
Machine learning–based diagnostic evaluation of shear-wave elastography in BI-RADS category 4 breast cancer screening: a multicenter, retrospective study
Introduction
Breast cancer is the most common cancer and the sixth leading cause of cancer death worldwide (1). Breast cancer screening and early diagnosis effectively prolong patients’ overall survival and quality of life (2). Therefore, they have clinically significant value. With the development and progress in medical imaging technology, breast imaging-based examinations demonstrate good efficacy in breast cancer diagnosis and play an important role in breast cancer screening (3).
Ultrasound is one of the most commonly used imaging methods in breast cancer screening due to its lack of radiation, noninvasiveness, and reasonable cost (4). The Breast Imaging Reporting and Data System (BI-RADS) developed by the American Society of Radiology is used to standardize the description of breast imaging terms and the potential malignancy of breast lesions (5). However, ultrasound alone lacks diagnostic power, especially in the differential diagnosis of BI-RADS category 4 lesions. The malignancy rate of BI-RADS category 4 lesions ranges between 3% and 94% (6). Therefore, further subcategories of 4A, 4B, and 4C were introduced with positive predictive values (PPVs) of 3.5–9.8%, 20.2–31.2%, and 71.2–88.5%, respectively (7). Because B-ultrasound is subjective, it is necessary to combine it with other imaging examination results such as mammography (MMG), magnetic resonance imaging (MRI), and pathology results from puncture biopsies to reach a precise diagnosis, which creates psychological stress, physical damage, and economic burden for patients (8). Therefore, there is a clinical imperative to build an objective, sensitive, and accurate diagnostic model with real clinical utility for the screening of BI-RADS 4 lesions.
Shear-wave elastography (SWE) is a novel, noninvasive ultrasound imaging method that can visualize and quantify tissue stiffness in vivo (9). By virtue of its quantification ability, it can overcome the influence of human factors, such as manual pressure and use frequency of the static/quasi-static elastic imaging probe. It also has the advantages of repeatability and high-speed compared with strain elastography (10,11). At present, shear-wave imaging is widely used to examine the thyroid, breast, liver, prostate, and other organs. Previous studies have shown that SWE can be used in the diagnosis of breast cancer and the evaluation of neoadjuvant chemotherapy efficacy (12,13). However, SWE has not been widely used for primary breast cancer screening in clinic settings and has not been recommended in the National Comprehensive Cancer Network (NCCN) guidelines (Version 4. 2021) (14). Therefore, the diagnostic efficacy of SWE requires comprehensive evaluation.
With the rapid development of machine learning and artificial intelligence (AI)–aided medicine, the construction and evaluation of clinical models have provided novel insights into understanding medical data and images. This study aims to explore the diagnostic value and clinical utility of shear-wave imaging in BI-RADS category 4 breast cancer screening using a machine learning–based diagnostic model.
Methods
Study design and patient enrollment
This research was designed as a retrospective cross-sectional study. The primary outcome was the area under the receiver operating characteristic (ROC) curve (AUC) of the predictive model. Other outcomes included recall sensitivity, specificity (SP), and accuracy. Data were retrospectively collected from April 2018 to May 2020. The sample size calculation was based on previously published studies, with an expected sensitivity of 0.8 and an expected specificity of 0.9 used in the final calculation (15-17). Alpha (α) was 0.05, with an allowable error of 10%. The calculation was made using PASS software (NCSS, version 24.0), and the calculated sample size was 70. The inclusion criteria were as follows: (I) patients with a breast lesion detected under ultrasound with a clinical diagnosis of BI-RADS category 4 (A/B/C) or 5 given by radiologists with over 10 years’ experience; (II) patients who had undergone thorough evaluations using B-mode ultrasound (B-US), color Doppler flow imaging (CDFI), and SWE; (III) patients who had undergone biopsy or surgery with a pathological diagnosis; and (IV) patients who had provided informed consent. Patients with any of the following conditions were excluded: (I) previously diagnosed breast cancer or other cancers; (II) receipt of neoadjuvant chemotherapy or radiotherapy; (III) patients who were pregnant or lactating; and (IV) patients who refused to give informed consent to participate in the study.
