
A deep learning-based post-processing method for automated pulmonary lobe and airway trees segmentation using chest CT images in PET/CT
Introduction
Positron emission tomography/computed tomography (PET/CT) provides functional and anatomical information about the human body in a single scan and has been widely used in clinical practice (1). Images from PET can yield the metabolic information with 18F-fluorodeoxyglucose (FDG) for tumor staging and other diagnostic information; however, its low spatial resolution and high noise level limit the ability to locate lesions precisely. It is a common approach to use CT images in PET/CT to acheive automatic organ segmentation. Automatic pulmonary region segmentation is a basic task in computer-aided diagnostic systems. The location and distribution of lesions can be a significant factor in determining the follow-up treatment. The National Emphysema Treatment Trial Research Group’s study (2) reported that locally distributed emphysema could be treated more effectively by lobar volume resection than homogeneously distributed emphysema. In addition, the early diagnosis of lung disease indicates a good prognosis. Recent advances in deep learning (DL), especially the deep convolutional neural network (CNN), have accelerated the development of automatic pulmonary nodule detection and classification system (3-7), which can assist radiologists in minimizing workloads. Precise segmentation of lung lobes and airway trees also contributes to the accurate identification of the lesion positioning, preventing the ignorance of location information during the direct lesion segmentation.
There have been some unsupervised models with traditional computer vision methods used in lung lobe segmentation, which usually include detecting fissures and locating bronchi and vessels (8,9). Common techniques are the main solution in the airway trees segmentation task (10-17). Other methods proposed in the past include rule-based methods (14,18), energy function minimization (17), and region of interest (ROI) modification-based techniques (19). Schlathoelter et al. (20) used a front-propagation algorithm for segmenting airway trees. Branch points are detected when the front splits up. With the development of computer technology, DL has emerged as the most promising technology. The CNN models have matured and now demonstrate considerable performance guarantees through many developmental milestones (21-26). Currently, research under the medical images domain widely leveraged the CNN models, including lesion detection, quantitative diagnosis, lesion segmentation, and so on (27-31).
The end-to-end trained fully convolutional networks (FCN) were initially developed as the solution for image segmentation problems in the computer vision domain (32,33). They were a popular model in the region and lesion segmentation task. In Gibson et al. (34), a new CNN architecture, dense V-network (DenseVNet), was reported and demonstrated in the multi-organ segmentation task on abdominal CT. Imran et al. (35) developed a new model for lung lobe segmentation using DenseVNet and progressive holistically nested networks on routine chest CT. Nadeem et al. (36) used the U-Net followed by a freeze-and-grow propagation algorithm to increase the completeness of the segmented airway trees iteratively. Garcia-Uceda et al. (37) developed a fully automatic and end-to-end optimized airway trees segmentation method for thoracic CT based on the U-Net architecture. However, most current algorithms are proposed only for lung lobe or pulmonary airway tree segmentation (20,21), and lack a suitable algorithm for simultaneous segmentation of pulmonary lobes and airway trees.
The focus of this study was to segment five lobes and airway trees, leveraging CT images in PET/CT scanning via DL combined with post-processing methods. We present the subsequent sections following the MDAR reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1116/rc).
Methods
Datasets
We retrospectively enrolled patients who underwent both PET/CT and separate CT at Peking Union Medical College Hospital. All cases were observed to have pulmonary nodules. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) under the approval of the Institutional Review Board of the Peking Union Medical College Hospital, and individual consent for this retrospective analysis was waived. The PET/CT scan was performed on a Siemens Biograph Truepoint PET/CT scanner (Siemens Healthineers, Erlangen, Germany). The whole-body PET/CT scanning protocol was described in a previous study (21). A separate chest CT imaging was also performed. The voltage output of the X-Ray generator was 120 kVp, and the X-Ray tube current was 300 mA. Using a standard reconstruction algorithm, the thin-slice reconstruction thickness was 1.25 mm, and the interval was 0.8 mm. The scanning range spanned from the apex of the lung to the diaphragm. CT images were recorded in the standard Digital Imaging and Communications in Medicine (DICOM) format with a voxel spacing 0.8×0.8×1 mm3 with dimensions of 512×512×337.
