
Clinical evaluation of a novel atlas-based PET/CT brain image segmentation and quantification method for epilepsy
Introduction
Epilepsy is a disorder that results from the disruption of central nervous system function. Between 0.5 and 1% of the global population suffers from epilepsy (1). Epilepsy usually causes the patient to experience recurrent seizures due to disrupted nerve cell activity. Most patients with epilepsy can control their condition with medication; however, one-third of patients have drug-refractory epilepsy (2). For these patients, accurate localization of the epileptogenic foci with medical imaging and subsequent neurosurgery can eliminate or mitigate seizures. As a functional imaging method, positron emission tomography (PET)/computed tomography (CT) has the unique capability to image cerebral metabolism (3). The most common PET/CT brain imaging agent, [18F]fluorodeoxyglucose {[18F]FDG}, uses parameters such as the metabolic rate of glucose in the brain and the standard uptake values to diagnose epilepsy.
In clinical practice, visual interpretation is widely used as a subjective method to evaluate PET/CT images in epilepsy. The results largely depend on the experience of the diagnostician; thus, this method has a number of disadvantages, including that it is highly subjective and difficult to explain (4). To achieve an objective evaluation, the intracerebral asymmetric index (AI) measurement method uses brain areas on the normal side of the same patient as a reference for assessing the epileptic status of the other side. Mirror-image regions of interest (ROIs) of the same size, shape, and area are drawn in the epileptic and normal areas of the brain, and their average standardized uptake value (SUV) and AI value are calculated (5). However, the AI method relies heavily on the physician’s diagnostic experience and has poor repeatability (5,6).
To address the limitations of the AI method, eliminate the subjective errors, and provide visual reports for comprehensive preoperative evaluation of patients with epilepsy, a series of methods based on standard brain mapping have been proposed. Currently, statistical parameter mapping (SPM), Scenium software, and MIMneuro software (7-9) are the most commonly used tools for brain image processing and analysis. Under these methods, a patient’s images are registered to a standard brain template, and the data are compared to data from a standard database to diagnose epilepsy. The voxel-based automatic processing and analysis of these methods have increased their objectivity and repeatability (10-12). However, due to the anatomical variation characteristics of individual brains, spatial mismatch problems occur often when these methods are used. In SPM and Scenium, such problems affect the subsequent quantitative analyses of sub-brain regions; however, the MIMnero software has the advantage of relying on standardized PET template registration and is thus a good choice for routine clinical studies, especially when individual magnetic resonance imaging (MRI) data are not available (7,13). Nevertheless, this quantification method is vulnerable to spillover effects, and it cannot completely avoid white matter interference (7).
In recent years, deep-learning and radiomics-based analysis and diagnostic methods have attracted extensive attention, owing to their ability to uncover the hidden information in digital images and potentially improve the accuracy of diagnosis, prediction, and classification (14). Abbasi et al. summarized machine-learning applications in the automatic detection of epilepsy, the analysis of image and clinical data, the localization of epilepsy, and the prediction of medical and surgical results (15). Studies have been conducted to establish radiation histological prediction models for low-grade glioma-related epilepsy based on MRI data, thus enabling the individualized treatment of patients with this type of epilepsy (16-19). Zhang et al. proposed a Siamese convolutional neural network based on cube pair for the accurate localization of epileptic foci, and then used the AI to automatically calculate and predict the degree of metabolic abnormalities of the foci (20). However, there are also several potential obstacles to the use of machine-learning applications, including the size of the training data set, the accuracy of the referenced label, confounded clinical variables, and variability in data collection and interpretation (21-23).
To meet the requirements for personalized whole-brain modeling, we previously developed an atlas-based method incorporating local anatomical landmarks and dual-modality information to segment brain volumes of interest (23). Under this method, the inaccurate registration caused by anatomical variations of individual brains could be avoided (24). To the best of our knowledge, this is the first study to investigate the effectiveness of this method for the localization and diagnosis of epilepsy using clinical PET brain imaging. To evaluate our proposed method, its registration and segmentation accuracy was compared against that of manual operation and SPM software, and its epileptic foci localization accuracy was compared to that of visual assessment by experienced physicians. We present the following article in accordance with the MDAR reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1005/rc).