External testing was completed using 2 independent cohorts collected under the same criteria within the 6 months. This study was approved by the Ethics Committees of Anhui Medical University, Jinan University, and Guangdong Key Laboratory of Traditional Chinese Medicine Research and Development. Individual consent for this retrospective analysis was waived. This study was reported according to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement (18).
Ultrasound examination
Ultrasound examinations were performed using an Aixplorer color Doppler ultrasound diagnostic instrument and an SL15-4 linear array probe (SuperSonic Imagine, Aix-en-Provence, France) with a 5–14 MHz frequency. Patients assumed the supine position to expose both breasts and armpits fully. The bilateral breast was firstly scanned by B-mode gray-scale ultrasound (B-US). The location, size, shape, edge, internal and posterior echo, calcification, and surrounding tissue of the mass were observed. Quantitative features were recorded, including the transverse diameter (T diameter) and anteroposterior diameter (AP diameter). The T:AP ratio was manually calculated. The blood flow in the lesion was displayed by CDFI. The resistance index (RI) and the maximum flow velocity of each vessel (Vmax) were measured and recorded. The classification of the lesions was made by experienced radiologists according to the BI-RADS fifth edition of the American College of Radiology and based on the above information combined with relevant medical history. For SWE, the regions of interest (ROIs) were manually depicted at the highest SWE velocity point within the lesion as previously described (19). Effective images were then captured in SWE, and the maximum and average elasticity values of the lesions were generated using the built-in quantitative analysis software “Q-boxTM Ratio”. Each lesion was measured 3 times, and the average value was recorded.
Pathological diagnosis
For all patients with lesions classified as BI-RADS category 4 or more, additional mammographic or MRI examinations were recommended. Clinical recommendations were given by surgeons based on the comprehensive evaluations from the radiologists. Histopathological results were obtained as the gold standard for patients who underwent biopsy or surgery. Immunohistochemical examinations were performed for malignant lesions only.
Propensity score matching (PSM)
Clinical characteristics were collected according to the 2012 World Health Organization (WHO) standard for breast tumor grading (20). PSM was used to match participants in the benign and malignant groups to maintain a similar baseline covariate distribution. Covariates were identified based on logistic regression with age, tumor size, and Vmax matched for balance. Patients from the cancer and benign group were 1:1 matched using nearest-neighbor matching. PSM was performed with the R package “MatchIt” (The R Foundation for Statistical Computing) (21).
Machine learning–based feature selection and diagnostic model construction
Continuous quantitative features were first estimated for collinearity, including age, T diameter, AP diameter, Vmax, RI, Emax, and Eave. For variables with identified collinearity, the data were converted into classified variables using cutoff values (RI: 0.7) (22). For multiclassification variables, dummy variables were introduced to transform multiclassification variables into multiple binary classification variables. We used both lasso regression and support vector machine-recursive feature elimination (SVM-RFE) algorithms for the feature selection. SVM models are a powerful tool to identify predictive models or classifiers because they can accommodate sparse data and can also classify groups or create predictive rules for data that linear decision functions cannot classify. The RFE algorithm for nonlinear kernels allows ranking of variables but does not compare the performance of all variables in a specific iteration. Lasso and SVM-RFE algorithms were combined to select features by both linear and nonlinear decision functions (23). In the lasso regression, the AUC was designated as the target parameter to minimize during the selection of a model for cross-validation, and the minimum lambda value was used for the feature selection. Patients were randomly assigned, with 70% composing the training set and 30% composing the validation set. Lasso regression was performed with the R package “glmnet” (24). In the SVM-RFE analysis, feature selection was obtained with 5-fold cross-validation using the R package “e1071” (23). All features were used to construct the models, and the generalization error of each model was estimated within 10-interval folds and 5-fold external cross-validation. The model with the highest accuracy and the lowest error was selected as the output. Features selected by both the lasso regression and SVM-RFE algorithms were used for the model construction. The stability of the model was estimated by comparing the training set with the validation set, and the model’s effectiveness was verified in independent external verification sets.
