
SMANet: multi-region ensemble of convolutional neural network model for skeletal maturity assessment
Introduction
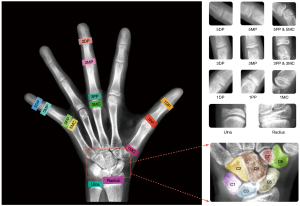
Skeletal maturation occurs through a series of discrete phases, particularly in the wrists and hands. Pediatric medicine uses progressive skeletal growth to assign a bone age and correlate it with the chronological age of the child. Discrepancies in these data facilitate the diagnosis of possible endocrine or metabolic disorders (1). Currently, the left-hand radiograph is widely used for bone age assessment (BAA). The morphological characteristics of bones such as the wrist and phalanges have a vital significance in BAA (2). Over the past decades, BAA has been performed manually using either the Greulich and Pyle (GP) method (3) or the Tanner-Whitehouse 3 (TW3) method (4). In the GP method, BAA is performed by comparing the left-hand radiograph with the GP atlas and evaluating the bone age by identifying the most visually similar bone samples in the atlas. However, interpretations may vary due to the subjective nature of this method (5). In the TW3 method, the maturity levels of 20 regions of interest (ROIs), comprising 13 radius, ulna, and short (RUS) bones and 7 carpal (C) bones are evaluated, and skeletal maturity scores are assigned to individual ROIs (Figure 1). These scores are then summed to obtain the total RUS maturity score and total C maturity score. Then, the scores are finally converted into a bone age using a bone age table. Therefore, the TW3 method is widely used for BAA due to its higher accuracy and interpretability. In the TW3 method, skeletal maturity is an intermediate variable for bone age; it describes the bone development level and is not affected by distinct racial and geographical differences. However, manual skeletal maturity assessment (SMA) is inefficient and depends greatly on the professional experience of radiologists. Therefore, an automatic method for SMA is urgently needed to aid clinicians.
In recent years, the development and application of deep learning techniques based on artificial neural networks has increased rapidly. Deep learning techniques have particularly exhibited superior results in medical image analysis (6-12). The present study established a novel, fully automated SMA method based on the deep learning algorithm that automated the entire process of the TW3 (both of RUS-series and C-series) BAA method.
In this paper, an SMA method was designed that incorporated the TW3 local ROIs with the overall image features of the left-hand radiographs. This method automatically assessed the skeletal maturity of the samples and derived a skeletal maturity score. The sample’s bone age was then derived from the corresponding bone age table. The method, again, was independent of racial and geographical variations and had a clear basis for clinical dissemination.
Related work
Many deep learning frameworks have been proposed for automatic BAA and can be divided into 2 categories. The first category comprises training of neural networks by extracting features of whole-hand images and focusing less on local regions. Spampinato et al. (13) trained and tested various deep learning networks (OverFeat, GoogLeNet, and OxfordNet) on a public dataset and exhibited an average difference of approximately 0.8 years between manual and automated assessments. Lee et al. (1) applied deep convolutional neural networks (CNNs) to eliminate background and detect the hand from radiographs, and subsequently selected 3 CNN models (AlexNet, GoogLeNet, and VGG-16) as regression networks for BAA. Larson et al. (14) trained models using CNNs that were compared by radiologists, exhibiting a mean difference of 0 years between the neural network models and the BAA of radiologists, with a mean effective value and mean absolute difference (MAD) of 0.63 and 0.50, respectively. He et al. (15) proposed a novel, end-to-end BAA approach that was based on lossless image compression and compression-stimulated deep residual networks. However, these methods omitted the inclusion of specific bone parts as ROIs. Consequently, their precision is limited.
The other category is the automatic BAA algorithm that combines local and global features and exhibits better performance. Cao et al. (16) proposed landmark-based, multi-region CNNs for automatic BAA based on whole-hand image and local ROIs. Using attention and recognition agent modules, Liu et al. (2) performed bone local landmark discrimination and extracted image features for bone age prediction, respectively. Wang et al. (17) proposed a new anatomical local awareness network (ALA-Net) for BAA. Chen et al. (18) proposed an attention-guided approach to obtain ROIs from whole-hand images and then aggregated these ROIs for BAA.