The CT dataset was randomly divided into the training, validation, and independent test sets. A total of 476 cases were used in the training model. The validation included 134 cases and was used for fine-tuning the hyperparameters and select for the best parameters, and the independent test dataset included 30 cases and was used to evaluate the performance of our methods.
Reference standard segmentations
All images were processed on an open-source platform named 3D Slicer (version 4.6) (38). The ground truth of pulmonary lobes and airway tree segmentation was marked by a nuclear medicine physician supervised by a senior physician with experience in chest diagnosis. The pulmonary region was labeled with six labels based on anatomy: three labels in the right lung, two labels in the left lung, and one label in the airway trees.
DL-based segmentation network
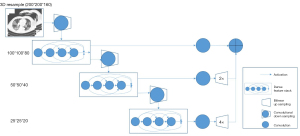
In this paper, we combined the DL method and traditional image post-processing methods to obtain the final segmentation of lung lobes and airway trees. The workflow is shown in Figure 1.
The DL method used the DenseVNet, and the architecture is shown in Figure 2. Before inputting the image to the DL model, the dimension of input data was resampled to 200×200×160 due to the high resolution of the medical three-dimensional (3D) images. Each convolutional kernel size in the dense feature block was 3×3×3. The max pooling method was used after the dense feature block ,and rectified linear unit (ReLU) was used to represent nonlinear functions. During the training procedure, the Dice score was used as the loss function, and Adam optimizer with lr=0.001 and mini batch size of 6 for 7,000 iterations were also used. Training each instance of the network took approximately 12 hours using v100 16G GPUs (NVIDIA Corp., Los Alamitos, CA, USA).
Post-processing methods
The traditional image post-processing methods were applied to increase the accuracy of segmentation results gained from DenseVNet. The region growing algorithm based on seed points is mainly used in the post-processing of airway trees, and it includes three steps. First, calculate the mean Hounsfield unit (HU) value (M) from the original airway trees label that comes from the DL segmentation model. Second, find the seed point on the largest connected area of the original airway trees label. Third, compute the final airway trees based on the seed point in the threshold (–1,024, M) range via the region growth method.
For the original pulmonary lobe labels from the DL segmentation model, two post-preprocessing methods were considered. Firstly, the center of the DL segmentation result was taken as the seed point, and the region growing method was used to eliminate redundant segmentation results; at the same time, a threshold of (–1,024, 400) was used to remove bone tissue parts in DL segmentation results; next, the closing method was used to obtain the final lung lobe labels.
Evaluation metrics
The results for pulmonary lobes and airway trees were compared to the ground truth segmentation by Dice coefficient, Jaccard coefficient, and Hausdorff distance, respectively (39,40). The Dice coefficient and Jaccard coefficient were used for the comparison between results and the ground truth via volumes. The Hausdorff distance was used to test the agreement of the boundaries.
Statistical analysis
Three essential metrics were used to assess the DenseVNet and the combined model. The average was used to summarize the evaluation metrics. The agreement between the results and the ground truth was calculated via the Bland–Altman plot. Student’s t-test or the Wilcoxon rank sum test was used to compute the differences between results and references, depending on if the data were normally distributed. Statistical analysis was performed on R software (version 3.5; R Foundationt for Statistical Computing, Vienna, Austria, http://www.r-project.org). A P value less than 0.05 was considered statistically significant.
Results
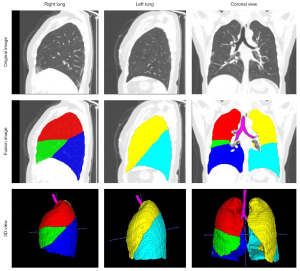
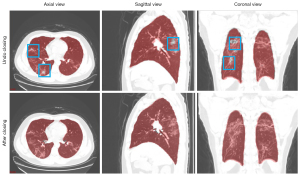
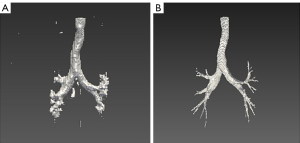
Figure 3 shows examples of results in pulmonary lobe segmentation and airway tree segmentation to demonstrate the effectiveness. The DL segmentation model successfully distinguished the pulmonary lobes and airway trees from all CT images. Still, there might have been missing parts in the cases with solid or part-solid lesions. The combined model results showed that the airway tree structure had a clearer display, and the missing part in the pulmonary lobes could be fixed using the closing method, as shown in Figure 4.