Decision curve analysis (DCA)
DCA was used to evaluate the diagnostic model's clinical benefit in identifying high-risk tumor patients for intervention while simultaneously avoiding overtreatment of low-risk patients (25). A clinical judgment was made of the relative value of benefit (treating a true-positive case) versus harm (treating a false-positive case) associated with the prediction models. As such, the preferences of patients or policy makers were accounted for by using threshold probability. A net benefit was then calculated for each possible threshold probability, which put benefits and harms on the same scale. A model’s clinical utility is estimated by determining if its net benefit is greater than that achieved by treating all patients or treating none (26). The bootstrap method was used to compare the confidence intervals of the 2 decision curves. DCA was performed using the R package “dca.r” with treatment reduction calculated per 200 patients.
Statistical analysis
All analyses were performed using RStudio version 1.2.5033 (RStudio, Boston, MA, USA) statistical software. For measurement data, variables with normal distribution are expressed as mean (SD) and were compared using the Student’s t-test. Variables with a skewed distribution are expressed as median (IQR) and were compared using the Wilcoxon rank-sum test. Multigroup comparisons were performed with one-way analysis of variance (ANOVA). For count data, variables are expressed as numbers and percentages and were compared using the chi-square test. The receiver operating characteristic curve (ROC) was drawn using the R package “pROC” (6). The area under the ROC AUC and 95% confidence intervals (95% CIs) were calculated and compared using the bootstrap method. A 2-tailed P value <0.05 was considered statistically significant.
Results
Basic clinical characteristics of enrolled patients
A total of 577 patients were retrospectively identified with breast masses in the training set, of whom 346 were excluded with a BI-RADS classification of 3 or lower. Of the remaining 231 patients, 127 patients underwent follow-up biopsy or surgery and had a pathological diagnosis. Four patients were excluded due to histories of contralateral breast cancer, and one patient had complications from other tumors. A final number of 122 patients met the eligibility criteria and were included in this study, comprising 51 benign lesions and 71 tumors (Figure 1A,1B). The median age was 50.61 years, with 56 patients younger than 50 years and 58 patients older than 50 years. The BI-RADS category of enrolled patients included 4A (n=36), 4B (n=21), 4C (n=35), and 5 (n=30), with 2 types of malignant pathological diagnoses [ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC)] and 5 types of benign lesions [benign phyllodes tumor (BPT), granulomatous mastitis (GLM), intraductal papilloma (IDP), fibroadenoma, and fibroadenosis]. For patients with pathologically confirmed breast cancer, the expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER-2), and Ki-67 were recorded, and pathological classifications were manually determined by experienced breast surgeons (Table 1).
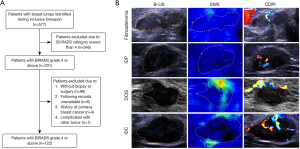
Table 1
| Characteristics | Statistics |
|---|---|
| Total (n) | 122 |
| Age, n (%) | 50.61 (14.73) |
| <50 | 56 (49.1) |
| ≥50 | 58 (50.9) |
| BI-RADS, n (%) | |
| 4A | 36 (29.5) |
| 4B | 21 (17.2) |
| 4C | 35 (28.7) |
| 5 | 30 (24.6) |
| Pathological diagnosis, n (%) | |
| Benign | 51 (41.8) |
| BPT | 1 (0.8) |
| Fibroadenoma | 17 (13.9) |
| Fibroadenosis | 17 (13.9) |
| GLM | 4 (3.3) |
| IDP | 12 (9.8) |
| Tumor | 71 (58.2) |
| DCIS | 10 (8.2) |
| IDC | 61 (50.0) |
| ER, n (%) | |
| Negative | 21 (30.9) |
| Positive | 47 (69.1) |
| PR, n (%) | |
| Negative | 23 (33.8) |
| Positive | 45 (66.2) |
| HER2, n (%) | |
| Indeterminate | 9 (13.2) |
| Negative | 46 (67.6) |
| Positive | 13 (19.1) |
| Ki-67, n (%) | |
| High | 41 (61.2) |
| Low | 26 (38.8) |
| Subtype, n (%) | |
| HER2 | 5 (7.4) |
| LumA | 12 (17.6) |
| LumB | 37 (54.4) |
| TNBC | 14 (20.6) |
For count data, variables are expressed as numbers and percentages. BI-RADS, Breast Imaging Reporting and Data System; BPT, benign phyllodes tumor; GLM, granulomatous mastitis; IDP, intraductal papilloma; DCIS, ductal carcinoma in situ; IDC, invasive ductal carcinoma; TNBC, triple-negative breast cancer.