Methods
Brain segmentation and quantification method
To achieve accurate head structure segmentation, we registered a high-quality mesh-based head anatomy template (25-27) from a fused PET/CT image of each participant and used this template to segment the head image accordingly. This template was in the form of a polygonal surface mesh and was converted into a volumetric label image for registration with the fusion image from dual-modality PET/CT.
If an atlas contains the whole head (i.e., the skin and skull structure in addition to the brain), then the skull structure information can be used to achieve a more accurate result during registration to the skull structure in the CT image. Thus, unlike conventional atlas-based image segmentation that uses only an image of the brain, our proposed method uses a head atlas containing the entire skull structure as the segmentation template to achieve more accurate and robust brain alignment. As none of the existing brain atlases include the skull structure, we used the freely available BodyParts3D (25) model for whole-head anatomy modeling and replaced its brain structures with the Chinese 2020 (26) atlas for accurate brain anatomy representation. The combined template was converted into a volumetric image with a 1.0-mm voxel size, and the voxel intensities were assigned according to the representative CT values of the head structures.
For image registration of the atlas template, the fused PET/CT images served as the target images, and the atlas images served as the moving images. Intensity-based registration can be used to accurately match the global organ structure while ignoring local anatomical details (28-30). Landmark-based registration can accurately register the image details around the anatomical landmark but does not use the intensity information of the image, which leads to inaccuracies in global registration. To examine both the global and local anatomical structures, a landmark intensity combination strategy that used both image intensity- and anatomical landmark-based registration was adopted. These two registration operations were performed separately to obtain both the global and local deformation fields, and then they were combined to generate the final deformation field that was used to warp the atlas template for brain area segmentation.
In relation to landmark definition, which is a prerequisite step for atlas registration, we used a Deep Q-Network (31-34) to detect the center points of the small brain regions and used them as key landmarks to guide the registration of the small brain structures. The following six ROIs were defined as landmarks in the central brain: the left and right thalamus (THA_L and THA_R), the left and right caudate nucleus (CAU_L and CAU_R), and the left and right putamen (PUT_L and PUT_R). The peripheral regions in the grey matter were not used as landmarks, as they had already been registered accurately through the dual-modality intensity-based registration.
After brain segmentation, a voxel-based PET quantitation database, which included the mean and standard deviation (SD) of the SUV, was used to quantify the PET images of the participants. The database was constructed based on brain PET image data collected from 116 patients, whose ages ranged from 24 to 81 years. None of the 116 patients had a previous history of brain disease or neurological disorder, and the PET/CT images of their brains were considered normal based on an analysis by a physician. To exclude diseased hypometabolic and activated pixels, the SUVs of the PET images of each patient were normalized to the same range based on 40–90% of the original SUVs. The mean and SD of the normalized SUV for each voxel were stored in the database for further evaluation. To test each voxel of each patient’s image, the SUV was compared to the mean value in the database. Voxels that had an SUV that deviated by >2.5 SD (35) were marked as abnormal.
Data preparation
We retrospectively evaluated our method in 36 randomly selected patients who underwent [18F]FDG-PET/CT scans at The First Affiliated Hospital of Fujian Medical University. All personal information of the patients was removed. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Institutional Review Board of The First Affiliated Hospital of Fujian Medical University. The requirement to obtain individual consent was waived for this retrospective analysis.
Of the 36 patients selected, 11 had pathologically confirmed epilepsy, and the other 25, who had no previous history of neurological disease and potential epilepsy, were treated as a reference group. The age of the patients in the reference group ranged from 18 to 83 years, and that of the patients in the epileptic group ranged from 9 to 44 years. All the patients fasted for at least 6 hours, and their blood glucose levels were monitored (<200 mg/dL). The patients were then injected with 3.7 MBq/kg [18F]FDG, and left to rest with their eyes closed in a dark and quiet environment for a 40-minute uptake period. The PET data were acquired on a clinical PET/CT system (SIEMENS Biograph64 mCT, Siemens Healthcare, Erlangen, Germany) with a head holder, using a 10-minute bed position and three-dimensional whole-head acquisition. The primary energy window was set between 435 and 650 keV. All the patients underwent unenhanced low-dose CT (120 kV, 150 mAs) for attenuation correction and anatomical reference. The PET images were reconstructed into coronal-transaxial-sagittal orientation slices (400×400×222 matrices with a voxel size of 0.102 cm) with vendor-provided software using a TrueX (high-definition PET) method that incorporated system spatial-resolution, scatter correction, random correction, decay correction, and CT-based attenuation correction.
