
TransConver: transformer and convolution parallel network for developing automatic brain tumor segmentation in MRI images
Introduction
Brain tumor segmentation is a critical step in the process of brain tumor diagnosis and treatment. With the help of multimodal brain images for tumor segmentation, doctors can conduct quantitative analysis of brain tumors to measure the maximum diameter, volume and quantity of brain lesions and formulate the best diagnosis and treatment plan for patients. However, brain segmentation usually relies on manual segmentation, which is time-consuming, labor-intensive and affected by personal experience. Therefore, finding an accurate and efficient automatic brain tumor segmentation method to reduce doctors’ workload and avoid subjective opinions is indispensable for research.
Early medical image segmentation systems were mainly based on conventional image segmentation algorithms (1), involving edge detection-based methods, threshold-based methods and region-based methods. However, medical images, especially MRI images, usually have characteristics with low contrast, complex texture and blurred boundary areas, which limit the effect and application of such image segmentation algorithms.
With the development of deep learning and the expansion of application fields, the accuracy of medical image segmentation methods based on deep learning has continuously improved. In current research, medical image segmentation algorithms (2,3) based on deep learning mainly fall into three categories: CNN-based methods, transformer-based methods, and methods based on both transformers and CNNs.
CNNs-based methods
In recent years, convolutional networks and other deep learning algorithms have been widely studied and applied because of their powerful nonlinear feature extraction capabilities. Among various CNN variants, existing medical image segmentation methods mainly rely on fully convolutional networks (4) (FCNs), which can accept input images of any size and use the deconvolution layer to upsample the feature map to restore it to the same size as the input image. In particular, since the introduction of U-Net (5), CNN-based networks with U-shaped structures [e.g., UNet++ (6), Attention U-Net (7), DenseUNet (8), 3D Unet (9), V-net (10)] have achieved state-of-the-art results and excellent segmentation potential on various 2D and 3D medical image segmentation tasks. However, the locality of convolution makes the model ignore the correlation of long-range information. Studies (11,12) show that good segmentation requires a model to extract local fine-grained details and interactions of global semantic information at the same time. To improve the locality of convolution, some studies (13-15) try to expand the receptive field by using an attention mechanism, image pyramids and superimposed convolution layers. Although these methods increase the receptive field, they still have limitations in modeling long-range contextual interactions and spatial dependencies.
Transformer-based methods
The major issue in the natural language processing (NLP) domain is how to model global and long-range contextual interactions and spatial dependencies in long sequences. Among the studies on this issue, the self-attention mechanism represented by transformer (16) has achieved great success on many NLP tasks. The key to the success of transformers lies in the modeling of the global semantic information interactions and spatial dependencies with the self-attention mechanism and feed-forward networks (FFNs). Motivated by the transformer’s success, many studies (17,18), especially the most representative vision transformer (ViT) (17), have also proven the feasibility and effectiveness of applying transformers to downstream tasks in computer vision, such as classification, detection and segmentation. Solving issues related to the complex calculations and huge training costs of transformers is the main challenge of applying transformers to medical image segmentation. Guo et al. (19) used external attention instead of a self-attention mechanism to reduce computational complexity. Liu et al. (18) proposed Swin Transformer, which uses window-based self-attention to reduce parameters and computation and uses a shifted windows mechanism to realize global dependency modeling. Furthermore, Cao et al. (20) and Lin et al. (21) proposed Swin-Unet and DS-TransUNet, respectively, both of which are Unet-like transformers for medical image segmentation that have achieved performance similar to the most advanced CNN-based methods. According to T2T-Vit (22), we visualized the feature maps of the first two layers of Swin-Unet (20) and U-Net (5), as shown in Figure 1. We observe that U-Net can effectively capture local structures (including edges, lines and textures) in low-level semantic information extraction, which is absent in Swin-Unet. In contrast to CNNs, transformers inevitably ignore local structures when directly splitting images into patches as tokens.
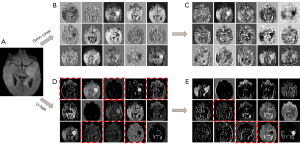
Transformer- and CNN-based methods
Local features and global representations are important counterparts (23). As shown in Figure 1, local features are compact vector representations of local image neighborhoods. Global representations include, but are not limited to, contour representations, shape descriptors, and object typologies at long distances. The main difference between global and local features is the size of receptive fields. Inspired by CNNs with strong local information extraction capacities and transformers with excellent global and long-range representation capacities, many concurrent works, such as CvT (24), PVT (25), TransUNet (26) and Segtran (12), try to combine convolution with transformers in different ways, hoping to combine the local advantages of convolution and the global advantages of transformers. For CvT and PVT, the linear projection of self-attention in the transformer is replaced by convolution. TransUNet and Segtran connect the transformer module behind multiple stacked convolution layers. Although transformer- and CNN-based methods have achieved remarkable results in medical segmentation, the performance of those methods mentioned can be further improved.
In medical image segmentation, the lack of fine-grained details will lead to fuzzy and inaccurate boundary segmentation of diseased tissues or organs, whereas the lack of long-distance information interaction will lead to segmentation failures when segmenting images with low contrast between organs. Therefore, how to combine CNN and transformer is an essential challenge for medical image segmentation. However, a simple combination of existing models (26,27) cannot effectively combine the advantages of convolution and transformers. Additionally, due to the ambiguity of semantic information between local features of convolution and global features of the transformer, combining local and global features directly in the channel dimension and then using Point-Wise Convolution for feature fusion is insufficient to fuse local and global features well. Furthermore, because of the quadratic computation complexity of the feature number, the calculation of the transformer is very complicated, and the complexity of the combination of the transformer and convolution is even more unaffordable. Finally, how to use convolution and transformers to design the encoder and decoder of medical image segmentation models to achieve a better training effect also remains a challenge.
