem cells in vitro and ex vivo. Although all techniques showed potential to quantify IONP-labeled stem cells in a phantom experiment, UTE-QSM demonstrated superior performance with high contrast in detecting stem cells injected into the different body parts of a mouse.
Acknowledgments
Funding: The study was supported by grants from the NIH (No. R01 AR078877), VA Clinical Science Research and Development (Merit Award I01CX002211), DFG (No. SE 3272/1-1), and GE Healthcare.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-654/coif). JA and MT are employees of VisiCELL Medical Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Institutional Animal Care and Use Committee of Lumigenics LLC (Hercules, CA), in compliance of Lumigenics institutional guidelines for care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lindvall O, Kokaia Z. Stem cells for the treatment of neurological disorders. Nature 2006;441:1094-6. [Crossref] [PubMed]
- Nedopil A, Klenk C, Kim C, Liu S, Wendland M, Golovko D, Schuster T, Sennino B, McDonald DM, Daldrup-Link HE. MR signal characteristics of viable and apoptotic human mesenchymal stem cells in matrix-associated stem cell implants for treatment of osteoarthritis. Invest Radiol 2010;45:634-40. [Crossref] [PubMed]
- Henning TD, Boddington S, Daldrup-Link HE. Labeling hESCs and hMSCs with iron oxide nanoparticles for non-invasive in vivo tracking with MR imaging. J Vis Exp 2008;685. [Crossref] [PubMed]
- Simon GH, von Vopelius-Feldt J, Fu Y, Schlegel J, Pinotek G, Wendland MF, Chen MH, Daldrup-Link HE. Ultrasmall supraparamagnetic iron oxide-enhanced magnetic resonance imaging of antigen-induced arthritis: a comparative study between SHU 555 C, ferumoxtran-10, and ferumoxytol. Invest Radiol 2006;41:45-51. [Crossref] [PubMed]
- Thu MS, Bryant LH, Coppola T, Jordan EK, Budde MD, Lewis BK, Chaudhry A, Ren J, Varma NR, Arbab AS, Frank JA. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nat Med 2012;18:463-7. [Crossref] [PubMed]
- Gutova M, Frank JA, D'Apuzzo M, Khankaldyyan V, Gilchrist MM, Annala AJ, Metz MZ, Abramyants Y, Herrmann KA, Ghoda LY, Najbauer J, Brown CE, Blanchard MS, Lesniak MS, Kim SU, Barish ME, Aboody KS, Moats RA. Magnetic resonance imaging tracking of ferumoxytol-labeled human neural stem cells: studies leading to clinical use. Stem Cells Transl Med 2013;2:766-75. [Crossref] [PubMed]
- Khurana A, Nejadnik H, Chapelin F, Lenkov O, Gawande R, Lee S, Gupta SN, Aflakian N, Derugin N, Messing S, Lin G, Lue TF, Pisani L, Daldrup-Link HE. Ferumoxytol: a new, clinically applicable label for stem-cell tracking in arthritic joints with MRI. Nanomedicine (Lond) 2013;8:1969-83. [Crossref] [PubMed]
- Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. J Comput Assist Tomogr 2003;27:825-46. [Crossref] [PubMed]
- Holmes JE, Bydder GM. MR imaging with ultrashort TE (UTE) pulse sequences: Basic principles. Radiography 2005;11:163-74. [Crossref]
- Liu F, Kijowski R. Assessment of different fitting methods for in-vivo bi-component T2(*) analysis of human patellar tendon in magnetic resonance imaging. Muscles Ligaments Tendons J 2017;7:163-72. [Crossref] [PubMed]
- Jang H, McMillan AB, Ma Y, Jerban S, Chang EY, Du J, Kijowski R. Rapid single scan ramped hybrid-encoding for bicomponent T2* mapping in a human knee joint: A feasibility study. NMR Biomed 2020;33:e4391. [Crossref] [PubMed]
- Williams A, Qian Y, Golla S, Chu CR. UTE-T2* mapping detects sub-clinical meniscus injury after anterior cruciate ligament tear. Osteoarthritis Cartilage 2012;20:486-94. [Crossref] [PubMed]