The performance of the models assessed by three metrics on the test set are summarized in Table 1. In the test set, the DL segmentation model demonstrated promising performance in pulmonary lobes segmentation. The all-lobes overall Dice coefficient, Hausdorff distance, and Jaccard coefficient were 0.951±0.013, 112.69±18.877 mm, and 0.914±0.023, respectively. The DL segmentation model could not obtain a clear airway tree structure in airway tree segmentation; the Dice coefficient, Hausdorff distance, and Jaccard coefficient were 0.512±0.042, 96.466±23.210 mm, and 0.393±0.050, respectively. The DL segmentation and post-processing model had a higher performance in pulmonary lobes and airway trees, especially in airway tree segmentation. The all-lobes overall Dice coefficient, Hausdorff distance, and Jaccard coefficient were 0.972±0.013, 12.025±3.67 mm, and 0.948±0.024, respectively, and the airway-tree Dice coefficient, Hausdorff distance, and Jaccard coefficient were 0.849±0.039, 32.076±12.528 mm, and 0.815±0.048, respectively. The Dice coefficient and Jaccard coefficient of the DL model and the combined model were not statistically different, while the Hausdorff distance was significantly different between the two models.
Table 1
| Model | Right upper lobe | Right middle lobe | Right lower lobe | Left upper lobe | Left lower lobe | All-lobe | Airway |
|---|---|---|---|---|---|---|---|
| Dice coefficient (range 0–1) | |||||||
| Dense V-network model | 0.954 | 0.925 | 0.964 | 0.945 | 0.967 | 0.951 | 0.512 |
| Combined model | 0.976 | 0.964 | 0.98 | 0.965 | 0.976 | 0.972 | 0.849 |
| Hausdorff distance (mm) | |||||||
| Dense V-network model | 94.097 | 140.218 | 98.512 | 138.661 | 91.961 | 112.690 | 96.466 |
| Combined model | 7.128 | 12.214 | 9.367 | 24.898 | 6.519 | 12.025 | 32.076 |
| Jaccard coefficient (range 0–1) | |||||||
| Dense V-network model | 0.918 | 0.873 | 0.934 | 0.906 | 0.939 | 0.914 | 0.393 |
| Combined model | 0.956 | 0.932 | 0.962 | 0.936 | 0.955 | 0.948 | 0.815 |
Combined model: dense V-network combined with post-processing methods. DenseVNet, dense V-Network.
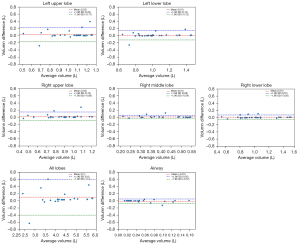
We further used Bland-Altman plots to evaluate the agreement between the combined model (DL model with post-processing method) and ground truth segmentations based on the voxel volumes in the test set. Good agreement was observed between our segmentation in every plot because 95% of cases in the dataset fell within two standard deviations (within an interval between two dotted lines), as shown in Figure 5.
Discussion
We showed that this automatic DL-based post-processing method could be a valuable tool for the simultaneous segmentation of the lung lobes and airway tree. In our study, we chose independent CT images other than the fusion PET/CT scans to do the segmentation due to the high image quality of the chest CT scans. This study serves as a preliminary attempt at the feasibility of the segmentation methods, and further works will focus on the additional functional information from PET images.