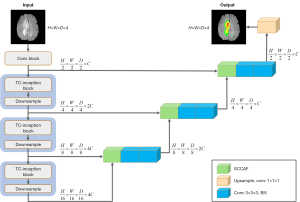
To address the aforementioned issues, as shown in Figure 2, we propose TransConver, an efficient encoder-decoder network that mainly combines the advantages of CNN and transformer for automatic medical image segmentation. Instead of using the serial connection structure of convolution and transformer as encoder, our TransConver adopts stacked Transformer-Convolution Inception (TC-Inception), which is a parallel module based on convolution and transformer inspired by Inception structure in GoogLeNet (28). To achieve a better feature fusion effect, motivated by dynamic fusion with intra and inter-modality attention flow (29) (DFAF) and cross-modal attention (30) success, we propose cross-attention fusion with global and local features (CAFGL) to alleviate the semantic difference between the local features extracted by convolution and the global features extracted by transformer. Considering that the feature fusion in skip connection will be affected by the semantic differences between different scale feature maps, we propose skip connection with cross-attention fusion mechanism (SCCAF), which can help feature fusion between semantically misaligned feature maps to effectively improve segmentation results. Based on the above components, we construct a U-shaped medical image segmentation network model named TransConver and design 2D-TransConver and 3D-TransConver for 2D and 3D tumor segmentation tasks, respectively.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-919/rc).
Methods
Model architecture
In this section, the overall architecture of our 3D-TransConver is introduced in detail as shown in Figure 2. TransConver is a U-like network based on an encoder-decoder structure and skip connection. The encoder is composed of a Conv Block, TC-Inception Block and Downsample Block. The encoder is mainly responsible for feature extraction and downsampling. Given a medical image with an input size of H×W×D×4, we first extract features and downsample through the Conv Block to obtain a feature map with a resolution of , where H, W, and D are the three dimensions of the 3D image and C is the channel dimension. Then, hierarchical feature representation is generated by the hierarchical architecture consisting of three stacked TC-Inception Blocks and Downsample Blocks, in which the resolution of the output feature map of each layer is reduced by half compared to the previous layer, and the number of channels is doubled while the channel on the last layer remains unchanged. Corresponding to the encoder, the decoder is mainly responsible for mapping the low-resolution feature map obtained by the encoder to the pixel-level prediction. The skip connection structure is one of the keys to the success of U-Net. In this work, we utilize SCCAF to fuse features of different scales to improve the semantic loss in upsampling. Finally, at the output of the decoder, we obtain a feature map with the same resolution as the input image and classify the pixels via convolution with a 1×1×1 filter. We will describe the main components in detail below.
TC-inception module
Motivated by the inception structure of GoogLeNet (28), we propose the TC-inception module, as shown in Figure 3. The Inception of GoogLeNet is designed to extract multiple features with different receptive fields to be able to obtain features with richer semantic information. Therefore, we design a convolution and transformer parallel module, which extracts local and global features by the convolution block and transformer block, respectively. At the same time, to fuse local features and global features with large semantic differences more effectively, we propose a cross-attention fusion with global and local features (CAFGL) mechanism, inspired by the success of cross-attention (29,31).
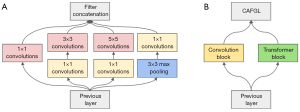
Global feature extraction via transformer block
Vision transformer (ViT) (17) is a pioneering work for transformers applied directly to images. In the multi-head self-attention (MSA) of the standard transformer, the computational complexity is proportional to the number of tokens quadratically, which is the main reason for the large computational cost of the transformer. To reduce the computational complexity of the 2D and 3D models during extraction of global features, we use the 2D Swin Transformer (18) and 3D Swin Transformer (32) instead of ViT in the 2D model and 3D model respectively, as shown in Figure 4.
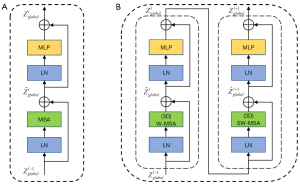
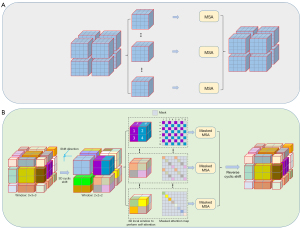
In contrast to the standard transformer, the Swin transformer utilizes window-based MSA (W-MSA) to reduce the computation. Moreover, it utilizes shifted window-based MSA (SW-MSA) in the next continuous layer to maintain cross-window connection and ensure global context information interaction, as shown in Figure 5. In layer i, the feature map is partitioned into 2×2×2 regular nonoverlapping windows, and self-attention is computed within each window. For layer i+1, the window partitioning is shifted by patches, resulting in new 3×3×3 irregular windows, to realize the interaction between adjacent windows in layer i. With the 3D W-MSA and 3D SW-MSA approaches, consecutive 3D Swin Transformer layers can be written as:
where and represent the output global features of the 3D(S)W-MSA module and MLP module in the lth 3D Swin Transformer block, respectively. MLP and LN denote the multilayer perceptron module and layer normalization, respectively. 3D W-MSA and 3D SW-MSA denote window-based multi-head self-attention using regular and shifted window partitioning configurations, respectively.
Through the mechanism of shifted windows, an increase in the window number will inevitably lead to an increase of computational complexity, as shown in Figure 6. Therefore, the 3D Swin Transformer utilizes a 3D cyclic-shifting and masking mechanism to efficiently compute batches for shifted configurations. As illustrated in Figure 6B, to keep the number of calculation windows consistent with W-MSA, the feature map is shifted cyclically by tokens along the axial, coronal and sagittal directions, while the shifted window partition stays consistent with the window partition in WSA. After this cyclic shift, a batched window may be composed of several nonadjacent subwindows in the initial feature map. Therefore, the masking mechanism is adopted to limit the self-attention computation to each subwindow. With the masked attention map Wmask, the masking self-attention in 3D SW-MSA can be formulated as:
where WmAttn and ZmSA denote the masking attention matrix and the result of 3D SW-MSA, respectively. Q, K, V are the query, key and value matrices. d is the Q/K dimension. With the above batch computation approach, 3D SW-MSA can retain efficient computational complexity consistent with 3D W-MSA.
