
Fully automated radiomic screening pipeline for osteoporosis and abnormal bone density with a deep learning-based segmentation using a short lumbar mDixon sequence
Introduction
Osteoporosis is a systemic disease characterized by low bone mass and microarchitectural deterioration of bone tissue. These qualities lead to enhanced bone fragility. It has become a major public health problem worldwide. After 50 years of age, the risk of osteoporotic fractures is 50% for women and 20% for men among many western populations (1). Patients with osteoporosis are at increased risk of fractures with subsequent complications such as pain and immobilization. Therefore, it is critically important to identify patients at high risk of osteoporosis in an earlier stage.
Osteoporosis can be diagnosed based on bone mineral density (BMD). Dual energy X-ray absorptiometry (DXA) is the standard of reference to quantify BMD. However, BMD measurement by DXA is affected by osteophytes, facet joint degeneration, and vertebral concavity. Recently, quantitative computed tomography (QCT) as a truly 3D technique has been used to quantify BMD. In contrast to DXA, QCT can avoid BMD overestimation due to spinal degeneration, abdominal aortic calcification, and other sclerotic lesions. Despite these advantages, QCT is not yet widely available. Furthermore, this method carries a non-negligible radiation dose. It is beneficial to investigate the role of alternative techniques for accurate prediction of osteoporosis with low or even no exposure to ionizing radiation, such as magnetic resonance imaging (MRI).
MRI has been demonstrated to assess osteoporosis by measuring bone marrow fat fraction (BMFF). Recent studies have highlighted the essential role of BMFF in bone health using chemical shift-based water-fat separation sequences, such as modified Dixon (mDixon) (2-5).
mDixon has several advantages, including good spatial coverage, short acquisition time, and simple technical requirement. It provides accurate fat quantification due to its fat spectrum modelling and built-in R2* correlation (6,7). There are many existing lumbar MRI scans performed on patients for a slew of indications, which can be used for “opportunistic osteoporosis screening”, without ionizing radiation exposure and substantial costs. Since mDixon is a fast, simple, and non-invasive method (16 seconds as used in this study), it can be easily added to conventional lumbar spinal MRI to obtain BMFF maps. An automatic opportunistic osteoporosis screening tool based on a simple additional chemical shift-based water-fat separation sequence will benefit patients with low back pain or lower limb symptoms.
A previous study demonstrated the capability of BMFF extracted using mDixon to predict abnormal bone density (ABD) (8). However, the clinical utility is limited by the requirement of manual segmentation, and the predictive power of more quantitative features beyond simple BMFF average remains to be explored. Convolutional neural network (CNN) has been applied in the context of radiomics to provide the possibility of fully automated segmentation pipelines. While CNNs have demonstrated comparable performances to those of manual segmentation in cancer applications, reports on the fully automated radiomics pipelines for bone disease prediction is still lacking. Furthermore, radiomics exploits high throughput features from medical images to aid in clinical decision making and personalized medicine. While radiomic analysis is commonly used in clinical oncology applications, only a limited number of studies investigated this approach on bone diseases. Rastegar et al. (9) developed classification models to predict osteoporosis and osteopenia using radiomics on bone mineral densitometry images, achieving area under the receiver operator characteristic curve (AUC) values from 0.50 to 0.78. Zhou et al. found that deep learning can provide automated segmentation of vertebral bodies using water-fat MR (Dixon) images to qualify bone marrow fat recently (10). However, there is no study to date reporting the use of water-fat MRI radiomics for osteoporosis diagnosis.
In this study, we demonstrated a fully automated end-to-end radiomics pipeline using reliable segmentation via CNN (11). The developed pipeline was further evaluated using a temporal validation set. Temporal validation is considered to be stronger evidence compared to random split (12). External data obtained with a different vendor scanner was used to evaluate the performance of the pipeline. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/qims-21-587).
Methods
Subjects
The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of The Third Affiliated Hospital of Southern Medical University and The Fifth Affiliated Hospital of Sun Yat-sen University, and informed consent was taken from all individual participants. This study is a retrospective study.