These methods provided direct predictions of bone age through an end-to-end architecture. However, Zhang et al. (19) reported racial and geographical variations in the status of pediatric development. Children from different races and geographic regions with the same skeletal maturity exhibit distinct variations in bone ages owing to differences in their developmental status, which to some extent limits the diffusion of relevant automatic BAA algorithms. Furthermore, end-to-end predictive BAA algorithms are limited in clinical dissemination because of the lack of a clear clinical basis.
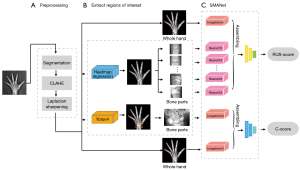
In this paper, a skeletal assessment method based on a multi-region ensemble of CNNs integrating with clinically interpretable methods was proposed to automate the entire process of the TW3 BAA method. In our method, the first step was to automatically remove the image background and enhance the hand bone contrast to pre-process the left-hand radiographs, and then the localization network and object detection network were trained to extract the RUS-ROIs and carpal region. Finally, we trained a feature extraction network to extract the features of both the whole left-hand and the ROIs to assess skeletal maturity scores and bone age. We constructed a database of left-hand radiographs of children with TW3-method annotation for the training and testing of the proposed algorithm. Experimental results on the database demonstrated that our method had an accuracy comparable to that of experienced endocrinologists and radiologists, with even greater stability and efficiency. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1158/rc).
Methods
Overall framework
The architecture of our proposed method comprised 3 parts (Figure 2).
Datasets
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Jishuitan Hospital (Approval No. 201907-11). The present study was conducted on 4,861 left-hand radiographic images of children of different ages obtained from the Beijing Jishuitan Hospital. Each image was labelled in detail by 5 experienced radiologists with 20 epiphyseal scores based on the TW3 scoring method, including 13 RUS-ROIs skeletal maturity scores for the RUS-series and 7 C-ROIs skeletal maturity scores for the C-series. Additionally, the bone age and gender of each child were included in the label. The result was accepted as the ground truth only when the same result was obtained by at least 3 radiologists. Otherwise, an image would be presented to the committee of experts for further validation and determination. Of the total 5,300 radiographs which were acquired between 26 August, 2019 and 31 December, 2019, 439 images with hand deformities and right-hand images were excluded. Thus, the present study was conducted using the remaining 4,861 images. The age distribution of the 4,861 images is illustrated in Figure 3A, with 49% of the images of female origin and 51% of male. The distribution of the skeletal maturity scores of the 4,861 images is illustrated in Figure 3B. The original dataset was unevenly distributed across the skeletal maturity stages (Figure 3B). Therefore, the dataset was equalized based on data augmentation algorithms (Figure 3C). Detailed sources of data and a description of the experimental setup of this study are displayed in the Appendix 1 and Figure S1.
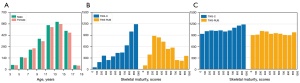
Image preprocessing
Background elimination
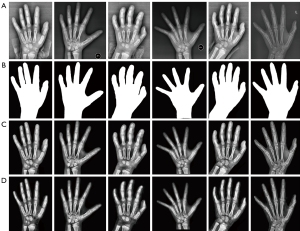
The images in the database were obtained through different devices, and their quality varied greatly due to the differing parameters and radiation doses. A reliable hand segmentation technique was required to extract the hand mask and remove the irrelevant regions. The present study used a variant network of U-Net (20), Attention U-Net (21), which incorporated the additive attention gate module on the traditional U-Net to improve the sensitivity of the model to the foreground pixels of an image. After removing the background, the left-hand radiographs were processed for enhancement with contrast-limited, adaptive histogram equalization and Laplace sharpening algorithms. The pre-processed images are illustrated in Figure 4.