Univariate and multivariate logistic regression
We used PSM to fully explore the diagnostic value of SWE. The variables matched for balance were age, tumor size, and Vmax. A total of 84 patients remained after PSM (42 tumors, 42 benign lesions). Univariate and multivariate logistic regressions were used to select the diagnostic variables (Table 2). In the univariate analysis, identical results were found in the direct comparisons between benign and cancer patients, except for Eave [odds ratio (OR) =1.02; 95% CI: 0.98–1.06; P=0.287]. The variables included in the multivariate logistic regression were selected by randomized gradient descent methods, which excluded the RI and T diameters. Among the variables included, only the BI-RADS category and Emax were significant, demonstrating their putative value in diagnosing breast cancer.
Table 2
| Characteristics | Univariate logistic regression | Multivariate logistic regression | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | ||
| Age | 1.07 | 1.03–1.11 | <0.001 | 1.04 | 0.99–1.1 | 0.096 | |
| ≥50 years | 1 | NA | |||||
| <50 years | 0.21 | 0.08–0.5 | <0.001 | ||||
| BI-RADS | |||||||
| 4A | 1 | NA | |||||
| 4B | 15.45 | 3.44–111.29 | 0.001 | 7.59 | 1.36–63.76 | 0.032 | |
| 4C | 82.17 | 18.67–604.56 | <0.001 | 17.37 | 2.73–162.07 | 0.005 | |
| Tumor size (mm) | |||||||
| T diameter | 1.07 | 1.03–1.13 | 0.002 | ||||
| AP diameter | 1.2 | 1.09–1.34 | <0.001 | 0.9 | 0.75–1.06 | 0.202 | |
| T/AP ratio | 1.37 | 0.73–2.66 | 0.331 | ||||
| Vmax (m/s) | 2.1 | 0.8–9.01 | 0.186 | ||||
| RI | |||||||
| >0.7 | 1 | NA | |||||
| ≤0.7 | 0.15 | 0.06–0.39 | <0.001 | ||||
| Elasticity value (kPa) | |||||||
| Emax | 1.03 | 1.02–1.04 | <0.001 | 1.03 | 1.01–1.06 | 0.005 | |
| Eave | 1.02 | 0.98–1.06 | 0.287 | ||||
For both univariate and multivariate logistic regression, a 2-tailed P value <0.05 was considered statistically significant. BI-RADS, Breast Imaging Reporting and Data System; T diameter, transverse diameter; AP diameter, anteroposterior diameter; Vmax, the maximum velocity of blood flow; RI, resistance index; OR, odds ratio; 95% CI: 95% confidence interval.
Machine learning–based construction and validation of the diagnostic model
We conducted a machine learning–based construction of the diagnostic model with variables selected using both lasso regression and SVM-RFE (Figure 2). Variables previously analyzed as both measurement and count data were included only once to avoid multiple comparisons. With age used as measurement data and RI as count data, 9 variables were included. Decisions were made based on the logistic regression results. Among all features included, only BI-RADS category 4A and Emax were selected by lasso regression as having the highest AUC.

Meanwhile, variables were ranked by SVM-RFE. The models constructed with 6 variables demonstrated the highest accuracy and the lowest error. The variables selected included Emax, BI-RADS category 4A, T/AP ratio, T diameter, BI-RADS category 4C, and age, with the sequence of variables ordered accordingly. Only Emax and BI-RADS category 4A were chosen by 2 methods and were included for model construction.