- Du J, Bydder GM. Qualitative and quantitative ultrashort-TE MRI of cortical bone. NMR Biomed 2013;26:489-506. [Crossref] [PubMed]
- Jang H, Ma Y, Carl M, Lombardi AF, Chang EY, Du J. Feasibility of an Inversion Recovery-Prepared Fat-Saturated Zero Echo Time Sequence for High Contrast Imaging of the Osteochondral Junction. Front Endocrinol (Lausanne) 2021;12:777080. [Crossref] [PubMed]
- Lombardi AF, Jang H, Wei Z, Jerban S, Wallace M, Masuda K, Ma YJ. High-contrast osteochondral junction imaging using a 3D dual adiabatic inversion recovery-prepared ultrashort echo time cones sequence. NMR Biomed 2021;34:e4559. [Crossref] [PubMed]
- Ma YJ, Searleman AC, Jang H, Fan SJ, Wong J, Xue Y, Cai Z, Chang EY, Corey-Bloom J, Du J. Volumetric imaging of myelin in vivo using 3D inversion recovery-prepared ultrashort echo time cones magnetic resonance imaging. NMR Biomed 2020;33:e4326. [Crossref] [PubMed]
- Jang H, Ma YJ, Chang EY, Fazeli S, Lee RR, Lombardi AF, Bydder GM, Corey-Bloom J, Du J. Inversion Recovery Ultrashort TE MR Imaging of Myelin is Significantly Correlated with Disability in Patients with Multiple Sclerosis. AJNR Am J Neuroradiol 2021;42:868-74. [Crossref] [PubMed]
- Wei Z, Jang H, Bydder GM, Yang W, Ma YJ. Fast T(1) measurement of cortical bone using 3D UTE actual flip angle imaging and single-TR acquisition (3D UTE-AFI-STR). Magn Reson Med 2021;85:3290-8. [Crossref] [PubMed]
- Jang H, von Drygalski A, Wong J, Zhou JY, Aguero P, Lu X, Cheng X, Ball ST, Ma Y, Chang EY, Du J. Ultrashort echo time quantitative susceptibility mapping (UTE-QSM) for detection of hemosiderin deposition in hemophilic arthropathy: A feasibility study. Magn Reson Med 2020;84:3246-55. [Crossref] [PubMed]
- Lu X, Jang H, Ma Y, Jerban S, Chang EY, Du J. Ultrashort Echo Time Quantitative Susceptibility Mapping (UTE-QSM) of Highly Concentrated Magnetic Nanoparticles: A Comparison Study about Different Sampling Strategies. Molecules 2019; [Crossref] [PubMed]
- Girard OM, Du J, Agemy L, Sugahara KN, Kotamraju VR, Ruoslahti E, Bydder GM, Mattrey RF. Optimization of iron oxide nanoparticle detection using ultrashort echo time pulse sequences: comparison of T1, T2*, and synergistic T1- T2* contrast mechanisms. Magn Reson Med 2011;65:1649-60. [Crossref] [PubMed]
- Lu X, Ma Y, Chang EY, He Q, Searleman A, von Drygalski A, Du J. Simultaneous quantitative susceptibility mapping (QSM) and R2* for high iron concentration quantification with 3D ultrashort echo time sequences: An echo dependence study. Magn Reson Med 2018;79:2315-22. [Crossref] [PubMed]
- Jang H, Lu X, Carl M, Searleman AC, Jerban S, Ma Y, von Drygalski A, Chang EY, Du J. True phase quantitative susceptibility mapping using continuous single-point imaging: a feasibility study. Magn Reson Med 2019;81:1907-14. [Crossref] [PubMed]
- von Drygalski A, Barnes RFW, Jang H, Ma Y, Wong JH, Berman Z, Du J, Chang EY. Advanced magnetic resonance imaging of cartilage components in haemophilic joints reveals that cartilage hemosiderin correlates with joint deterioration. Haemophilia 2019;25:851-8. [Crossref] [PubMed]
- Strobel K, Hoerr V, Schmid F, Wachsmuth L, Löffler B, Faber C. Early detection of lung inflammation: exploiting T1-effects of iron oxide particles using UTE MRI. Magn Reson Med 2012;68:1924-31. [Crossref] [PubMed]
- Gharagouzloo CA, McMahon PN, Sridhar S. Quantitative contrast-enhanced MRI with superparamagnetic nanoparticles using ultrashort time-to-echo pulse sequences. Magn Reson Med 2015;74:431-41. [Crossref] [PubMed]