Analysis of the segmentation results
To investigate the accuracy of the brain region segmentation, sex regions (THA_L, THA_R, PUT_L, PUT_R, CAU_L, and CAU_R) were selected as the ROIs for all 36 patients. The six regions were manually delineated using ITK-SNAP software (36) (version 3.6.0) by a nuclear medicine physician who is a specialist in epilepsy diagnosis and has extensive neuroimaging experience. Manual segmentation was conducted by an operator, who was blinded to the other information, and the results served as the gold standard for evaluating both our proposed method and the SPM software (9) (version 12). For the SPM process, we first co-registered the CT images with the PET images of each patient, and co-registered the “transm” template (37) with the Neuromorphometrics atlas (38), which was provided by the SPM software. The “transm” template was aligned and registered to the CT image of each patient to generate the deformation information between them using mutual information as the objective function of the registration. The deformation information was then used on the Neuromorphometrics atlas to co-register it with the PET and CT images. As a result, we obtained specifically matched segmentation results for the PET images.
The dice coefficient of each selected brain region was used to quantify the segmentation accuracy of the proposed method and SPM results using the manual delineation as the reference standard. The following formula was used:
where X is the mask of the specific brain region of the different methods (i.e., the proposed method or SPM), and R is the mask of the referenced manual method. For both the reference and epileptic groups, the dice results of each method in each region were averaged to obtain an overall evaluation metric (DICE-A), and a paired t-test was conducted on the DICE-A to assess the statistical significance of the proposed and reference methods using the following formula:
where x1, x2, and s1, s2 are the mean value and SD value of the two different paired groups, and n is the number of each group. The P value was derived from the t-distribution table after the t-value had been obtained.
Additionally, for all three methods, we calculated the average SUV (SUV-A) of each ROI. The SUV of each ROI was averaged for both patient groups to obtain the SUV-A to evaluate quantification efficacy of the three methods.
Analysis of diagnostic results
The diagnostic and localization results obtained using the proposed method were compared to those of visual assessment by physicians and the pathological results for the 11 patients who had a clear diagnosis of epilepsy. The visual assessments were conducted by two nuclear medicine physicians who have extensive neuroimaging experience. Areas of low metabolic intensity in the PET images were considered abnormal, and their locations were recorded. The pathological results were derived from the results of brain samples taken after comprehensive clinical diagnosis (e.g., by stereoelectroencephalography or intracranial electroencephalography) and served as the gold standard for diagnosis. For the proposed method, a voxel-based quantitation analysis of each PET/CT image was performed, and voxels with an SUV deviation of >2.5 from the mean value of the standard database were marked as abnormal. The [18F]FDG-PET images were also visually assessed by two experienced physicians, who marked the brain regions they believed to have abnormalities as a result of epileptic defects.
Results
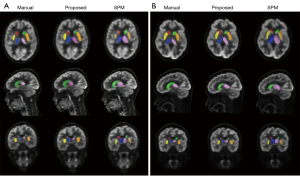
Figure 1 shows the segmentation results of the six selected brain regions from one sample patient with epilepsy and one sample reference patient using our proposed method, SPM, and the referenced manual method. Both the proposed and the SPM method located the six brain regions correctly, but the segmentation results of the proposed method were more similar to those of the referenced manual method. The segmented caudate nucleus and shell nucleus regions were smaller with SPM than with the manual and proposed methods, whereas the segmented thalamus region was bigger.