In both the training and validation sets, the combination of Emax and BI-RADS category 4A demonstrated outstanding and stable diagnostic efficacy, with the AUC of both sets above 0.85 (training set: AUC =0.92, 95% CI: 0.843−0.998; validation set: AUC =0.897, 95% CI: 0.765−1; P=0.761). Furthermore, the model showed high recall sensitivities in both the training and validation sets (training set: 0.9643; validation set: 0.9), which is essential for cancer screening. Comparatively, the combination of Emax and BI-RADS category 4A had a significantly higher AUC than did either measure used alone (Emax: AUC =0.863, 95% CI: 0.781−0.944, P=0.046; BI−RADS 4A: AUC =0.783, 95% CI: 0.701−0.866, P<0.001). The combined efficacy of Emax and BI-RADS category 4A was further tested in 2 independent cohorts. A total of 77 patients were enrolled in test cohort 1; the diagnostic efficacy was validated using the same model, and the AUC was 0.908 (95% CI: 0.834−0.982), with a recall sensitivity of 0.945. Comparisons between models also showed a significantly better efficacy with Emax and BI-RADS category 4A considered together than did the BI-RADS category 4A used alone (P=0.003). Similar results were seen in the test cohort 2 in which 55 patients enrolled; the AUC was 0.939 (95% CI: 0.874−1), and the recall sensitivity was 0.977. Moreover, compared with BI-RADS categories used alone, the addition of Emax showed significantly better efficacy, demonstrating the good predictive value of combining Emax with BI-RADS category 4A in the screening of breast cancer.
As the BI-RADS category 4 contains 3 levels, extrapolative comparisons were made between the 4A, 4B, and 4C subgroups. As shown in Figure 2, BI-RADS category 4A had the highest AUC among all 3 subgroups. However, the 4B and 4C categories also showed good predictive value, and comparisons between subgroups revealed no significant differences. Therefore, we speculated that Emax had significant diagnostic value in patients with BI-RADS category 4 lesions. Comparisons between models confirmed the significantly better efficacy of Emax combined with BI-RADS category 4 than of the BI-RADS category alone (BI−RADS+Emax: AUC =0.908, 95% CI: 0.842−0.974; BI−RADS AUC =0.862, 95% CI: 0.784−0.94; P=0.024).
The clinical benefit of Emax in BI-RADS category 4 breast cancer screening
To estimate the clinical benefits of the model, DCA was used to compare the combination of Emax and BI-RADS category 4 with either used alone. In the BI-RADS category 4A subgroup, adding Emax to the breast cancer screening benefited patients and showed a greater absolute benefit than did the BI-RADS category alone in patients with a higher probability of cancer (>0.403; Figure 3A). The clinical benefit of the model demonstrated similar efficacy when used in all BI-RADS category 4 lesions, with a higher absolute benefit seen in patients with a threshold probability of over 0.389 (Figure 3B).
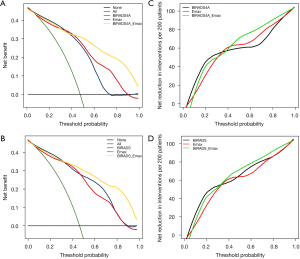
Further evaluations were made regarding the reduction of overtreatment. Intriguingly, in patients with a threshold probability of less than 0.403, the BI-RADS category was significantly valuable in reducing overdiagnosis and treatment. For patients with a higher probability, the combination of Emax and BI-RADS category outperformed the BI-RADS category alone, with the highest reduction of 50% per 200 patients (Figure 3C,3D). This demonstrated the significant value of adding Emax to BI-RADS category 4 lesions in breast cancer screening in ensuring clinical benefit to patients and reducing overtreatment.
Discussion
We constructed a machine learning–based diagnostic model for breast cancer screening with data collected from 3 independent centers. Analyses using multivariate logistic regressions confirmed Emax and the BI-RADS category as independent diagnostic features. Emax and BI-RADS category 4A were selected as the diagnostic variables to build the model using both lasso regression and an SVM-RFE algorithm and demonstrated significantly better efficacy than did the BI-RADS category alone in 2 independent validation cohorts. Furthermore, DCA analysis showed that adding Emax to the screening of BI-RADS category 4 patients could reduce overdiagnosis and treatment, indicating the clinical value of Emax in the screening and early diagnosis of BI-RADS category 4 breast cancer.