- Nguyen KL, Yoshida T, Kathuria-Prakash N, Zaki IH, Varallyay CG, Semple SI, et al. Multicenter Safety and Practice for Off-Label Diagnostic Use of Ferumoxytol in MRI. Radiology 2019;293:554-64. [Crossref] [PubMed]
- Yin Y, Zhou X, Guan X, Liu Y, Jiang CB, Liu J. In vivo tracking of human adipose-derived stem cells labeled with ferumoxytol in rats with middle cerebral artery occlusion by magnetic resonance imaging. Neural Regen Res 2015;10:909-15. [Crossref] [PubMed]
- Vandergriff AC, Hensley TM, Henry ET, Shen D, Anthony S, Zhang J, Cheng K. Magnetic targeting of cardiosphere-derived stem cells with ferumoxytol nanoparticles for treating rats with myocardial infarction. Biomaterials 2014;35:8528-39. [Crossref] [PubMed]
- Su Z, Wang X, Zheng L, Lyu T, Figini M, Wang B, Procissi D, Shangguan J, Sun C, Pan L, Qin L, Zhang B, Velichko Y, Salem R, Yaghmai V, Larson AC, Zhang Z. MRI-guided interventional natural killer cell delivery for liver tumor treatment. Cancer Med 2018;7:1860-9. [Crossref] [PubMed]
- Ma YJ, Zhao W, Wan L, Guo T, Searleman A, Jang H, Chang EY, Du J. Whole knee joint T(1) values measured in vivo at 3T by combined 3D ultrashort echo time cones actual flip angle and variable flip angle methods. Magn Reson Med 2019;81:1634-44. [Crossref] [PubMed]
- Ma YJ, Lu X, Carl M, Zhu Y, Szeverenyi NM, Bydder GM, Chang EY, Du J. Accurate T(1) mapping of short T(2) tissues using a three-dimensional ultrashort echo time cones actual flip angle imaging-variable repetition time (3D UTE-Cones AFI-VTR) method. Magn Reson Med 2018;80:598-608. [Crossref] [PubMed]
- Chang EY, Du J, Iwasaki K, Biswas R, Statum S, He Q, Bae WC, Chung CB. Single- and Bi-component T2* analysis of tendon before and during tensile loading, using UTE sequences. J Magn Reson Imaging 2015;42:114-20. [Crossref] [PubMed]
- Fessler JA. On NUFFT-based gridding for non-Cartesian MRI. J Magn Reson 2007;188:191-5. [Crossref] [PubMed]
- Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med 2000;43:682-90. [Crossref] [PubMed]
- Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR, Wang Y. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage 2012;59:2560-8. [Crossref] [PubMed]
- Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed 2011;24:1129-36. [Crossref] [PubMed]
- Cusack R, Papadakis N. New robust 3-D phase unwrapping algorithms: application to magnetic field mapping and undistorting echoplanar images. Neuroimage 2002;16:754-64. [Crossref] [PubMed]
- Yang B, Parsha K, Schaar K, et al. Various Cell Populations Within the Mononuclear Fraction of Bone Marrow Contribute to the Beneficial Effects of Autologous Bone Marrow Cell Therapy in a Rodent Stroke Model. Transl Stroke Res 2016;7:322-30. [Crossref] [PubMed]
- Kurebayashi Y, Choyke PL, Sato N. Imaging of cell-based therapy using (89)Zr-oxine ex vivo cell labeling for positron emission tomography. Nanotheranostics 2021;5:27-35. [Crossref] [PubMed]
- Herrmann KH, Krämer M, Reichenbach JR. Time Efficient 3D Radial UTE Sampling with Fully Automatic Delay Compensation on a Clinical 3T MR Scanner. PLoS One 2016;11:e0150371. [Crossref] [PubMed]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Baron CA, Dwork N, Pauly JM, Nishimura DG. Rapid compressed sensing reconstruction of 3D non-Cartesian MRI. Magn Reson Med 2018;79:2685-92. [Crossref] [PubMed]
- Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487-92. [Crossref] [PubMed]