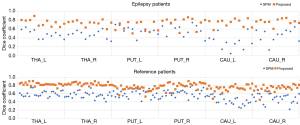
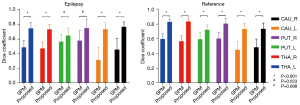
The dice coefficient results are set out in Figure 2, and the averaged DICE-A results are set out in Figure 3. In general, the dice results of the proposed method were higher than those of SPM, with the average dice values for the proposed and SPM methods being 0.78 and 0.55 in the reference group, respectively (P<0.001), and 0.73 and 0.48 in the epileptic group, respectively (P<0.001). Compared to SPM, the proposed method improved the dice value by 41.31% and 52.18% in the reference and epileptic groups, respectively. In terms of the different brain regions, the DICE-A results of the proposed method were significantly higher (P<0.05) than the SPM results for all regions except for the left putamen region in the epileptic group, for which the proposed method had higher DICE-A results but the difference was not significant (P>0.05). As Figure 2 shows, the dice values for both methods were higher in the reference group than the epileptic group. The distribution of the dice results for SPM was more discrete in the caudate nucleus region than in the other regions for both epileptic and reference groups, while the results of the proposed method were distributed more consistently across all six regions.
The SUV-A results are shown in Figure 4. In the epilepsy group, the SUV differences for the SPM method relative to the referenced manual method were 25.33%, 28.94%, 15.14%, 14.17%, 38.71%, and 34.54% for the CAU_R, CAU_L, PUT_R, PUT_L, THA_R and THA_L regions, respectively, and those for the proposed method were 8.35%, 8.80%, 7.55%, 3.68%, 3.17%, and 3.03%, respectively. In the reference group, the SUV differences for the SPM method relative to the referenced manual method were 27.25%, 22.20%, 7.95%, 11.21%, 31.81%, and 22.40% for the CAU_R, CAU_L, PUT_R, PUT_L, THA_R, and THA_L regions, respectively, and those for the proposed method were 6.70%, 1.57%, 0.91%, 0.21%, 7.01%, and 7.01%, respectively. The quantification results of the proposed method were close to those of the referenced method, especially in the PUT regions, while the SPM method results deviated more from those of the reference method, especially in the THA regions.
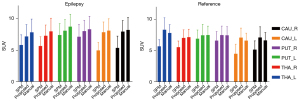
Figure 5 shows a sample diagnostic result using the proposed method; the blue region is the abnormal area suggested by the proposed algorithm. Table 1 shows the diagnostic results using the proposed method, visual assessment, and pathological examination. Notably, among the 11 patients with epilepsy, all the pathologically reported epileptic defects were detected by our proposed method, whereas visual assessment missed epileptic defects in three patients (patients no. 2, no. 4, and no. 5).
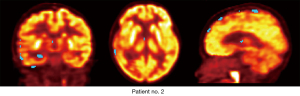
Table 1
| Patient no. | Location | Visual assessment | Pathology | Proposed method |
|---|---|---|---|---|
| 1 | Left hippocampus | √ | √ | √ |
| Left temporal lobe | √ | |||
| 2 | Right occipital lobe | √ | ||
| Right parietal lobe | √ | |||
| Right frontal lobe | √ | √ | ||
| Left Hippocampus | √ | |||
| Left temporal lobe | √ | |||
| 3 | Right hippocampus | √ | √ | √ |
| Right temporal lobe | √ | √ | √ | |
| Right parietal lobe | √ | √ | √ | |
| Right occipital lobe | √ | |||
| Left temporal lobe | √ | |||
| Right thalamus | √ | |||
| 4 | Left frontal lobe | √ | √ | |
| Left temporal lobe | √ | √ | ||
| Lenticular nucleus | √ | |||
| Right frontal lobe | √ | |||
| Right temporal lobe | √ | |||
| 5 | Right frontal lobe | √ | ||
| Right parietal lobe | √ | √ | √ | |
| Right temporal lobe | √ | √ | √ | |
| Right hippocampus | √ | √ | ||
| 6 | Right frontal lobe | √ | √ | √ |
| Right temporal lobe | √ | |||
| Cerebellum | √ | |||
| 7 | Left occipital lobe | √ | √ | √ |
| Left parietal lobe | √ | √ | ||
| Left temporal lobe | √ | √ | ||
| Right temporal lobe | √ | |||
| 8 | Right hippocampus | √ | √ | √ |
| Right temporal lobe | √ | √ | ||
| Right occipital lobe | √ | |||
| 9 | Left temporal lobe | √ | √ | √ |
| Left parietal lobe | √ | √ | √ | |
| Left occipital lobe | √ | √ | √ | |
| 10 | Right temporal lobe | √ | √ | √ |
| Right frontal lobe | √ | |||
| Right hippocampus | √ | √ | √ | |
| Cerebellum | √ | |||
| 11 | Right frontal lobe | √ | √ | √ |
| Right parietal lobe | √ | √ | ||
| Right temporal lobe | √ | √ | √ | |
| Right thalamus | √ | |||
| Right hippocampus | √ | √ | √ |
Discussion
Images obtained by PET/CT play an important role in the diagnosis and localization of epilepsy. Accurate brain region segmentation and precise detection and localization of metabolic abnormalities on PET/CT images, aided by image processing algorithms, are key steps in epilepsy diagnosis and treatment. However, current clinical software for such processing is limited in terms of its accuracy for brain region segmentation and its precision for abnormal region localization. The present study sought to investigate the effectiveness of applying our previously proposed atlas-based method for brain region segmentation in the PET/CT-based diagnosis of epilepsy by evaluating its ability to accurately segment brain regions and automatically identify metabolically abnormal brain regions.