RUS-ROI extraction
The TW3-related ROIs, including RUS and C series, of the left-hand radiographs were acquired and used to assess the skeletal maturity scores. The 13 RUS-ROIs were extracted and utilized to assess the RUS scores. The present study initially located 37 keypoints on the left-hand radiographs and used them as reference marks to cut out the desired 13 RUS-ROIs. Such keypoint localization methods usually require complex network structures and a large amount of training data to provide high accuracy (22). A total of 5 high-performance, keypoint detection CNNs (22-25) were considered candidate networks. Payer et al. (22) conducted a comparative study of these candidate networks. The localization results of these networks were triple cross-validated on 895 left-hand radiographs. The point-to-point error (PEall) and the number of outliers at different error radii (#Or) were used as measures of the keypoint localization error. Spatial Configuration-Net (SCN) exhibited the best results in terms of both local accuracy and robustness to keypoint error identification. Therefore, SCN was selected as the network for keypoint detection in the present experiment. Furthermore, a technique called positive mining, an iterative process, was employed to mitigate the labelling cost. In the positive mining method, manual labelling was combined with automatic processing; hence, this method allowed the rapid acquisition of accurate labels for all images in the training set.
After obtaining the image keypoints (Figure 5), 13 RUS keypoints were selected, and the cropped region was determined on the basis of these keypoints to obtain the RUS-ROIs.
C-ROIs extraction
Detecting specific carpal bones individually is relatively challenging because the bones have a gradual ossification process with differing morphological characteristics at different times. Therefore, the You Only Look Once (YOLO)_v. 4 (26) object detection network was utilized to identify the entire carpal bone region. Then, the entire carpal bone image was cropped out as the input for the C-series score evaluation.
Multi-region ensemble networks in SMA
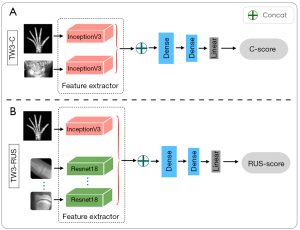
Many advanced backbone networks, such as Inception_v3 (27), ResNet (28), and GoogLeNet (29), have emerged in the field of computer vision. The addition of more fully connected sub-networks to these backbone networks and changing the loss function of the networks can enable regression or classification tasks (30). The Inception_v3 network was chosen as the feature extraction network for the entire hand and wrist. A lightweight ResNet_18 network was used to extract small-sized patches from ROIs. The network adopted the mean absolute error (MAE) as the loss function of this regression task, as follows:
where m denotes the number of data in the set, yi denotes the value of the ith number, and denotes the average of all numbers.
The C-series SMA model architecture comprised 2 separate Inception_v3 sub-networks (Figure 6A), which were used to extract whole-hand and C-ROIs features. The RUS-series SMA model architecture comprised one Inception_v3 network and 13 ResNet_18 networks (Figure 6B), where these networks were responsible for extracting the whole-hand features and RUS-ROIs features, respectively. Furthermore, 2 completely connected layers and a linear layer were added as additional sub-networks to extract more useful features and reduced the feature space dimension to save computational effort.
Results
Image pre-processing
The accuracy of keypoint localization and carpal bone region detection directly affects the effectiveness of ROIs extraction. To compare the effects of different pre-processing methods on keypoint localization and carpal bone region detection, comparison experiments were conducted. As shown in Table 1, the keypoint localization and carpal bone region detection models performed better after background elimination and contrast enhancement were applied to the left-hand radiographs. In this study, the accurate extraction of ROIs determined the reasonableness and accuracy of the skeletal maturity assessment network (SMANet) prediction scores. Therefore, the performance of the pre-processing methods was closely related to the prediction results of the final model.
Table 1
| Method | RUS-ROIs (SCN) | C-ROIs (YOLO_v4) | ||||||
|---|---|---|---|---|---|---|---|---|
| PEall | Outliers (>10 mm) (%) | Accuracy (%) | Recall (%) | Precision (%) | F1 (%) | |||
| Median (mm) | Mean ± SD (mm) | |||||||
| Original image | 0.92 | 0.97±1.01 | 17 | 95.83 | 98.09 | 97.63 | 97.86 | |
| Enhanced contrast | 0.87 | 0.91±0.96 | 12 | 96.83 | 99.07 | 97.70 | 98.38 | |
| Background elimination | 0.63 | 0.80±0.93 | 8 | 97.11 | 99.22 | 97.85 | 98.53 | |
| Background elimination and enhanced contrast | 0.42 | 0.54±0.60 | 6 | 97.30 | 99.34 | 97.29 | 98.30 | |
RUS-ROIs, radius, ulna, and short bones regions of interest; C-ROIs, carpal bones regions of interest; SCN, Spatial Configuration-Net; YOLO, You Only Look Once; SD, standard deviation; PEall, point-to-point error; ; ; ; . TP, true positives; FP, false positive; FN, false negatives; TN, true negatives.