Our results support adding SWE to B-US, which has a well-established clinical utility in detecting, screening, and diagnosing breast cancer. Quantitative SWE parameters used alone have been able to classify breast lesions with a specificity of 86% and a sensitivity of 84% (27). Our results also agree with a previous study that found Emax to be the best-performing parameter in classifying breast lesions, achieving the highest AUC of 0.90 (95% CI: 0.77–1.00) (15). Similarly, the integration of SWE and B-US has been shown to improve diagnostic efficacy in breast cancer screening, particularly in specificity (28). A previously published meta-analysis focusing on the comparison of the pooled diagnostic accuracy of combined SWE and B-US to that of B-US alone revealed significantly elevated pooled specificity in all SWE parameters (SWE+B-US: AUC =0.85, 95% CI: 0.77–0.90, B-US: AUC =0.61, 95% CI: 0.42–0.78, P=0.009) (29).
However, the studies mentioned above used the retrospective data of relatively small sample sizes from either single centers or limited centers, which significantly reduced the quality of these studies. The methods adopted lacked statistical rigor considering the information provided was often based on a limited sample size without calculation of the required sample size, and the methods chosen were restricted to logistic regression with multiple comparisons where baseline biases were unadjusted.
Comparatively, the sample size of our study was both scientifically and statistically acceptable. In this study, we calculated the required sample size for statistical reliability, which has rarely been done in previous research. The sample size reported in our study included samples enrolled in the training cohort only (n=122). Furthermore, there were 77 patients enrolled in the external validation cohort 1 and 55 in the external validation cohort 2, making a total of 259 patients in our study. Compared to previously published studies, Zhang et al. conducted a retrospective study of 291 women from 2 centers to compare the diagnostic performance between B-US and SWE in classifying breast masses (15). Patients were divided into a training cohort (n=198), an independent validation cohort (n=65), and an external test cohort (n=28). However, among the patients enrolled, only 87 patients were pathologically diagnosed with malignant masses. Moreover, Yang et al. reported a high diagnostic performance of SWE with only 63 patients (malignant:benign ratio =33:30) (16), and Ranjkesh et al. enrolled 104 women in their study with 110 breast lesions, of which 77 were benign and 33 malignant (17).
Despite the perspectives and advantages mentioned above, this study has several limitations, one of which is the absence of diverse pathological types, particularly for cancer samples. Among the enrolled patients, only DCIS and IDC were included, and discussions were limited to subtypes only. Comprehensive evaluations with all pathological types should be conducted for further validation.
A recent study by Wang et al. demonstrated that with the assistance of AI, a number of unnecessary biopsies can be avoided (30). AI-based stratification systems significantly reduced the biopsy rate in BI-RADS 4 lesions from 100% to 67.4% without missing biopsy cases. Another study developed a dual-modal neural network model to characterize ultrasound images of breast masses (31) that demonstrated significant clinical utility, with an AUC of 0.982, 95% CI: 0.961–0.993, a specificity of 88.7%, 95% CI: 0.86–0.92, and an accuracy of 92.6%, 95% CI: 0.90–0.94. The implementation of computer-aided diagnostic systems has validated the clinical benefits and potency of the ultrasound-based diagnosis of breast lesions, providing novel perspectives to the use of ultrasound in breast cancer screening (32). Combined with the present results, the implications for machine learning in the interpretation of radiomic images and data suggest a promising future for further explorations into the medical image-based diagnosis of breast cancer.
In conclusion, we constructed a machine learning–based diagnostic model for BI-RADS category 4 breast cancer screening with an integrative combination of Emax and the BI-RADS category that demonstrated significantly better efficacy than did the BI-RADS category alone. This indicates the clinical value of SWE in BI-RADS category 4 breast cancer screening in reducing overdiagnosis and unnecessary biopsies.