Training and temporal validation cohort
A total of 222 participants were recruited through poster and email announcement approved by local institution review board between October 2016 and November 2017. Before the examination we obtained written consent from all participants. In this population, a 3T Philips MR scanner was used. Inclusion criteria: adults between the ages of 20 and 80. Exclusion criteria: spinal tumor, history of trauma, spine fracture, dysplasia, spinal surgery, metabolic bone diseases such as hyperparathyroidism and renal osteopathy, hormone therapy and contraindication for magnetic resonance examination such as cardiac pacemaker or claustrophobia.
Additional exclusion after MRI images were acquired: end-plate Modic changes, acute lumbar fracture, and inflammations were seen on T1-weighted and T2-weighted sequences. The image-based exclusion was performed jointly by two musculoskeletal radiologists (YZ and XZ) with 5 and 7 years of experience, respectively.
In total, 206 subjects out of the 222 participants were included in the final analysis (140 females and 66 males). The age ranged from 20 to 78 years old (mean, 48.8±14.9 years old) with body mass index (BMI) ranging from 16.6 to 32.9 kg/m2 (mean, 23.0±3.1 kg/m2).
These 206 subjects were temporally split into the training cohort and the temporal validation cohort. The training cohort consisted of 142 subjects (50 osteopenia, 24 osteoporosis and 68 normal, examined between July 2017 and December 2017), while 64 subjects (17 osteopenia, 9 osteoporosis and 38 normal, examined between November 2016 and January 2017) were assigned to the temporal validation (testing) cohort. The distribution of the training and temporal validation regions of interest (ROIs) was summarized in Table 1, and the final diagnosis label of each subject was determined using the mean BMD of L1, L2 and L3. The splitting ratio is chosen according to Dobbin et al. (13) Temporal splitting was used since it is superior to random splitting, according to the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) (12) guidelines.
Table 1
| Cohort | Time | Total | Normal | Osteopenia | Osteoporosis |
|---|---|---|---|---|---|
| Training (ROIs) | 2017.7–2017.12 | 426 (n=142) | 204 (48%) | 149 (35%) | 73 (17%) |
| Temporal validation (ROIs) | 2016.11–2017.1 | 192 | 114 (59%) | 51 (27%) | 27 (14%) |
| Temporal validation (subjects) | 64 | 38 | 17 | 9 | |
| External validation (subjects) | 2020.09–2020.12 | 25 | 9 | 9 | 7 |
ROIs, regions of interest.
External validation cohort
Additionally, external data from a 3T GE SIGNA Pioneer Scanner was also included in this study as external validation. The inclusion and exclusion criteria are the same as the training and temporal validation cohort. Total of 25 subjects (9 osteopenia, 7 osteoporosis, examined between September 2020 and December 2020) were included in the analysis. (14 females and 11 males). The age ranged from 40 to 74 years old (mean. 56.1±8.8 years old) with BMI ranging from 18.4 to 28.4 kg/m2 (mean, 22.9±2.6 kg/m2). The distribution of the external validation cohort was used for external validation and its ROIs were summarized in Table 1.
Image acquisition and segmentation
QCT examination
In order to obtain BMD of three most superior lumbar vertebrae (L1 to L3), a multidetector CT scanner (Aquilion64, Toshiba, Tokyo, Japan) was used with a synchronously calibrated phantom (Mindways, Austin, TX) placed under each participant’s lumbar spine (14). For all QCT scans, the following protocol was used: 120 kvp, 75 mAs, 2.0-mm reconstruction slice thickness, 65-cm table height, and large field of view covering the five density columns. Lumbar spine volume was analyzed by a computer workstation (Mindways QCT Pro, Austin, TX). A radiologist with 12 years of experience (XZ) used elliptical shapes on the midplane of L1, L2, and L3 vertebrae to delineate the separate ROI; these ROIs were then utilized to calculate the trabecular BMD. The BMD results were expressed in mg/cm3 of calcium hydroxyapatite (15). The CT scanner transformed HU into bone mineral equivalents (mg/cm3) by using an appropriate bone mineral phantom which contain various concentrations of material with similar X-ray attenuation characteristics to bone in the scan field through QCT. The calibrated phantom is stable with solid material (hydroxyapatite). BMD can be obtained based on the regression of attenuation and concentration of the calibration substance. The mean BMD of L1, L2 and L3 were used to determine final reference diagnosis. The thresholds to classify mean BMD into reference diagnosis were in line with the International Society for Clinical Densitometry (ISCD) in 2007 (16) and the American College of Radiology in 2013 (17), characterizing the groups as: osteoporosis, <80 mg/cm3; osteopenia, 80 to 120 mg/cm3; normal, >120 mg/cm3. In the training cohort, the 3 ROIs of each subject were separately classified.