To our best knowledge, DL algorithms are currently widely used in the field of medical image segmentation, especially in CNN. In the lung segmentation task, a model that automatically segments the lung lobes and lung airway trees can help radiologists improve efficiency and accuracy and optimize the diagnosis process in actual clinical applications. In previous studies, the use of DL algorithms to train the model is more common for the segmentation of pulmonary lobes, with an average accuracy of over 0.9 (38,41-44). In pulmonary airway tree segmentation, some traditional image processing algorithms are usually used, such as the region growing algorithm. Due to the complexity and instability of the airway tree segmentation issues, few DL models can solve the segmentation problem of lung lobes and trachea at the same time. Besides, some partially solid nodules and other lesions also cause inaccurate lung segmentations. Our study leveraged the DenseVNet with the post-processing method into the pulmonary lobes and airway trees segmentation task, which would improve the overall segmentation accuracy.
The improved DenseVNet exhibited better performance in segmenting multi-organs on abdominal CT compared with previous models (34). The shallow V-network architecture and densely linked layers are important factors in this network. Normal organs can achieve accurate segmentation results, but our datasets’ occupancy lesions like lung nodules lead to unsatisfactory results. After using the post-processing method in the original results of pulmonary lobes and airway trees, the accuracy of the model could be increased. Figure 4 shows that the missing parts of the pulmonary lobes are filled in after using the closing method in the segmentation of the pulmonary lobes. Figure 6 demonstrates clearer airway tree segmentation using the seed point in the threshold (−1,024, M) range via the region growth method.
In our combined model, the average Dice coefficient of the five lung lobes was more than 0.97 on the test set, which shows that DenseVNet combined with post-processing methods has a high performance in segmenting pulmonary lobes and airway trees without interactive manual correction. The Dice coefficient and Jaccard coefficient of the DL model and the combined model were not statistically different, but the Hausdorff distance was significantly different between the two models. The possible reason for this was the small sample size of the test set, and the Dice coefficient and Jaccard coefficient of the DL model were also relatively high. In the case of lung disease, the automatic segmentation method can potentially improve disease detection and treatment strategies. Our automatic segmentation model was mainly used for the automatic segmentation of lesions rather than disease identification and diagnosis. Therefore, the advantage for users was to shorten segmentation time and increase accuracy.
In the previous studies of pulmonary lobe segmentation, there has been some difficulty in segmenting the right middle lobe, with a Dice coefficient ranging from 0.85 to 0.94 when employing a DL algorithm (38,41-44) and the traditional image processing approaches to gain a lower performance in the right middle lobe (45). Our current model has a high Dice coefficient in segmenting the right middle lobe, 0.964 in the test set. This result illustrates the generalizability and robustness of our model.
There were some limitations to our study. First, this was a single-center study; the training and test data set were from a single hospital, and the final model should be validated and tested on multi-center data sets. Second, patients with lung nodules were involved in this study, and it is necessary to include other lung disease data to improve the model’s generalization in future studies. Overall, we proposed a combined DL-based post-processing method that had good performance for segmenting the lung lobes and airway trees. The efficiency based on a single DL and joint model was compared. Our results indicate this combined model can be helpful in precise anatomical information for further PET/CT lesion location issues.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81501513), the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (No. 2017-I2M-1-001), CAMS Innovation Fund for Medical Sciences (No. CIFMS 2021-I2M-1-002), and the National Key Research and Development Program of China (No. 2016YFC0901500).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1116/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1116/coif). XZ, YN, and SW are employed at GE Healthcare, whose products or services may be related to the subject matter of the article, although these authors provided technical support only. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Peking Union Medical College Hospital, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li L, Zhao X, Lu W, Tan S. Deep Learning for Variational Multimodality Tumor Segmentation in PET/CT. Neurocomputing 2020;392:277-95. [Crossref] [PubMed]
- Fishman A, Fessler H, Martinez F, McKenna RJ Jr, Naunheim K, Piantadosi S, Weinmann G, Wise R. Patients at high risk of death after lung-volume-reduction surgery. N Engl J Med 2001;345:1075-83. [Crossref] [PubMed]
- Zhu W, Liu C, Fan W, Xie X. Deeplung: 3d deep convolutional nets for automated pulmonary nodule detection and classification. arXiv preprint arXiv:170905538 2017.
10.1101/189928 10.1101/189928 - Tang H, Kim DR, Xie X, editors. Automated pulmonary nodule detection using 3D deep convolutional neural networks. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018: IEEE.