Acknowledgments
Funding: This study was funded by the Medical Scientific Research Foundation of Guangdong Province (No. B2020184).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-341). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committees of Anhui Medical University, Jinan University, and Guangdong Key Laboratory of Traditional Chinese Medicine Research and Development. Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Leone JP, Leone BA, Tayob N, Hassett MJ, Leone J, Freedman RA, Tolaney SM, Winer EP, Vallejo CT, Lin NU. Twenty-year risks of breast cancer-specific mortality for stage III breast cancer in the surveillance, epidemiology, and end results registry. Breast Cancer Res Treat 2021;187:843-52. [Crossref] [PubMed]
- Lee SH, Park H, Ko ES. Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review. Korean J Radiol 2020;21:779-92. [Crossref] [PubMed]
- Cozzi A, Schiaffino S, Giorgi Rossi P, Sardanelli F. Breast cancer screening: in the era of personalized medicine, age is just a number. Quant Imaging Med Surg 2020;10:2401-7. [Crossref] [PubMed]
- Magny SJ, Shikhman R, Keppke AL. Breast Imaging Reporting and Data System. Treasure Island (FL): StatPearls, 2020.
- Sedgwick E. The breast ultrasound lexicon: breast imaging reporting and data system (BI-RADS). Semin Roentgenol 2011;46:245-51. [Crossref] [PubMed]
- Spinelli Varella MA, Teixeira da Cruz J, Rauber A, Varella IS, Fleck JF, Moreira LF. Role of BI-RADS Ultrasound Subcategories 4A to 4C in Predicting Breast Cancer. Clin Breast Cancer 2018;18:e507-11. [Crossref] [PubMed]
- Bulliard JL, Beau AB, Njor S, Wu WY, Procopio P, Nickson C, Lynge E. Breast cancer screening and overdiagnosis. Int J Cancer 2021; Epub ahead of print. [Crossref] [PubMed]
- Sigrist RMS, Liau J, Kaffas AE, Chammas MC, Willmann JK. Ultrasound Elastography: Review of Techniques and Clinical Applications. Theranostics 2017;7:1303-29. [Crossref] [PubMed]
- Mesurolle B, El Khoury M, Chammings F, Zhang M, Sun S. Breast sonoelastography: Now and in the future. Diagn Interv Imaging 2019;100:567-77. [Crossref] [PubMed]
- Chamming's F. Hangard C, Gennisson JL, Reinhold C, Fournier LS. Diagnostic Accuracy of Four Levels of Manual Compression Applied in Supersonic Shear Wave Elastography of the Breast. Acad Radiol 2021;28:481-6. [Crossref] [PubMed]
- Kim JY, Shin JK, Lee SH. The Breast Tumor Strain Ratio Is a Predictive Parameter for Axillary Lymph Node Metastasis in Patients With Invasive Breast Cancer. AJR Am J Roentgenol 2015;205:W630-8. [Crossref] [PubMed]
- Evans A, Whelehan P, Thompson A, et al. Identification of pathological complete response after neoadjuvant chemotherapy for breast cancer: comparison of greyscale ultrasound, shear wave elastography, and MRI. Clin Radiol 2018;73:910.e1-910.e6. [Crossref] [PubMed]
- Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, et al. NCCN Guidelines® Insights: Breast Cancer, Version 4.2021. J Natl Compr Canc Netw 2021;19:484-93. [Crossref] [PubMed]
- Zhang X, Liang M, Yang Z, Zheng C, Wu J, Ou B, Li H, Wu X, Luo B, Shen J. Deep Learning-Based Radiomics of B-Mode Ultrasonography and Shear-Wave Elastography: Improved Performance in Breast Mass Classification. Front Oncol 2020;10:1621. [Crossref] [PubMed]
- Yang H, Xu Y, Zhao Y, Yin J, Chen Z, Huang P. The role of tissue elasticity in the differential diagnosis of benign and malignant breast lesions using shear wave elastography. BMC Cancer 2020;20:930. [Crossref] [PubMed]