Similarly, QCT of the external data were acquired using 128-channel multidetector CT scanners (uCT 760, United Imaging Healthcare) to obtain the BMD of lumbar vertebrae with an asynchronously calibrated phantom. The CT parameters were set as follows: 120 kVp; automatic tube current, 2.0 mm reconstruction slice thickness, 83.6-cm table height, and large field of view covering the five density columns. Lumbar spine volume was analyzed (18) by a computer workstation (Mindways QCT Pro, Austin, TX, USA). A radiologist with 10 years of experience (XC) calculate the trabecular BMD following the same procedure used in training/temporal cohort.
MRI examination
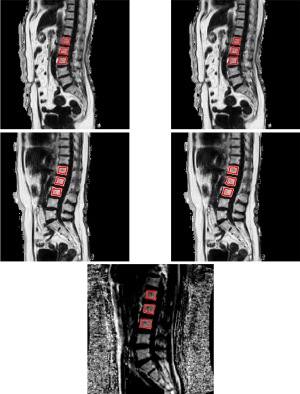
MRI scans of the lumbar spine were acquired using a 3T MR scanner (Ingenia, Phillips, Amsterdam, Netherlands) with posterior coil was done after the QCT scan on the same day. The MRI protocols are summarized in Table 2. Conventional sagittal lumbar spine T1-weighted and T2-weighted images were used for anatomical and morphological assessments. A mDixon Quant sequence with six-echo, seven fat peaks modeling, and T2* correction was used to quantify the vertebral fat fraction. ROIs were then manually drawn on the mid-sagittal view of each vertebral body (L1-L3) on the fat fraction maps by the same radiologist (XZ). Figure 1 shows some examples of the ROIs. As the goal of this study is to develop an automated opportunistic osteoporosis screening tool that can be added to every clinical lumbar MRI protocol, we kept the mDixon acquisition extremely short at 16 seconds. The MRI scan of the external data was performed using another 3T GE MR scanner (SIGNA Pioneer, GE Healthcare, Waukesha, WI, USA). The MRI protocol is summarized in Table 2. The resolution is consistent within training and temporal validation cohort. The resolution for external validation cohort is different from training and temporal cohort but consistent within the cohort itself. There is no adverse event happened from performing QCT or MRI examination during the study.
Table 2
| Sequence | TE1/TR (ms) | FOV (mm2) | Matrix size | Voxel size (mm2) | Slice thickness (mm) | Gap (mm) | Acquisition time (s) | ΔTE (ms) | Flip angle | |
|---|---|---|---|---|---|---|---|---|---|---|
| Philips mDixon | 3D-FFE | 0.96/5.6 | 400×350 | 268×208 | 1.5×1.7 | 3 | 0 | 16 | 0.7 | 3° |
| GE IDEAL IQ | 3D-IDEAL IQ | 1.1/7.3 | 320×320 | 160×160 | 2.0×2.0 | 8 | 0 | 16 | 0.8 | 4° |
MRI, magnetic resonance imaging; mDixon, modified Dixon; FOV, field of view; GE, General Electric Co.