Local feature extraction via convolution block
Similar to the convolution block of U-Net (5), we use two consecutive convolutions with a 3×3×3 filter size to extract local features and add batch normalization and ReLU (33) after each convolution for normalization and nonlinear transformation, as illustrated in Figure 7. The Convolution Block can be expressed as:
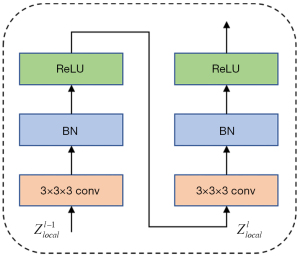
where represent the output local features of the convolution module in the lth convolution layer, and BN denotes batch normalization.
Cross-attention fusion with global and local features
Inspired by several studies (29-31) on multimodal feature fusion with an attention mechanism, we propose cross-attention fusion with global and local feature structures to fuse global and local features with semantic differences as shown in Figure 8. Cross-attention includes master input and auxiliary input. The Cross-Attention can be formulated as follows.
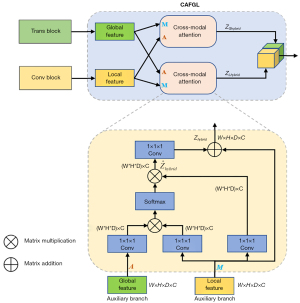
where A and M are the auxiliary and master input features, Mattn denotes the attention matrix, f1(·), f2(·), f3(·) and f4(·) are all linear projections with depthwise convolution, and d is the channel number of A/M. Therefore, the attention matrix Mattn reflects the importance of the master features to the auxiliary features and then obtains hybrid features by Eq. [9]. To allow efficient backpropagation during training, we finally obtain the output by summing M and . In CAFGL, the global and local features are input into two cross-attention as master branches and auxiliary branches in turn, and then the global hybrid features ZGhybrid and local hybrid features ZLhybrid are obtained. Finally, we concatenate them in the channel dimension.
Skip connection with cross-attention fusion mechanism
The skip connection structure is one of the keys to the success of U-Net (5). Skip connections can compensate for the details lost due to upsampling. The conventional skip connection structure always directly upsamples the decoder’s feature map to the resolution as one of the encoder’s feature maps and concatenates them in the channel dimension. As the literature (34) mentioned, the feature mapping from the encoder and decoder subnetworks in the skip connection operation has a large semantic difference. In addition, Wang et al. (35) also conduct a detailed analysis of skip connections. The features of the encoder and decoder are inconsistent, and not all simple skip connections are effective for the segmentation model. In general, the two features participating in skip connection are different in size. Before the fusion of two features, the small size feature is usually upsampled. Most of the commonly used upsampling methods use bilinear interpolation or bicubic interpolation. However, these methods will lead to blurred images, particularly when upsampling low-resolution features.
Considering the semantic difference between the encoder’s features and decoder’s features and the pixel misalignment caused by rough upsampling, we proposed a structure of skip connection with cross-attention fusion (SCCAF) mechanism to better integrate encoder features and decoder features with different scales, as shown in Figure 9. By using the SCCAF structure, irrelevant or noisy pixels in two inputs of skip connections can be masked, and more useful pixels can be highlighted.
2D-TransConver and 3D-TransConver
In this work, we also build a 2D-TransConver model to solve various 2D medical segmentation tasks. We transform the above 3D-TransConver into a 2D-TransConver through the following two steps:
- Transformation of data dimensions. The size of the input will be reduced from 3D to 2D, that is, from H×W×D×4 to H×W×4. Meanwhile, the output size is also reduced from H×W×D×4 to H×W×4.
- Transformation of model dimensions. In our proposed methods, the dimension reduction of the model is relatively direct. Specifically, all 3D convolution and 3D normalization operations in the whole network are replaced by 2D convolution and 2D normalization operations, respectively. For the TC-Inception module, we use a 2D Swin Transformer instead of a 3D Swin Transformer in the Transformer Block.
Loss function
The cross-entropy loss function is a basic loss function of image segmentation. However, cross entropy cannot evaluate the contour similarity between the ground truth and the predicted result. Meanwhile, it has high susceptibility when the data classes are imbalanced. The Dice loss function (10) can effectively alleviate these two problems. Therefore, the loss function we use is composed of Dice and cross entropy, which can be formulated as follows:
where I is the number of voxels, and J is the number of classes. Gi,j and Yi,j denote the one-hot encoded ground truth and predicted value for class j at voxel i, respectively.
Experimental framework
Datasets
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The 3D MRI image data for brain tumor segmentation in our experiments were provided by the Brain Tumor Segmentation 2018 (BraTS2018) challenge (36-38). Each BraTS2018 dataset consists of four modalities of MRI scans, namely, native T1-weighted (T1), postcontrast T1-weighted (T1ce), T2-weighted (T2) and fluid attenuated inversion recovery (FLAIR), and each modality’s size is 240×240×155. The labels for tumor segmentation involve four classes: whole tumor (WT), tumor core (TC), enhancing tumor (ET) and background. The dataset contains 285 examplar patient images for training and 66 examplars for validation. The Dice scores and Hausdorff distance (39) (95%) metrics of ET, WT and TC on the validation scans were obtained from the official evaluation server. Another 3D MRI dataset was provided by the Brain Tumor Segmentation 2019 (BraTS2019) challenge, including 335 examplars for training and 125 examplars for validation.