- Ranjkesh M, Hajibonabi F, Seifar F, Tarzamni MK, Moradi B, Khamnian Z. Diagnostic Value of Elastography, Strain Ratio, and Elasticity to B-Mode Ratio and Color Doppler Ultrasonography in Breast Lesions. Int J Gen Med 2020;13:215-24. [Crossref] [PubMed]
- Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Diabet Med 2015;32:146-54. [Crossref] [PubMed]
- Golatta M, Pfob A, Büsch C, et al. The Potential of Shear Wave Elastography to Reduce Unnecessary Biopsies in Breast Cancer Diagnosis: An International, Diagnostic, Multicenter Trial. Ultraschall Med. 2021; Epub ahead of print. [Crossref] [PubMed]
- Lebeau A, Denkert C. Updated WHO classification of tumors of the breast: the most important changes. Pathologe 2021;42:270-80. [Crossref] [PubMed]
- Weyland MS, Fellermann H, Hadorn M, Sorek D, Lancet D, Rasmussen S, Füchslin RM. The MATCHIT automaton: exploiting compartmentalization for the synthesis of branched polymers. Comput Math Methods Med 2013;2013:467428. [Crossref] [PubMed]
- Kapetas P, Woitek R, Clauser P, Bernathova M, Pinker K, Helbich TH, Baltzer PA. A Simple Ultrasound Based Classification Algorithm Allows Differentiation of Benign from Malignant Breast Lesions by Using Only Quantitative Parameters. Mol Imaging Biol 2018;20:1053-60. [Crossref] [PubMed]
- Sanz H, Valim C, Vegas E, Oller JM, Reverter F. SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinformatics 2018;19:432. [Crossref] [PubMed]
- Engebretsen S, Bohlin J. Statistical predictions with glmnet. Clin Epigenetics 2019;11:123. [Crossref] [PubMed]
- Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA 2015;313:409-10. [Crossref] [PubMed]
- Van Calster B, Wynants L, Verbeek JFM, Verbakel JY, Christodoulou E, Vickers AJ, Roobol MJ, Steyerberg EW. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur Urol 2018;74:796-804. [Crossref] [PubMed]
- Huang R, Jiang L, Xu Y, Gong Y, Ran H, Wang Z, Sun Y. Comparative Diagnostic Accuracy of Contrast-Enhanced Ultrasound and Shear Wave Elastography in Differentiating Benign and Malignant Lesions: A Network Meta-Analysis. Front Oncol 2019;9:102. [Crossref] [PubMed]
- Hao Y, Ren G, Yang W, Zheng W, Wu Y, Li W, Li X, Li Y, Guo X. Combination diagnosis with elastography strain ratio and molecular markers effectively improves the diagnosis rate of small breast cancer and lymph node metastasis. Quant Imaging Med Surg 2020;10:678-91. [Crossref] [PubMed]
- Park SY, Kang BJ. Combination of shear-wave elastography with ultrasonography for detection of breast cancer and reduction of unnecessary biopsies: a systematic review and meta-analysis. Ultrasonography 2021;40:318-32. [Crossref] [PubMed]
- Wang XY, Cui LG, Feng J, Chen W. Artificial intelligence for breast ultrasound: An adjunct tool to reduce excessive lesion biopsy. Eur J Radiol 2021;138:109624. [Crossref] [PubMed]
- Qian X, Zhang B, Liu S, Wang Y, Chen X, Liu J, Yang Y, Chen X, Wei Y, Xiao Q, Ma J, Shung KK, Zhou Q, Liu L, Chen Z. A combined ultrasonic B-mode and color Doppler system for the classification of breast masses using neural network. Eur Radiol 2020;30:3023-33. [Crossref] [PubMed]
- Yang Y, Hu Y, Shen S, Jiang X, Gu R, Wang H, Liu F, Mei J, Liang J, Jia H, Liu Q, Gong C. A new nomogram for predicting the malignant diagnosis of Breast Imaging Reporting and Data System (BI-RADS) ultrasonography category 4A lesions in women with dense breast tissue in the diagnostic setting. Quant Imaging Med Surg 2021;11:3005-17. [Crossref] [PubMed]