Automated ROI delineation using CNN
As demonstrated in Figure 2, a U-Net was developed using the training cohort and the corresponding manual ROIs. The U-Net was developed to segment L1-L3 from fat fraction maps. Fat-water ratio images were provided as inputs, which were consistent with manual segmentation. The network was trained using Adam optimizer with a learning rate of 0.001, first-moment exponential decay rates of 0.9 and second-moment exponential decay rates of 0.999 (19). All weights were initialized using the Glorot-uniform method (20), and biases were initialized to zero. The network was trained in 2D. The minibatches of individual slices were randomly selected from subjects in the training cohort. The training objective was to minimize the mean squared logarithmic error between the network’s output with radiologists’ manually-drawn segmentations. The cost function was averaged over all slices in each minibatch. Batch normalization was implemented after each layer.
The CNN output was in the range of 0 and 1. An optimal threshold for binarizing the CNN output was determined using training cohort to achieve highest average Dice coefficient at subject level. The binarized image was then processed by labeling connected pixels to acquire all connected components. The three components with highest volume were selected as L1, L2 and L3. The CNN performance was assessed using Dice coefficient between network segmentations and manual segmentations at subject level. The data used for the evaluation is the temporal validation cohort.
Data calibration
As the external dataset was collected using an MRI scanner from a different vendor, the BMFF maps from the two vendors have different dynamic range. The BMFF maps for the external data were calibrated to be in the same range as the training/temporal validation cohorts (21).
Feature extraction
Radiomic features were extracted from L1-L3 on fat fraction using an in-house MATLAB software (22), including first order, 2D shape, texture [grey level co-occurrence matrix (GLCM) and grey-level run length matrix (GLRLM)] features. 2D features were extracted on the center slice for each vertebral body. A total of 56 features were extracted compliant with the Imaging Biomarker Standardization Initiative (IBSI) (23) standard as shown in Table 3. The features extracted were used for both the ABD prediction model and the osteoporosis prediction model.
Table 3
| Feature family (settings) | Number | Features |
|---|---|---|
| First order | ||
| Intensity-based statistics | 11 | Energy, excess kurtosis, maximum intensity, mean deviation, minimum intensity, mean intensity, median intensity, intensity range, intensity root mean square, intensity skewness, intensity variance |
| Intensity histogram (mean ± 3 SD, 64 bins) | 2 | Discretised intensity entropy and discretised intensity uniformity (energy) |
| Shape (2D) | 10 | Mesh surface, pixel area, perimeter, perimeter to surface ratio, sphericity, spherical disproportion, maximum 2D diameter, major axis length, minor axis length, elongation |
| Texture | ||
| GLCM | 22 | Autocorrelation, cluster prominence, cluster shade, cluster tendency, contrast, correlation, difference entropy, dissimilarity, energy, joint entropy, inverse difference, homogeneity, informational measure of correlation 1, informational measure of correlation 2, inverse difference moment normalized, inverse difference normalized, inverse variance, joint maximum, sum entropy, sum variance, joint variance |
| GLRLM | 11 | Short-run emphasis, long-run emphasis, low gray-level run emphasis, high gray-level run emphasis, short-run low gray-level emphasis, short-run high gray-level emphasis, long-run low gray-level emphasis, long-run high gray-level emphasis, gray-level non-uniformity for run, run length non-uniformity, and run percentage |
MRI, magnetic resonance imaging; GLCM, grey level co-occurrence matrix; GLRLM, grey-level run length matrix.
Feature selection
Feature selection was then conducted separately for ABD (osteopenia and osteoporosis) and osteoporosis prediction.
To reduce feature dimensionality, Wilcoxon rank sum test was performed with a threshold P value of 0.05 between subjects with or without ABD in the training cohort (24). Next, feature pairs with Spearman correlation |ρ| >0.8 were identified for multicollinearity check (25). Among these pairs, only the feature with higher discriminative power (i.e., AUC) in univariate analysis was selected for model development.
Prediction model development and validation
Demographic characteristics (age, sex and BMI) were pooled with image features with reduced dimension to build the model. Least absolute shrinkage and selection operator (LASSO) with 3-fold cross validation was used to select up to 10 features to avoid overfitting (25). Using the selected features, classification models were constructed to predict ABD and osteoporosis with coefficients obtained from logistic regression. The optimal threshold was determined in the training cohort by maximizing Youden’s Index (26). Receiver operating characteristic (ROC) analysis was performed to assess model performance.