From the evaluation results, we noted that the dice coefficients obtained with our proposed method were generally higher than those obtained with the SPM software in all selected regions in both the reference and epileptic groups. The proposed method had relatively high dice coefficient values in both the epileptic and reference groups, with improvements on the average dice values of 51.63% and 36.52%, respectively. However, in the reference group, the distribution of the dice results for SPM was more discrete in the caudate nucleus region than in the other regions. The thin and long structure of the caudate nucleus may be the cause of this phenomenon. In the epileptic group, the dice values showed broad dispersion across all six brain regions with a wide range of distribution, which demonstrates that SPM lacks robustness for images with abnormalities in the intensity of brain regions.
Given the good performance of our proposed method in brain region segmentation, the integration of dual-modality image registration and landmark constraint is potentially an effective way to improve the accuracy and robustness of brain structure segmentation (24). In relation to dual-modality atlas registration, our segmentation method takes advantage of both the prior anatomical knowledge in the atlas and the dual-modality information in the target image. The use of the atlas guarantees a robust performance, even when the image includes abnormal brain uptake (e.g., uptake in patients with epilepsy), and the combination of dual-modality atlas registration helps to achieve more accurate alignment at the skull-brain border. Further, the entire workflow is fully automatic and does not require any manual intervention, which increases its convenience and the ease with which it can be used in clinical applications.
To analyze the performance of our proposed method with regard to the image intensity-based automatic detection and localization of epileptic foci, we used a database constructed from normal patients as a reference and assessed abnormalities in the images of patients with epilepsy by examining the SUV deviations. From the results of the 11 test patients, all the epileptic foci were detectable using our method, while missed diagnoses (false negatives) occurred using the manual visual judgment method in three patients. We checked the pathology results of these three patients and observed that their epileptogenic foci were relatively small, which may be why they were missed in the visual assessments. The incorrect identification of non-epileptic areas as epileptic areas occurred with both our proposed method and the visual assessment method (e.g., patients no. 7 and no. 8). Both visual assessment and our method had better detection rates than did detection based on pathological results. This finding may be due to the fact that the size of abnormal metabolic regions in PET is usually larger than the size of the true lesion, and because not all abnormal metabolic regions are necessarily epileptic lesions, which can lead to some bias between visual assessment and image intensity-based automatic diagnosis and the true pathological results. In clinical diagnosis, while a false positive is also a misdiagnosis, unlike a false negative, it ensures that all epileptic areas are detected and the patient is referred to the physician for further confirmation. For this reason, we ensured that there were no false negatives in the present study by adjusting the selection range of the SD during the dataset comparison.
As can be seen in Table 1, only the lobes of the patients with epilepsy were indicated in the results, but the results of the brain regions are not shown in more detail. The reason for this is that the study was designed to analyze the effectiveness of the different methods in diagnosing epilepsy, and the physicians were only asked to label the lobes in which epilepsy was observed when analyzing the results. The results in the Table 1 also show the differences in the diagnostic results between the methods. If the epilepsy occurred at the junction of two lobes, both lobes were marked in the table. Notably, such results do not show the epileptic areas in more detail. Thus, in future studies, we will compare and analyze the effectiveness of different methods for diagnosing and localizing epilepsy in a more detailed manner.