At the same time, to verify the validity of 2D-TransConver, we sliced the training sets in BraTS2018 and BraTS2019 to obtain corresponding 2D data with labels. The 3D MRI images, are sliced along the axial view to obtain 155 2D images. However, not all 155 2D slice images are selected as experimental images since the original 3D image contains slices that cover background areas exclusively. We select the slices with the lesion region and discard the slices that only have background to avoid class imbalance. The data are divided into 18923 examplars for training and 3219 examplars for validation. The same evaluation metric is used with that of 3D MRI data.
Data preprocessing and augmentation
Before inputting the data into the network for training or inference, we first combine 4 images with different modalities (T1, T1ce, T2, FLAIR) into a 3D voxel with 4 channels and then apply Z Score normalization for each image, that is, for each pixel, subtract the mean value from the image value and divide it by the standard deviation of the non-background region of the image. Since 2D data are obtained from 3D data, they share the same preprocessing technique.
Using data augmentation can effectively increase the diversity of training data and improve the generalization ability of the model. Thus, in the training process, we adopt the following data augmentation methods: (I) randomly crop the data from 240×240×155 to 128×128×128 voxels; (II) randomly shift intensity between [−0.1, 0.1] and randomly scale intensity between the range [0.9, 1.1]; (III) randomly mirror flip across each 3D axis with a probability of 50%.
All experiments (including the cited experiments) in this paper are based on the above data preprocessing and augmentation steps.
Evaluation metric
To compare and evaluate the segmentation results quantitatively, we calculate two widely used metrics in segmentation research, namely, the Dice metric and Hausdorff distance.
The Dice metric evaluates the degree of pixel overlap between the ground truth and prediction results and is calculated as follows:
where TP is the number of pixels that are correctly classified as a given class, FP is the number of pixels that are incorrectly classified as a given class, and FN is the number of pixels that are incorrectly classified as nonlabels in the predicted results.
The Hausdorff distance calculates the maximum distance between the contours of the ground truth and predicted results, which can be formulated as follows:
where G and P denote the contours of the ground truth and predicted results, respectively, and h(G,P) denotes the unidirectional Hausdorff distance from G to P.
Results
Implementation details
TransConver is implemented in PyTorch and trained with 2 parallel NVIDIA GeForce 2080Ti GPUs. We trained 400 epochs of 3D-TransConver and 2D-TransConver with batch sizes of 4 and 16, respectively. We adopted the Adam optimizer with an initial learning rate =0.001, momentum =0.9 and weight decay =0.0001.
Comparison with state-of-the-art network architectures
Comparative experiment of 3D segmentation approaches
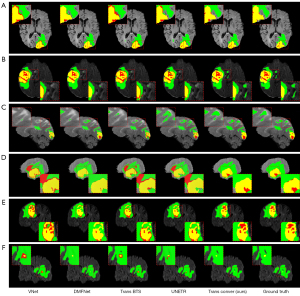
To verify the effectiveness of TransConver, as shown in Table 1, we compare the performance of 3D-TransConver against the baselines of one-stage CNN-based networks and Transformer- and CNN-based networks on the BraTS2019 dataset. It should be noted that due to the limitation of our computer’s computational power, the parameters (including the dimensions of the hidden layer and feedforward layer) of UNETR had to be adjusted to a suitable size to make the model train successfully on our computer in this experiment. The quantitative results demonstrate that our 3D-TransConver network achieves the best segmentation performance with segmentation accuracy of 83.72% (Average Dice of ET, WT and TC) and 4.741 mm (Average Hausdorff of ET, WT and TC) in the one-stage methods. Among all the one-stage methods, compared with CNN-based networks [3D U-Net (9), V-Net (10), Att-Unet (7), DMFNet (41)], transformer- and CNN-based networks [UNETR (40), TransBTS (42), 3D-TransConver] have obvious improvement in segmentation performance. This demonstrates that the combination of transformer and CNN is effective and significant. In other words, the ability of long-range contextual information interaction in the model is beneficial to improve the segmentation result. As shown in Figure 10, we show a visual comparison of the brain tumor segmentation results of various methods, including CNN-based networks [3D U-Net (9) and V-Net (10)] and transformer- and CNN-based networks [UNETR (40), TransBTS (42) and our method, TransConver]. Because the validation set provided by the BraTS dataset does not provide ground truth, we conducted a fivefold cross-validation evaluation on the training set for all methods. Compared with other models, our method is more complete and more accurate in boundary segmentation and category prediction. This shows the great potential of transformer- and convolution-based networks in medical image segmentation.
Table 1
| Method | Dice (%) ↑ | Hausdorff 95% (mm) ↓ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg | ET | WT | TC | Avg | ET | WT | TC | ||
| 3D U-Net (9) | 76.21 | 72.21±5.2 | 85.36±2.1 | 71.05±5.1 | 12.936 | 10.262±3.4 | 12.393±3.7 | 16.155±4.1 | |
| V-Net (10) | 77.02 | 72.43±5.2 | 85.18±2.0 | 73.46±4.3 | 8.916 | 7.076±2.0 | 8.893±2.6 | 10.779±2.4 | |
| Att-Unet (7) | 79.53 | 74.51±5.0 | 87.24±1.9 | 76.85±4.0 | 7.613 | 6.183±1.9 | 8.042±2.6 | 8.615±2.5 | |
| UNETR (40) | 82.25 | 77.37±4.7 | 88.56±2.2 | 80.81±3.9 | 7.071 | 5.846±2.0 | 7.296±2.7 | 8.071±2.7 | |
| DMFNet (41) | 82.60 | 76.88±4.4 | 89.38±1.5 | 81.55±3.5 | 5.375 | 4.508±1.9 | 5.036±1.4 | 6.581±1.5 | |
| TransBTS (42) | 82.32 | 77.58±4.6 | 88.42±2.0 | 80.96±3.5 | 7.171 | 6.213±2.1 | 7.632±1.9 | 7.668±2.0 | |
| TransConver | 83.72 | 78.40±4.4 | 90.19±1.2 | 82.57±3.4 | 4.741 | 3.414±0.8 | 4.848±1.2 | 5.962±1.8 | |
Data with 95% confidence interval in Dice and Hausdorff. ↑ indicates that the higher the value is, the better the result is. ↓ indicates that the lower the value is, the better the result is. Avg, average. ET, enhancing tumor; WT, whole tumor; TC, tumor core.