The extraction of ROIs
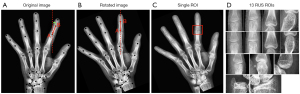
The SCN network was evaluated on the test sets, and the median of the outliers radii was statistically obtained as 0.42 mm. The median standard deviation of the outliers radius was 0.54±0.60 mm, and the outliers larger than 10 mm accounted for about 6% of the overall keypoints. After locating the keypoint, the tilt angle of the bone patches was calculated using the 2 adjacent keypoints. The whole-hand image was then rotated to ensure that the bone patches were in a vertical position. The process is illustrated in Figure 7. If the vertical image of the A joint had to be acquired, the tilt angle of the line between point A and point B was initially calculated, and then the image was rotated to ensure that the AB line was in a vertical state. The bone patch was subsequently cropped (Figure 7C), with A as the center. Eventually, 13 epiphyseal images were acquired, as shown in Figure 7D.
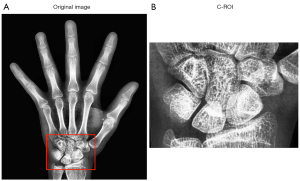
For the carpal bone region detection task, many advanced object detection networks, such as Mask R-CNN (31), Fast R-CNN (32), and YOLO_v. 4, were available. A comparison experiment was conducted separately to identify the network that matched well with our dataset. The YOLO_v. 4 network performed better on our dataset and was therefore selected to detect the carpal bone region. The cropped carpal bone region is shown in Figure 8.
SMANet performance
The SMANet network was constructed using the PyTorch framework and trained separately for the C-series and RUS-series. The RUS-ROIs images were resized to 128×128 pixels, whereas the whole carpal image was resized to 448×668 pixels. The 4,861 sets of image data were divided into the training set, validation set, and test set in a ratio of 8:1:1. The mini-batch size was set to 16 and the epoch size was set to 500. The learning rate was set to 0.001 and decreased in steps by a factor of 5 every 100 iterations.
The SMANet performance was evaluated on 486 left-hand radiographs, and SMANet obtained a skeletal maturity MAE of 29.9±0.21 points and bone age MAE of 0.43±0.17 years for the RUS-series and a skeletal maturity MAE of 31.4±0.19 points and bone age MAE of 0.45±0.13 years for the C-series (Table 2). The statistical error plots of the RUS-series and C-series were determined (Figure 9). No significant outlier points were observed in both the RUS-series and C-series, reflecting the strength of the generalizability of our network. Additionally, the MAE of the skeletal maturity scores among the different age groups was compared (Table 3). The features of the TW3-related image were not obvious due to the incomplete development of the carpal and finger bones in the early period. Therefore, the MAE score was relatively large. However, as the carpal and finger bones gradually reach complete development with age, the features of the TW3-related image became more obvious, resulting in a lower MAE.
Table 2
| Network | TW3-RUS | TW3-C | |||
|---|---|---|---|---|---|
| Skeletal maturity (score) | Bone age (year) | Skeletal maturity (score) | Bone age (year) | ||
| SMANet | 29.9±0.21 | 0.43±0.17 | 31.4±0.19 | 0.45±0.13 | |
MAE, mean absolute error; SMANet, skeletal maturity assessment network; TW3-RUS, Tanner-Whitehouse 3 radius, ulna, and short bones; TW3-C, Tanner-Whitehouse 3 carpal bones.
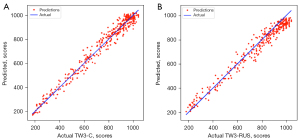
Table 3
| Age (years) | TW3-RUS (scores) | TW3-C (scores) |
|---|---|---|
| 3–5 | 34.9±0.32 | 36.1±0.26 |
| 5–7 | 33.3±0.19 | 35.7±0.22 |
| 7–9 | 30.1±0.21 | 32.7±0.23 |
| 9–11 | 29.3±0.11 | 32.1±0.18 |
| 11–13 | 27.9±0.22 | 32.5±0.13 |
| 13–15 | 28.1±0.25 | 28.3±0.19 |
| 15–17 | 27.8±0.15 | 28.1±0.17 |
| 17–18 | 27.4±0.21 | 25.8±0.16 |
| Average | 29.9±0.21 | 31.4±0.19 |
MAE, mean absolute error; TW3-RUS, Tanner-Whitehouse 3 radius, ulna, and short bones; TW3-C, Tanner-Whitehouse 3 carpal bones.