Our study had several limitations. First, for the segmentation accuracy assessment, only six central brain regions were evaluated; whole brain areas were not evaluated due to the lack of clarity in other brain regions and the low resolution and contrast of PET/CT images. Further, the database used for epilepsy diagnosis and localization comprised data collected from one center, while the patient data used as the testing data were collected from a different center. The mismatch between the data from the different centers may have affected the accuracy of the quantification results (39). The implementation of a multi-center effect compensation algorithm to improve the accuracy of the quantification results is an issue that needs to be addressed in the future. Additionally, there was a large mismatch between the age of the patients used to create the database and the age of the test patients with epilepsy; this age difference may also have led to inaccurate quantification. The collection of more data to create a larger database is a future research task. Furthermore, cases in which the distance between the skull and the brain parenchyma was significantly larger due to brain atrophy were not included in our study; thus, the effectiveness of the proposed method has not been validated in such patients. In future studies, we will investigate cases with structural differences due to brain atrophy or traumatic brain injury to further improve our method. Moreover, the application of deep-learning techniques for the diagnosis and localization of epilepsy has been proposed by another research team (20). However, the training labels used in their deep-learning method were obtained by manually labeling the areas with visually low metabolic activity on PET/CT images without comparison to the gold standard of pathological results. In a future study, we plan to further combine the pathological results with those of manual recognition to perform a comparison of our proposed method with deep-learning algorithms to investigate how to achieve more accurate localization and diagnosis of epilepsy.
Conclusions
Our proposed atlas-based approach enables the accurate segmentation of [18F]FDG-PET/CT brain regions and the accurate and effective identification and localization of epileptic foci. This method has great potential value for future application in the image processing and clinical diagnosis of patients with epilepsy.
Acknowledgments
Funding: This work was supported by research grants from the National Natural Science Foundation of China (No. 62101510) and the Natural Science Foundation of Fujian Province (No. 2021J01707).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1005/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1005/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of The First Affiliated Hospital of Fujian Medical University, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Michael-Titus A, Revest P, Shortland P. 13 - Epilepsy. In: Michael-Titus A, Revest P, Shortland P. editors. The Nervous System. 2nd ed. London: Churchill Livingstone; 2010:237-50.
- Engel J Jr, Wiebe S, French J, Sperling M, Williamson P, Spencer D, Gumnit R, Zahn C, Westbrook E, Enos BAmerican Association of Neurological Surgeons. Practice parameter: temporal lobe and localized neocortical resections for epilepsy: report of the Quality Standards Subcommittee of the American Academy of Neurology, in association with the American Epilepsy Society and the American Association of Neurological Surgeons. Neurology 2003;60:538-47. [Crossref] [PubMed]
- Su H, Zuo C, Zhang H, Jiao F, Zhang B, Tang W, Geng D, Guan Y, Shi S. Regional cerebral metabolism alterations affect resting-state functional connectivity in major depressive disorder. Quant Imaging Med Surg 2018;8:910-24. [Crossref] [PubMed]
- Zhu Y, Feng J, Wu S, Hou H, Ji J, Zhang K, Chen Q, Chen L, Cheng H, Gao L, Chen Z, Zhang H, Tian M. Glucose Metabolic Profile by Visual Assessment Combined with Statistical Parametric Mapping Analysis in Pediatric Patients with Epilepsy. J Nucl Med 2017;58:1293-9. [Crossref] [PubMed]
- Gaillard WD, Weinstein S, Conry J, Pearl PL, Fazilat S, Fazilat S, Vezina LG, Reeves-Tyer P, Theodore WH. Prognosis of children with partial epilepsy: MRI and serial 18FDG-PET. Neurology 2007;68:655-9. [Crossref] [PubMed]