- Weisman AJ, Kieler MW, Perlman SB, Hutchings M, Jeraj R, Kostakoglu L, Bradshaw TJ. Convolutional Neural Networks for Automated PET/CT Detection of Diseased Lymph Node Burden in Patients with Lymphoma. Radiol Artif Intell 2020;2:e200016. [Crossref] [PubMed]
- Bianconi F, Fravolini ML, Pizzoli S, Palumbo I, Minestrini M, Rondini M, Nuvoli S, Spanu A, Palumbo B. Comparative evaluation of conventional and deep learning methods for semi-automated segmentation of pulmonary nodules on CT. Quant Imaging Med Surg 2021;11:3286-305. [Crossref] [PubMed]
- Ni Y, Xie Z, Zheng D, Yang Y, Wang W. Two-stage multitask U-Net construction for pulmonary nodule segmentation and malignancy risk prediction. Quant Imaging Med Surg 2022;12:292-309. [Crossref] [PubMed]
- Kuhnigk JM, Dicken V, Zidowitz S, Bornemann L, Kuemmerlen B, Krass S, Peitgen HO, Yuval S, Jend HH, Rau WS, Achenbach T. Informatics in radiology (infoRAD): new tools for computer assistance in thoracic CT. Part 1. Functional analysis of lungs, lung lobes, and bronchopulmonary segments. Radiographics 2005;25:525-36. [Crossref] [PubMed]
- Lassen B, van Rikxoort EM, Schmidt M, Kerkstra S, van Ginneken B, Kuhnigk JM. Automatic segmentation of the pulmonary lobes from chest CT scans based on fissures, vessels, and bronchi. IEEE Trans Med Imaging 2013;32:210-22. [Crossref] [PubMed]
- Chiplunkar R, Reinhardt JM, Hoffman EA, editors. Segmentation and quantitation of the primary human airway tree. Medical Imaging 1997: Physiology and Function from Multidimensional Images; 1997: International Society for Optics and Photonics.
- Tozaki T, Kawata Y, Niki N, Ohmatsu H, Kakinuma R, Eguchi K, Kaneko M, Moriyama N. Pulmonary organs analysis for differential diagnosis based on thoracic thin-section CT images. IEEE Transactions on Nuclear Science 1998;45:3075-82. [Crossref]
- Law TY, Heng P, editors. Automated extraction of bronchus from 3D CT images of lung based on genetic algorithm and 3D region growing. Medical Imaging 2000: Image Processing; 2000: International Society for Optics and Photonics.
- Mori K, Hasegawa J, Suenaga Y, Toriwaki J. Automated anatomical labeling of the bronchial branch and its application to the virtual bronchoscopy system. IEEE Trans Med Imaging 2000;19:103-14. [Crossref] [PubMed]
- Sonka M, Sundaramoorthy G, Hoffman EA, editors. Knowledge-based segmentation of intrathoracic airways from multidimensional high-resolution CT images. Medical imaging 1994: physiology and function from multidimensional images; 1994: International Society for Optics and Photonics.
- Aykac D, Hoffman EA, McLennan G, Reinhardt JM. Segmentation and analysis of the human airway tree from three-dimensional X-ray CT images. IEEE Trans Med Imaging 2003;22:940-50. [Crossref] [PubMed]
- Bilgen D. Segmentation and analysis of the human airway tree from 3D X-ray CT images: University of Iowa; 2000.
- Kiraly AP. 3D image analysis and visualization of tubular structures. The Pennsylvania State University; 2003.
- Park W, Hoffman EA, Sonka M. Segmentation of intrathoracic airway trees: a fuzzy logic approach. IEEE Trans Med Imaging 1998;17:489-97. [Crossref] [PubMed]
- Kitasaka T, Mori K, Hasegawa J-i, Suenaga Y, Toriwaki J-i, editors. Extraction of bronchus regions from 3D chest X-ray CT images by using structural features of bronchus. International Congress Series; 2003: Elsevier.