In addition, we also carried out the same comparative experiment on the BraTS 2018 dataset. As shown in Table 2, our network achieves the best segmentation performance with segmentation accuracy of 86.32% (Average Dice of ET, WT and TC) and 4.23 mm (Average Hausdorff of ET, WT and TC).
Table 2
| Method | Dice (%) ↑ | Hausdorff 95% (mm) ↓ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg | ET | WT | TC | Avg | ET | WT | TC | ||
| 3D U-Net | 78.49 | 74.36±4.8 | 88.34±1.8 | 72.79±4.2 | 11.35 | 5.98±1.9 | 15.62±2.4 | 12.47±2.6 | |
| V-Net | 80.59 | 78.84±4.9 | 88.42±1.9 | 74.51±4.3 | 10.29 | 6.34±2.0 | 13.59±2.5 | 10.95±2.4 | |
| UNETR | 83.85 | 79.46±4.6 | 89.16±2.3 | 82.93±4.1 | 5.92 | 4.76±1.9 | 6.34±2.1 | 6.67±2.1 | |
| DMFNet | 84.76 | 80.09±3.9 | 89.86±1.5 | 84.35±4.1 | 4.74 | 3.14±1.1 | 4.61±1.4 | 6.46±2.0 | |
| TransBTS | 85.36 | 81.09±4.1 | 90.82±1.8 | 84.16±3.8 | 5.41 | 4.06±1.1 | 5.79±1.9 | 6.37±2.0 | |
| Ours | 86.32 | 81.73±4.1 | 91.57±1.8 | 85.68±3.3 | 4.23 | 3.27±1.0 | 3.74±1.3 | 5.68±1.4 | |
Data with 95% confidence interval in Dice and Hausdorff. ↑ indicates that the higher the value is, the better the result is. ↓ indicates that the lower the value is, the better the result is. Avg, average; ET, enhancing tumor; WT, whole tumor; TC, tumor core.
Meanwhile, as shown in Table 3, we quantitatively analyze the parameters and computational complexity of all the models in the 3D segmentation experiment. Compared with other methods, TransConver uses relatively small parameters and computational power, although it achieves high accuracy.
Table 3
| Method | Parameter (M) | FLOPs (G) |
|---|---|---|
| 3D U-Net | 2.4 | 162.7 |
| V-Net | 37.7 | 396.3 |
| Att-Unet | 6.4 | 151.2 |
| UNETR | 102.8 | 2198.5 |
| DMFNet | 3.8 | 27.0 |
| TransBTS | 30.6 | 263.8 |
| Ours | 9.0 | 66.7 |
Furthermore, we compare the results of the ensemble methods on the BraTS2019 dataset. The two-stage cascaded U-Net (43) with an ensemble of 12 models is the best ranked method on the BraTS2019 Challenge. The experimental results demonstrate that such model ensemble strategy and postprocessing also work well in our proposed network. As shown in Table 4, we improve the final result via model ensemble strategy and post-processing.
Table 4
| Method | Dice (%) ↑ | Hausdorff 95% (mm) ↓ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg | ET | WT | TC | Avg | ET | WT | TC | ||
| Two-stage U-Net | 85.26 | 83.27 | 88.80 | 83.70 | 3.799 | 2.650 | 4.618 | 4.130 | |
| Ours w/o ensemble | 83.72 | 78.40 | 90.19 | 82.57 | 4.741 | 3.414 | 4.848 | 5.962 | |
| Ours w/ensemble | 85.32 | 81.69 | 90.53 | 83.74 | 4.027 | 3.081 | 4.839 | 4.162 | |
The data of Two-stage U-Net is from (43). ↑ indicates that the higher the value is, the better the result is. ↓ indicates that the lower the value is, the better the result is. Avg, average; ET, enhancing tumor; WT, whole tumor; TC, tumor core.
For model ensemble, we choose 5 models by the 5-fold cross validation method just like the two-stage U-Net. Then, during the inference phase, the five cross-validated models were ensembled by averaging the probabilities, and we adopted test time augmentation to enhance the prediction results. For postprocessing, to alleviate the situation in which the model predicted a few voxels with an enhanced tumor but there were no voxels in the ground truth, we applied the following postprocessing strategy. For each connected region composed of an enhanced tumor, if the number of voxels was less than 16 and the mean probability was less than 0.9, we replaced its class with a necrotic tumor. Afterward, if overall voxels number of an enhanced tumor was less than 73 and the mean probability was less than 0.9, we replaced its class with a necrotic tumor. The results show that network architecture, postprocessing and model ensembles are equally important for medical image segmentation.
Comparative experiment of 2D segmentation approaches
Actually, the contrast experiment of 2D approaches is almost the same as those of 3D approaches, except that the dimensions of input, output and model are converted from 3D to 2D. As shown in Table 5, we compare the performance of 2D-TransConver against baselines of CNN-based networks and Transformer- and CNN-based networks on the sliced 2D image data of the BraTS 2019 validation dataset. Our approach achieves the best average Dice score of 82.87% and an average Hausdorff distance of 2.2951 mm. This proves that our methods can be applied not only to 3D models but also to 2D models.