- Didelot A, Mauguière F, Redouté J, Bouvard S, Lothe A, Reilhac A, Hammers A, Costes N, Ryvlin P. Voxel-based analysis of asymmetry index maps increases the specificity of 18F-MPPF PET abnormalities for localizing the epileptogenic zone in temporal lobe epilepsies. J Nucl Med 2010;51:1732-9. [Crossref] [PubMed]
- Choi WH, Um YH, Jung WS, Kim SH. Automated quantification of amyloid positron emission tomography: a comparison of PMOD and MIMneuro. Ann Nucl Med 2016;30:682-9. [Crossref] [PubMed]
- Mendes Coelho VC, Morita ME, Amorim BJ, Ramos CD, Yasuda CL, Tedeschi H, Ghizoni E, Cendes F. Automated Online Quantification Method for 18F-FDG Positron Emission Tomography/CT Improves Detection of the Epileptogenic Zone in Patients with Pharmacoresistant Epilepsy. Front Neurol 2017;8:453. [Crossref] [PubMed]
- Ashburner J. SPM: a history. Neuroimage 2012;62:791-800. [Crossref] [PubMed]
- Li L, Song M, Zhang C, Qian Z, Li Y, Li R, Li C, Yang Z, Zhou D. Hemangiopericytomas: Spatial Intracranial Location in a Voxel-Based Mapping Study. J Neuroimaging 2020;30:370-7. [Crossref] [PubMed]
- Akman CI, Ichise M, Olsavsky A, Tikofsky RS, Van Heertum RL, Gilliam F. Epilepsy duration impacts on brain glucose metabolism in temporal lobe epilepsy: results of voxel-based mapping. Epilepsy Behav 2010;17:373-80. [Crossref] [PubMed]
- Kim YK, Lee DS, Lee SK, Kim SK, Chung CK, Chang KH, Choi KY, Chung JK, Lee MC. Differential features of metabolic abnormalities between medial and lateral temporal lobe epilepsy: quantitative analysis of (18)F-FDG PET using SPM. J Nucl Med 2003;44:1006-12. [PubMed]
- Guo K, Yuan M, Wei L, Lu J. Epileptogenic zone localization using a new automatic quantitative analysis based on normal brain glucose metabolism database. Int J Neurosci 2021;131:128-34. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Abbasi B, Goldenholz DM. Machine learning applications in epilepsy. Epilepsia 2019;60:2037-47. [Crossref] [PubMed]
- Liu Z, Wang Y, Liu X, Du Y, Tang Z, Wang K, Wei J, Dong D, Zang Y, Dai J, Jiang T, Tian J. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. Neuroimage Clin 2018;19:271-8. [Crossref] [PubMed]
- Sun K, Liu Z, Li Y, Wang L, Tang Z, Wang S, Zhou X, Shao L, Sun C, Liu X, Jiang T, Wang Y, Tian J. Radiomics Analysis of Postoperative Epilepsy Seizures in Low-Grade Gliomas Using Preoperative MR Images. Front Oncol 2020;10:1096. [Crossref] [PubMed]
- Wang Y, Wei W, Liu Z, Liang Y, Liu X, Li Y, Tang Z, Jiang T, Tian J. Predicting the Type of Tumor-Related Epilepsy in Patients With Low-Grade Gliomas: A Radiomics Study. Front Oncol 2020;10:235. [Crossref] [PubMed]
- Yang Q, Zhang H, Xia J, Zhang X. Evaluation of magnetic resonance image segmentation in brain low-grade gliomas using support vector machine and convolutional neural network. Quant Imaging Med Surg 2021;11:300-16. [Crossref] [PubMed]
- Zhang Q, Liao Y, Wang X, Zhang T, Feng J, Deng J, et al. A deep learning framework for 18F-FDG PET imaging diagnosis in pediatric patients with temporal lobe epilepsy. Eur J Nucl Med Mol Imaging 2021;48:2476-85. [Crossref] [PubMed]
- Mo J, Liu Z, Sun K, Ma Y, Hu W, Zhang C, Wang Y, Wang X, Liu C, Zhao B, Zhang K, Zhang J, Tian J. Automated detection of hippocampal sclerosis using clinically empirical and radiomics features. Epilepsia 2019;60:2519-29. [Crossref] [PubMed]
- Wang Y, Hu Q, Zhu P, Li L, Lu B, Garibaldi JM, Li X. Deep fuzzy tree for large-scale hierarchical visual classification. IEEE Transactions on Fuzzy Systems 2019;28:1395-406. [Crossref]
- Wang Y, Wang Z, Hu Q, Zhou Y, Su H. Hierarchical Semantic Risk Minimization for Large-Scale Classification. IEEE Trans Cybern 2021; Epub ahead of print. [Crossref] [PubMed]
- Chen Z, Qiu T, Tian Y, Feng H, Zhang Y, Wang H. Automated brain structures segmentation from PET/CT images based on landmark-constrained dual-modality atlas registration. Phys Med Biol 2021; [Crossref] [PubMed]
- Mitsuhashi N, Fujieda K, Tamura T, Kawamoto S, Takagi T, Okubo K. BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res 2009;37:D782-5. [Crossref] [PubMed]
- Liang P, Shi L, Chen N, Luo Y, Wang X, Liu K, Mok VC, Chu WC, Wang D, Li K. Construction of brain atlases based on a multi-center MRI dataset of 2020 Chinese adults. Sci Rep 2015;5:18216. [Crossref] [PubMed]
- Chen Z, Qiu T, Huo L, Yu L, Shi H, Zhang Y, Wang H. Inter-subject shape correspondence computation from medical images without organ segmentation. IEEE Access 2019;7:130772-81.