Discussion
SMANet compared with other networks
The information of the bone age and gender of each individual in the Jishuitan database was also included in the annotation file. Moreover, as skeletal maturity scores can be converted into a bone age using a bone age table, the official model of BoNet (33), SIMBA (34), and Yitu-AICARE (35) were trained and tested on the Jishuitan database. The best performing models on the validation set were selected to conduct the comparative experiments. The comparisons between SMANet, BoNet, Yitu-AICARE, and SIMBA are presented in Table 4. As shown in Table 4, SMANet outperformed BoNet, Yitu-AICARE, and SIMBA on test sets, which reflected a positive correlation between the skeletal maturity MAE (score) values and bone age MAE (year) values.
Table 4
| Network | Mean absolute error (years) | Probability for less than 1 year |
|---|---|---|
| BoNet (33) | 0.65±0.14 | 93.7% |
| Yitu-AICARE (35) | 0.57±0.21 | 94.2% |
| SIMBA (34) | 0.48±0.19 | 96.1% |
| SMANet | 0.43±0.17 | 97.3% |
BoNet, Hand pose estimation for pediatric bone age assessment; Yitu-AICARE, diagnostic performance of convolutional neural network-based Tanner-Whitehouse 3 bone age assessment system; SIMBA, specific identity markers for bone age assessment; SMANet, skeletal maturity assessment network.
Comparison of the performance of different network combinations
A cross-sectional comparison was conducted to compare the performance of different network architectures. The different SMA network architectures utilizing the input of ROIs patches, whole hand images, and a combination of both were trained and tested, respectively (Table 5). The evaluation metric was defined as the MAE between the ground truth and the estimated skeletal maturity score and the estimated bone age. The combination of ROIs and whole hand performed optimally on the top of the model (Figure 10).
Table 5
| Category | Networks | MAE (scores) | MAE (years) | Probability for less than 1 year |
|---|---|---|---|---|
| TW3-C | Whole hand | 35.1±0.26 | 0.51±0.17 | 93.7% |
| C-ROIs | 40.7±0.31 | 0.59±0.21 | 91.4% | |
| Whole hand + ROIs | 31.4±0.19 | 0.45±0.13 | 96.9% | |
| TW3-RUS | Whole hand | 33.9±0.27 | 0.51±0.16 | 94.1% |
| RUS-ROIs | 37.1±0.18 | 0.55±0.23 | 92.2% | |
| Whole hand + ROIs | 29.9±0.21 | 0.43±0.17 | 97.3% |
MAE, mean absolute error; TW3-C, Tanner-Whitehouse 3 carpal bones; TW3-RUS, Tanner-Whitehouse 3 radius, ulna, and short bones; ROIs, regions of interest; C-ROIs, carpal bones regions of interest; RUS-ROIs, radius, ulna, and short bones regions of interest.
The effect of gender on SMANet
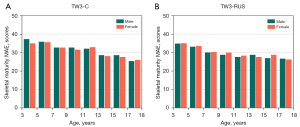
Our model predicted the bone maturity score, which was independent of gender. A standard clinical BAA process is that the physician first derives a skeletal maturity score from the left-hand radiographs and then derives the bone age from the bone age table, which corresponds to a different bone age table for each gender. Therefore, differences in bone age by gender are only reflected in the bone age table. The method we proposed was fully compatible with the clinical reading process. In addition, we performed statistics on the experimental results based on different genders. As shown in the statistical distribution (Figure 11), the SMA model did not differ significantly by gender, which also indicated that the SMA model was more robust to gender differences.
The limitations of SMANet
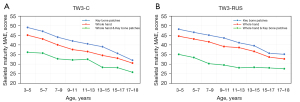
In the initial data screening stage, 439 images with hand deformities and right-hand images were excluded. Therefore, the final model lacked the ability to discriminate these abnormal images and therefore had some limitations.