Table 5
| Method | Dice (%) ↑ | Hausdorff 95% (mm) ↓ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg | ET | WT | TC | Avg | ET | WT | TC | ||
| Unet++ | 81.46 | 77.46±0.27 | 84.23±0.27 | 82.69±0.35 | 2.4115 | 2.8456±1.02 | 2.6393±0.98 | 1.7496±1.56 | |
| DenseUNet | 81.71 | 78.09±0.31 | 84.59±0.26 | 82.46±0.30 | 2.3485 | 2.7571±1.13 | 2.6140±0.91 | 1.6746±1.47 | |
| Att-UNet | 82.01 | 78.23±0.42 | 84.66±0.31 | 83.15±0.39 | 2.4122 | 2.8783±1.31 | 2.6714±1.04 | 1.6869±1.58 | |
| TransUNet | 82.19 | 78.49±0.40 | 84.76±0.32 | 83.33±0.31 | 2.3851 | 2.8126±1.08 | 2.6883±0.93 | 1.6546±1.46 | |
| SwinUnet | 82.25 | 78.53±0.41 | 84.78±0.32 | 83.45±0.33 | 2.3783 | 2.8334±1.13 | 2.6571±1.07 | 1.6445±1.26 | |
| 2D-TransConver | 82.87 | 78.93±0.38 | 85.94±0.28 | 83.76±0.31 | 2.2951 | 2.6915±1.09 | 2.5865±0.96 | 1.6073±1.31 | |
We report the results of mean ± standard deviation by fixed the training set and run three different seeds. ↑ indicates that the higher the value is, the better the result is. ↓ indicates that the lower the value is, the better the result is. Avg, average; ET, enhancing tumor; WT, whole tumor; TC, tumor core.
Ablation study
TC-inception module
To verify the effectiveness of our proposed TC-Inception module for the ablation study, we replaced the TC-Inception module with a convolution block and transformer block separately. Table 6 shows the results of different feature extraction modules in the encoder. As expected, the complete TC-Inception module with convolution block, transformer block and CAFGL achieves the best result. Table 6 reports the Dice metric of ET, WT, TC of different feature extraction modules in 2D-TransConver. The Dice metric of ET is 77.39, 78.44, 78.51 for the 2D-TransConver with only Convolution Block, the 2D-TransConver with only Trans Block, the 2D-TransConver without CAFGL structure, respectively, and 78.93 for the complete model (P values: <0.1, <0.1, >0.1). The Dice metric of WT is 84.23, 84.89, 85.34 for the 2D-TransConver with only Convolution Block, the 2D-TransConver with only Trans Block, the 2D-TransConver without CAFGL structure, respectively, and 85.94 for the complete model (P value: <0.05, <0.05, <0.05). The dice metric of TC is 82.41, 83.13, 83.46 for 2D-TransConver with only Convolution Block, 2D-TransConver with only Trans Block, and 2D-TransConver without the CAFGL structure, respectively, and 83.76 for the complete model (P values: >0.1, >0.1, >0.1). The difference between TC-Inception and other feature extraction modules in TransConver on the metrics of WT was statistically significant (P value <0.05).
Table 6
| Module | Dice (%) ↑ | |||
|---|---|---|---|---|
| Avg | ET | WT | TC | |
| Convolution Block | 81.34 | 77.39±0.42 | 84.23±0.35 | 82.41±0.41 |
| Transformer Block | 82.15 | 78.44±0.43 | 84.89±0.33 | 83.13±0.40 |
| without CAFGL | 82.43 | 78.51±0.40 | 85.34±0.29 | 83.46±0.34 |
| TC-Inception | 82.87 | 78.93±0.38 | 85.94±0.28 | 83.76±0.31 |
We report the results of mean ± standard deviation by fixed the training set and run three different seeds. CAFGL, cross-attention fusion with global and local features mechanism. ↑ indicates that the higher the value is, the better the result is. Avg, average; ET, enhancing tumor; WT, whole tumor; TC, tumor core.
According to the experimental results of the model using convolution blocks and the model using transformer blocks, the latter can achieve higher accuracy in brain tumor segmentation. In addition, by removing CAFGL in TC-Inception and using a simple concatenation instead, we verify that the semantic gap between local features and global features can be narrowed by a cross-attention mechanism. As shown in Figure 11, we observe that TransConver w/o CAFGL relies on local features for segmentation, especially when segmenting tumor images with low contrast. Meanwhile, we found that the training process of the model using CAFGL is more stable than that of the model without CAFGL. The result suggests that model extraction of local features and global features at the same time can improve the segmentation accuracy.
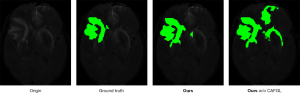
Skip connection with cross-attention fusion mechanism
Skip connections are important components of networks with encoder-decoder structures. Many studies (6,7) are devoted to the study of an effective skip connection to better fuse encoder features and decoder features to achieve remarkable results. In this work, we compare the SCCAF with the skip connection structure of U-Net. Table 7 reports the Dice metric of ET, WT, TC of different skip connection structures in 2D-TransConver. By comparing the simple concatenation with the SCCAF structure in 2D-TransConver, the Dice metric ranged from 78.52 to 78.93 for ET (P value: >0.1) and from 85.74 to 85.94 for WT (P value: >0.1) and from 83.62 to 83.76 for TC (P value: >0.1). The difference between SCCAF and simple concatenation structure in TransConver on the metrics was not statistically significant (P value <0.05). The purpose of using skip connections is to restore the details of the image by combining the high-resolution features of the encoder during upsampling. Through the visualization results shown in Figure 12, we observe that the model with SCCAF is beneficial for segmenting detailed features such as the edge of the tumor. This result shows that SCCAF can reduce the influence of pixel misalignment caused by upsampling.
Table 7
| Skip connection method | Dice (%) ↑ | |||
|---|---|---|---|---|
| Avg | ET | WT | TC | |
| Simple concatenation | 82.62 | 78.52±0.39 | 85.74±0.30 | 83.62±0.31 |
| SCCAF | 82.87 | 78.93±0.38 | 85.94±0.28 | 83.76±0.31 |
We report the results of mean ± standard deviation by fixed the training set and run three different seeds. SCCAF, skip connection with cross-attention fusion mechanism. ↑ indicates that the higher the value is, the better the result is. Avg, average; ET, enhancing tumor; WT, whole tumor; TC, tumor core.