- Zhang D, Pretorius PH, Ghaly M, Zhang Q, King MA, Mok GSP. Evaluation of different respiratory gating schemes for cardiac SPECT. J Nucl Cardiol 2020;27:634-47. [Crossref] [PubMed]
- Zhang D, Ghaly M, Mok GSP. Interpolated CT for attenuation correction on respiratory gating cardiac SPECT/CT - A simulation study. Med Phys 2019;46:2621-8. [Crossref] [PubMed]
- Zhang D, Sun J, Pretorius PH, King M, Mok GSP. Clinical evaluation of three respiratory gating schemes for different respiratory patterns on cardiac SPECT. Med Phys 2020;47:4223-32. [Crossref] [PubMed]
- Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, Graves A, Riedmiller M, Fidjeland AK, Ostrovski G, Petersen S, Beattie C, Sadik A, Antonoglou I, King H, Kumaran D, Wierstra D, Legg S, Hassabis D. Human-level control through deep reinforcement learning. Nature 2015;518:529-33. [Crossref] [PubMed]
- Ghesu FC, Georgescu B, Zheng Y, Grbic S, Maier A, Hornegger J, Comaniciu D. Multi-Scale Deep Reinforcement Learning for Real-Time 3D-Landmark Detection in CT Scans. IEEE Trans Pattern Anal Mach Intell 2019;41:176-89. [Crossref] [PubMed]
- Ghesu FC, Georgescu B, Mansi T, Neumann D, Hornegger J, Comaniciu D. An artificial agent for anatomical landmark detection in medical images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer, 2016:229-37.
- Alansary A, Oktay O, Li Y, Folgoc LL, Hou B, Vaillant G, Kamnitsas K, Vlontzos A, Glocker B, Kainz B, Rueckert D. Evaluating reinforcement learning agents for anatomical landmark detection. Med Image Anal 2019;53:156-64. [Crossref] [PubMed]
- Ih G, Waxman A, Rowe C, Minoshima S. The determination of specificity of PET and SPECT brain scans using a quantitative normal database approach. J Nucl Med 2011;52:abstr 1244.
- Yushkevich PA, Gao Yang, Gerig G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. Annu Int Conf IEEE Eng Med Biol Soc 2016;2016:3342-5. [Crossref] [PubMed]
- Rorden C, Bonilha L, Fridriksson J, Bender B, Karnath HO. Age-specific CT and MRI templates for spatial normalization. Neuroimage 2012;61:957-65. [Crossref] [PubMed]
- Kang D, Yarach U. In MH, Gray EM, Trzasko JD, Jo HJ, Shu Y, Huston J 3rd, Bernstein MA. The effect of spiral trajectory correction on pseudo-continuous arterial spin labeling with high-performance gradients on a compact 3T scanner. Magn Reson Med 2020;84:192-205. [Crossref] [PubMed]
- Zhang D, Pretorius PH, Lin K, Miao W, Li J, King MA, Zhu W. A novel deep-learning-based approach for automatic reorientation of 3D cardiac SPECT images. Eur J Nucl Med Mol Imaging 2021;48:3457-68. [Crossref] [PubMed]