Conclusions
In the present study, skeletal maturity instead of bone age was used as an evaluation indicator of development in children, which can potentially overcome the problem of racial and geographical variations, to some extent. The study discussed the application of multi-region ensemble networks for automatic SMA. The accuracy of automatic SMA could be similar to that of radiologists. Moreover, by comparing our method with the proposed popular BAA method, we found that the predictive accuracy of our method was superior to that of all other methods. To the best of our knowledge, SMANet is a novel network that can automatically implement an end-to-end SMA. This is crucial for the dissemination of an automated TW3 (both of RUS-series and C-series) BAA method across different ethnicities and geographic regions.
Acknowledgments
Funding: This research was funded by National Natural Science Foundation of China (No. 61971445), Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support (No. ZYLX202107), and Criminal Technology Double Ten Plan of the Ministry of Public Security (No. 2019SSGG0401).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1158/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1158/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work and ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Jishuitan Hospital (Approval No. 201907-11).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee H, Tajmir S, Lee J, Zissen M, Yeshiwas BA, Alkasab TK, Choy G, Do S. Fully Automated Deep Learning System for Bone Age Assessment. J Digit Imaging 2017;30:427-41. [Crossref] [PubMed]
- Liu C, Xie H, Liu Y, Zha Z, Lin F, Zhang Y. Extract Bone Parts Without Human Prior: End-to-end Convolutional Neural Network for Pediatric Bone Age Assessment. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science, vol 11769. Cham: Springer, 2019. doi:
10.1007/978-3-030-32226-7_74 .10.1007/978-3-030-32226-7_74 - Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. 2nd ed. Stanford: Stanford University Press, 1959.
- TTanner JM, Healy MJR, Goldstein H, et al. Assessment of skeletal maturity and prediction of adult height (TW3 method). 3rd ed. London: WB Saunders, 2001.
- Bae B, Lee J, Kong ST, Sung J, Jung KH. Manifold Ordinal-Mixup for Ordered Classes in TW3-Based Bone Age Assessment. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol 12266. Cham: Springer, 2020. doi:
10.1007/978-3-030-59725-2_64 .10.1007/978-3-030-59725-2_64 - Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Ghafoorian M, Karssemeijer N, Heskes T, van Uden IWM, Sanchez CI, Litjens G, de Leeuw FE, van Ginneken B, Marchiori E, Platel B. Location Sensitive Deep Convolutional Neural Networks for Segmentation of White Matter Hyperintensities. Sci Rep 2017;7:5110. [Crossref] [PubMed]
- Charbonnier JP, Rikxoort EMV, Setio AAA, Schaefer-Prokop CM, Ginneken BV, Ciompi F. Improving airway segmentation in computed tomography using leak detection with convolutional networks. Med Image Anal 2017;36:52-60. [Crossref] [PubMed]
- Wang S, Zhang R, Deng Y, Chen K, Xiao D, Peng P, Jiang T. Discrimination of smoking status by MRI based on deep learning method. Quant Imaging Med Surg 2018;8:1113-20. [Crossref] [PubMed]
- Li CS, Yao GR, Xu X, Yang L, Zhang Y, Wu TN, Sun JH. DCSegNet: Deep Learning Framework Based on Divide-and-Conquer Method for Liver Segmentation. IEEE Access 2020;8:146838-46.
- Bianconi F, Fravolini ML, Pizzoli S, Palumbo I, Minestrini M, Rondini M, Nuvoli S, Spanu A, Palumbo B. Comparative evaluation of conventional and deep learning methods for semi-automated segmentation of pulmonary nodules on CT. Quant Imaging Med Surg 2021;11:3286-305. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Spampinato C, Palazzo S, Giordano D, Aldinucci M, Leonardi R. Deep learning for automated skeletal bone age assessment in X-ray images. Med Image Anal 2017;36:41-51. [Crossref] [PubMed]
- Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology 2018;287:313-22. [Crossref] [PubMed]
- He J, Jiang D. Fully Automatic Model Based on SE-ResNet for Bone Age Assessment. IEEE Access 2021;9:62460-6.