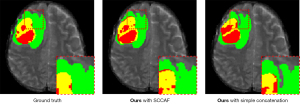
Discussion
Advantages of TransConver
Good segmentation requires a model to extract local fine-grained details and global semantic information simultaneously. Conventional CNNs have achieved remarkable performance in several medical image segmentation tasks and have good generalization ability. However, medical images usually contain many interrelated organs and tissues, so the size of the receptive field is particularly important in medical image segmentation. As shown in Figure 10C, the locality of CNNs (V-Net, DMFNet) becomes a limitation when segmenting an outlier region that is small in size and positioned far from the core of the disease. In contrast, the transformer and convolution-based networks (UNETR, TransConver) can successfully segment these regions. Although UNETR output is oversegmented, it also shows that the model has successfully delineated these regions through a transformer. This indicates clearly that long-range contextual interaction (i.e., global feature extraction) is essential for brain tumor segmentation. In addition, as illustrated in Figure 10B, other methods, including TransBTS and UNETR, still cannot segment these regions when segmenting images with low contrast between organs and targets. However, our proposed TransConver network can accurately fulfill the task. These results demonstrate that the combination method of transformer and convolution in the proposed network is better than other transformer- and convolution-based networks. Meanwhile, accurate classification of lesions can help doctors conduct quantitative analysis and clinical treatment more effectively. Our TransConver achieved the best performance in the classification of lesion categories, as illustrated in Figure 10D and Figure 10E. In summary, compared with state-of-the-art segmentation approaches, our network shows the best results in terms of the accuracy of tumor boundary segmentation, the integrity of tumor segmentation and the classification of lesion status. The structural characteristics and advantages of TransConver can be summarized in the following points:
- The structural design of TransConver follows the U-shaped network structure. The ability of U-shaped networks in medical image segmentation has been proven by many studies and experiments. Based on this network structure, we proposed a new tumor segmentation network with higher accuracy.
- We proposed a parallel module consisting of transformer and convolution, called TC-Inception, which performs convolution and transformer operations on the same feature map simultaneously and fuses local features of convolution and global features of transformer through the CAFGL mechanism. Through the TC-Inception module, we can effectively extract the local and global features of the image to accurately segment regions with low contrast and inconspicuous local features.
- We proposed skip connection with a cross-attention fusion mechanism to effectively fuse low-level semantics and high-level semantics having different scales and semantical misalignments.
Training from scratch
In general, transformer-based networks may achieve good performance only after a very large amount of data training or pretraining because of the lack of inductive bias for images. However, our TransConver model is trained from scratch. For our model to achieve the expected performance without a large amount of data for pretraining, we enhanced it with the following rationale:
- Swin Transformer Block. In this study (18), the author also expressed the view that inductive bias is still beneficial to visual modeling, especially in the task of object detection and segmentation. Because of the window-based self-attention mechanism, Swin Transformer introduces inductive bias of locality.
- Patchify stem replaced by the convolution stem. Xiao et al. (44) found that after the patchify stem is replaced by the convolution stem, the transformer performs more stably and converges faster in training. This is one of the reasons why our TransConver structure uses convolution block to extract features at the beginning.
- Data augmentation. Data augmentation is a very effective data expansion method. We adopt data augmentation methods to alleviate the problem of insufficient medical image data.
Limitations and further work
Compared with other computer vision tasks, such as face recognition and object detection, the primary limitation of medical image segmentation lies in insufficient datasets. In our work, we use a variety of data augmentation methods, including random cropping, random translation and random flipping, to increase the amount of data. However, these data augmentation methods cannot fundamentally solve this limitation. In recent years, semi-supervised and self-supervised learning methods have attracted much attention in the field of training with few samples, and many studies (45,46) have made remarkable achievements.
For future work, we intend to try to use semi-supervised or self-supervised learning methods in medical image segmentation tasks with transformer and convolution based networks.
Conclusions
In this paper, we proposed 3D and 2D medical image segmentation networks based on convolution and transformers, which can achieve high accuracy on 3D and 2D brain tumor segmentation, respectively. Differing from other transformer- and convolution-based networks, we designed a TC-inception module to parallelize the transformer and convolution operations. Moreover, inspired by many studies of multimodal feature fusion, we proposed CAFGL to effectively fuse global features and local features. In addition, we also used a cross-attention mechanism to improve the skip connection structure. The experimental results and ablation study on the BraTS 2018 and BraTS 2019 datasets show that our methods achieved superior Dice scores and Hausdorff distances in comparison with existing methods. Quantitative analysis of brain MRI based on our proposed model indicate that our method can segment tumor regions more accurately and improve clinical diagnosis.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Jiangxi Province, China (No. 20202BABL202028).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-919/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-919/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shan J, Cheng HD, Wang Y. Completely automated segmentation approach for breast ultrasound images using multiple-domain features. Ultrasound Med Biol 2012;38:262-75. [Crossref] [PubMed]
- Kim T, Hedayat M, Vaitkus VV, Belohlavek M, Krishnamurthy V, Borazjani I. Automatic segmentation of the left ventricle in echocardiographic images using convolutional neural networks. Quant Imaging Med Surg 2021;11:1763-81. [Crossref] [PubMed]
- Cai S, Tian Y, Lui H, Zeng H, Wu Y, Chen G. Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant Imaging Med Surg 2020;10:1275-85. [Crossref] [PubMed]
- Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; 2015: Springer.
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 2018;11045:3-11. [Crossref] [PubMed]
- Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:180403999 2018.
- Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging 2018;37:2663-74. [Crossref] [PubMed]
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, editors. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International conference on medical image computing and computer-assisted intervention; 2016: Springer.
- Milletari F, Navab N, Ahmadi S-A, editors. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 fourth international conference on 3D vision (3DV); 2016: IEEE.