- Schlathoelter T, Lorenz C, Carlsen IC, Renisch S, Deschamps T, editors. Simultaneous segmentation and tree reconstruction of the airways for virtual bronchoscopy. Medical imaging 2002: image processing; 2002: International Society for Optics and Photonics.
- LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998;86:2278-324. [Crossref]
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 2012;25:1097-105.
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556 2014.
- Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A, editors. Going deeper with convolutions. Computer Vision and Pattern Recognition (CVPR). 2015 IEEE Conference on; 2015: IEEE.
- He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016.
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, editors. Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017.
- Kisilev P, Walach E, Barkan E, Ophir B, Alpert S, Hashoul SY. From medical image to automatic medical report generation. IBM Journal of Research and Development 2015;59:2:1-2:7.
- Shen W, Zhou M, Yang F, Yang C, Tian J. Multi-scale Convolutional Neural Networks for Lung Nodule Classification. Inf Process Med Imaging 2015;24:588-99. [Crossref] [PubMed]
- Ding J, Li A, Hu Z, Wang L, editors. Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2017: Springer.
- Men K, Zhang T, Chen X, Chen B, Tang Y, Wang S, Li Y, Dai J. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med 2018;50:13-9. [Crossref] [PubMed]
- Norman B, Pedoia V, Majumdar S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology 2018;288:177-85. [Crossref] [PubMed]
- Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- Noh H, Hong S, Han B, editors. Learning deconvolution network for semantic segmentation. Proceedings of the IEEE international conference on computer vision; 2015.
- Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, Davidson B, Pereira SP, Clarkson MJ, Barratt DC. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans Med Imaging 2018;37:1822-34. [Crossref] [PubMed]
- Imran AAZ, Hatamizadeh A, Ananth SP, Ding X, Tajbakhsh N, Terzopoulos D. Fast and automatic segmentation of pulmonary lobes from chest CT using a progressive dense V-network. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization 2020;8:509-18. [Crossref]
- Nadeem SA, Hoffman EA, Sieren JC, Comellas AP, Bhatt SP, Barjaktarevic IZ, Abtin F, Saha PK. A CT-Based Automated Algorithm for Airway Segmentation Using Freeze-and-Grow Propagation and Deep Learning. IEEE Trans Med Imaging 2021;40:405-18. [Crossref] [PubMed]
- Garcia-Uceda A, Selvan R, Saghir Z, Tiddens HAWM, de Bruijne M. Automatic airway segmentation from computed tomography using robust and efficient 3-D convolutional neural networks. Sci Rep 2021;11:16001. [Crossref] [PubMed]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297-302. [Crossref]
- Rote G. Computing the minimum Hausdorff distance between two point sets on a line under translation. Information Processing Letters 1991;38:123-7. [Crossref]
- Zhang Z, Ren J, Tao X, Tang W, Zhao S, Zhou L, Huang Y, Wang J, Wu N. Automatic segmentation of pulmonary lobes on low-dose computed tomography using deep learning. Ann Transl Med 2021;9:291. [Crossref] [PubMed]
- Ferreira FT, Sousa P, Galdran A, Sousa MR, Campilho A, editors. End-to-End Supervised Lung Lobe Segmentation. 2018 International Joint Conference on Neural Networks (IJCNN); 2018 8-13 July 2018.
- Park J, Yun J, Kim N, Park B, Cho Y, Park HJ, Song M, Lee M, Seo JB. Fully Automated Lung Lobe Segmentation in Volumetric Chest CT with 3D U-Net: Validation with Intra- and Extra-Datasets. J Digit Imaging 2020;33:221-30. [Crossref] [PubMed]
- Wang X, Teng P, Lo P, Banola A, Kim G, Abtin F, Goldin J, Brown M, editors. High Throughput Lung and Lobar Segmentation by 2D and 3D CNN on Chest CT with Diffuse Lung Disease. Image Analysis for Moving Organ, Breast, and Thoracic Images; 2018 2018//; Cham: Springer International Publishing.
- Chen X, Zhao H, Zhou P. Lung Lobe Segmentation Based on Lung Fissure Surface Classification Using a Point Cloud Region Growing Approach. Algorithms 2020;13:263. [Crossref]