- Cao SM, Chen ZY, Li CS, Lv CF, Wu TN, Lv B. Landmark-based multi-region ensemble convolutional neural networks for bone age assessment. Int J Imaging Syst Technol 2019;29:457-64. [Crossref]
-
Wang D Zhang K Ding J Wang L Improve bone age assessment by learning from anatomical local regions. arXiv:2005.13452 [cs.CV];2020 .10.1007/978-3-030-59725-2_61 - Chen C, Chen Z, Jin X, Li L, Speier W, Arnold CW. Attention-Guided Discriminative Region Localization and Label Distribution Learning for Bone Age Assessment. IEEE J Biomed Health Inform 2022;26:1208-18. [Crossref] [PubMed]
- Zhang SY, Liu LJ, Wu ZL, Liu G, Ma ZG, Shen XZ, Xu RL. Standards of TW3 skeletal maturity for Chinese children. Ann Hum Biol 2008;35:349-54. [Crossref] [PubMed]
-
Ronneberger O Fischer P Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:1505.04597 [cs.CV];2015 .10.1007/978-3-319-24574-4_28 -
Oktay O Schlemper J Folgoc LL Lee MJ Heinrich M Misawa K Mori K McDonagh S Hammerla NY Kainz B Glocker B Rueckert D Attention U-Net: Learning Where to Look for the Pancreas. arXiv:1804.03999 [cs.CV];2018 . - Payer C, Štern D, Bischof H, Urschler M. Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Med Image Anal 2019;54:207-19. [Crossref] [PubMed]
- Urschler M, Ebner T, Štern D. Integrating geometric configuration and appearance information into a unified framework for anatomical landmark localization. Med Image Anal 2018;43:23-36. [Crossref] [PubMed]
- Payer C, Štern D, Bischof H, Urschler M. Regressing Heatmaps for Multiple Landmark Localization Using CNNs. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, vol 9901. Cham: Springer, 2016. doi:
10.1007/978-3-319-46723-8_2 .10.1007/978-3-319-46723-8_2 - Štern D, Ebner T, Urschler M. From Local to Global Random Regression Forests: Exploring Anatomical Landmark Localization. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, vol 9901. Cham: Springer, 2016. doi:
10.1007/978-3-319-46723-8_26 .10.1007/978-3-319-46723-8_26 - Xu C, Zhang Y, Fan X, Lan X, Ye X, Wu T. An efficient fluorescence in situ hybridization (FISH)-based circulating genetically abnormal cells (CACs) identification method based on Multi-scale MobileNet-YOLO-V4. Quant Imaging Med Surg 2022;12:2961-76. [Crossref] [PubMed]
- Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016. doi:
10.1109/CVPR.2016.308 .10.1109/CVPR.2016.308 - He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. doi:
10.1109/CVPR.2016.90 .10.1109/CVPR.2016.90 - Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015. doi:
10.1109/CVPR.2015.7298594 10.1109/CVPR.2015.7298594 - Liu Y, Pu HB, Sun DW. Efficient extraction of deep image features using convolutional neural network (CNN) for applications in detecting and analysing complex food matrices. Trends Food Sci Technol 2021;113:193-204. [Crossref]
- He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. 2017 IEEE International Conference on Computer Vision (ICCV); 22-29 Oct. 2017; Venice, Italy. New York: IEEE, 2017.
- Girshick RB. Fast R-CNN. Proceedings of the IEEE International Conference on Computer Vision (ICCV), 2015:1440-8.
- Escobar M, Gonzalez C, Torres F, Daza L, Triana G, Arbeláez P. editors. Hand Pose Estimation for Pediatric Bone Age Assessment. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science, vol 11769. Cham: Springer, 2019.
- González C, Escobar M, Daza L, Torres F, Triana G, Arbeláez P. SIMBA: Specific identity markers for bone age assessment. International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer, 2020:753-63.
- Zhou XL, Wang EG, Lin Q, Dong GP, Wu W, Huang K, Lai C, Yu G, Zhou HC, Ma XH, Jia X, Shi L, Zheng YS, Liu LX, Ha D, Ni H, Yang J, Fu JF. Diagnostic performance of convolutional neural network-based Tanner-Whitehouse 3 bone age assessment system. Quant Imaging Med Surg 2020;10:657-67. [Crossref] [PubMed]