- Yan H, Li Z, Li W, Wang C, Wu M, Zhang C. ConTNet: Why not use convolution and transformer at the same time? arXiv preprint arXiv:210413497 2021.
- Li S, Sui X, Luo X, Xu X, Liu Y, Goh RSM. Medical Image Segmentation using Squeeze-and-Expansion Transformers. arXiv preprint arXiv:210509511 2021.
10.24963/ijcai.2021/112 10.24963/ijcai.2021/112 - Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018;40:834-48. [Crossref] [PubMed]
- Xia H, Sun W, Song S, Mou X. Md-net: multi-scale dilated convolution network for CT images segmentation. Neural Processing Letters 2020;51:2915-27. [Crossref]
- Wang X, Girshick R, Gupta A, He K, editors. Non-local neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2018.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, editors. Attention is all you need. Advances in neural information processing systems; 2017.
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929 2020.
- Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B. Swin transformer: Hierarchical vision transformer using shifted windows. arXiv preprint arXiv:210314030 2021.
- Guo M-H, Liu Z-N, Mu T-J, Hu S-M. Beyond self-attention: External attention using two linear layers for visual tasks. arXiv preprint arXiv:210502358 2021.
- Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv preprint arXiv:210505537 2021.
- Lin A, Chen B, Xu J, Zhang Z, Lu G. DS-TransUNet: Dual Swin Transformer U-Net for Medical Image Segmentation. arXiv preprint arXiv:210606716 2021.
- Yuan L, Chen Y, Wang T, Yu W, Shi Y, Jiang Z, Tay FE, Feng J, Yan S. Tokens-to-token vit: Training vision transformers from scratch on imagenet. arXiv preprint arXiv:210111986 2021.
- Gulati A, Qin J, Chiu C-C, Parmar N, Zhang Y, Yu J, Han W, Wang S, Zhang Z, Wu Y. Conformer: Convolution-augmented transformer for speech recognition. arXiv preprint arXiv:200508100 2020.
10.21437/Interspeech.2020-3015 10.21437/Interspeech.2020-3015 - Wu H, Xiao B, Codella N, Liu M, Dai X, Yuan L, Zhang L. Cvt: Introducing convolutions to vision transformers. arXiv preprint arXiv:210315808 2021.
- Wang W, Xie E, Li X, Fan D-P, Song K, Liang D, Lu T, Luo P, Shao L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. arXiv preprint arXiv:210212122 2021.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:210204306 2021.
- Petit O, Thome N, Rambour C, Soler L. U-Net Transformer: Self and Cross Attention for Medical Image Segmentation. arXiv preprint arXiv:210306104 2021.
10.1007/978-3-030-87589-3_28 10.1007/978-3-030-87589-3_28 - Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A, editors. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- Gao P, Jiang Z, You H, Lu P, Hoi SC, Wang X, Li H, editors. Dynamic fusion with intra-and inter-modality attention flow for visual question answering. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2019.
- Song X, Guo H, Xu X, Chao H, Xu S, Turkbey B, Wood BJ, Wang G, Yan P. Cross-modal Attention for MRI and Ultrasound Volume Registration. arXiv preprint arXiv:210704548 2021.
10.1007/978-3-030-87202-1_7 10.1007/978-3-030-87202-1_7 - Xu X, Wang T, Yang Y, Zuo L, Shen F, Shen HT. Cross-Modal Attention With Semantic Consistence for Image-Text Matching. IEEE Trans Neural Netw Learn Syst 2020;31:5412-25. [Crossref] [PubMed]
- Liu Z, Ning J, Cao Y, Wei Y, Zhang Z, Lin S, Hu H. Video swin transformer. arXiv preprint arXiv:210613230 2021.
- Glorot X, Bordes A, Bengio Y, editors. Deep sparse rectifier neural networks. Proceedings of the fourteenth international conference on artificial intelligence and statistics; 2011: JMLR Workshop and Conference Proceedings.
- Cai Y, Wang Y. MA-Unet: An improved version of Unet based on multi-scale and attention mechanism for medical image segmentation. arXiv preprint arXiv:201210952 2020.
- Wang H, Cao P, Wang J, Zaiane OR. UCTransNet: Rethinking the Skip Connections in U-Net from a Channel-wise Perspective with Transformer. arXiv preprint arXiv:210904335 2021.
- Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [Crossref] [PubMed]
- Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:181102629 2018.
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993-2024. [Crossref] [PubMed]
- Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell 1993;15:850-63. [Crossref]
- Hatamizadeh A, Yang D, Roth H, Xu D. Unetr: Transformers for 3d medical image segmentation. arXiv preprint arXiv:210310504 2021.
- Chen C, Liu X, Ding M, Zheng J, Li J, editors. 3D dilated multi-fiber network for real-time brain tumor segmentation in MRI. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2019: Springer.
- Wang W, Chen C, Ding M, Li J, Yu H, Zha S. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. arXiv preprint arXiv:210304430 2021.
10.1007/978-3-030-87193-2_11 10.1007/978-3-030-87193-2_11 - Jiang Z, Ding C, Liu M, Tao D, editors. Two-stage cascaded u-net: 1st place solution to brats challenge 2019 segmentation task. International MICCAI Brainlesion Workshop; 2019: Springer.
- Xiao T, Singh M, Mintun E, Darrell T, Dollár P, Girshick R. Early convolutions help transformers see better. arXiv preprint arXiv:210614881 2021.
- Pandey P, Pai A, Bhatt N, Das P, Makharia G, AP P. Contrastive Semi-Supervised Learning for 2D Medical Image Segmentation. arXiv preprint arXiv:210606801 2021.
- Ahn E, Feng D, Kim J. A Spatial Guided Self-supervised Clustering Network for Medical Image Segmentation. arXiv preprint arXiv:210704934 2021.
10.1007/978-3-030-87193-2_36 10.1007/978-3-030-87193-2_36