Model evaluation was based on the temporal validation cohort, with radiomic features extracted from ROI generated by fully automated CNN. For comparison, the model was also evaluated based on the same validation cohort, with radiomic features extracted from manually segmented ROIs.
The external validation set was processed through the same feature extraction procedure. The model built using the training dataset was applied on external validation set to evaluate the performance of the model.
Results
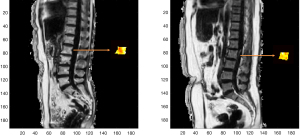
Some examples of the automated ROIs are shown alongside the manually drawn ROIs in Figure 1 for training and temporal validation cohort. In addition, one example of the automated segmentation result from the external validation cohort is also shown in Figure 1.
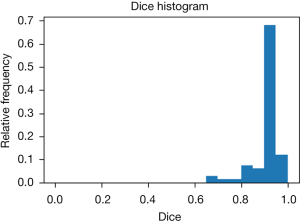
As shown in Figure 3, the U-Net achieved a mean Dice coefficient of 0.912±0.062 compared to the manual ROIs in the temporal validation cohort. The manual ROIs and corresponding automated ROIs of a representative subject is shown in Figure 3. This suggests the CNN based automated segmentation yields accurate ROI delineation.
For the prediction of ABD, the model yielded an AUC of 0.963 in the training cohort and the curve is shown in the left of Figure 4. The model selected nine radiomic features from fat fraction with one clinical variable (age), as displayed in the top of Figure 5. The t-SNE (t-distributed stochastic neighbor embedding) visualization of the features are shown in the left of Figure 6. When applied to the temporal validation cohort, this model achieved an AUC of 0.929/0.925 and an accuracy of 0.859/0.844 with manual/CNN segmented ROIs, respectively. In addition, it achieved a sensitivity of 0.923/0.923 and a specificity of 0.816/0.789 with manual/CNN segmented ROIs, respectively. The results are summarized in Table 4. The model achieved similar performance using CNN segmented ROIs compared with manual segmented ROIs. In particular, the model achieved excellent NPV at 0.939/0.938 for manual and CNN segmentation, respectively, which is clinically important for identifying patients with normal bone density as a screening tool. For comparison, a separate model was trained only with BMFF average and clinical variables. These results are summarized in Table 5, where the AUC in training cohort was 0.942 and in the temporal validation cohort was 0.929. The accuracy and the negative predictive value (NPV) of 0.828 and 0.935 were also lower in this model compared with model built in the automated pipeline.
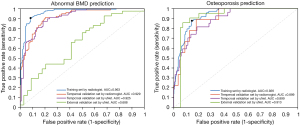
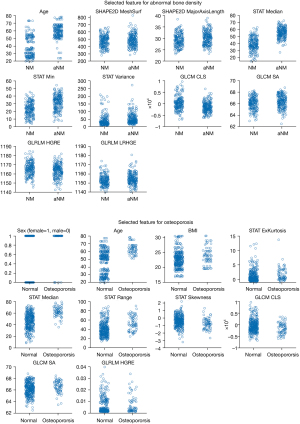
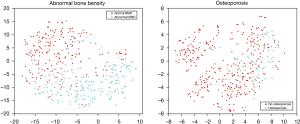
Table 4
| Abnormal BMD | AUC | sensitivity | specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|
| Training | 0.963 | 0.905 | 0.917 | 0.922 | 0.899 | 0.911 |
| 3-fold cross validation | 0.914 | 0.866 | 0.843 | 0.866 | 0.914 | 0.885 |
| Temporal validation | 0.929 | 0.923 | 0.816 | 0.774 | 0.939 | 0.859 |
| Validation uNet | 0.925 | 0.923 | 0.789 | 0.75 | 0.938 | 0.844 |
| External validation uNet | 0.688 | 0.786 | 0.545 | 0.688 | 0.667 | 0.68 |
BMD, bone mineral density; ROI, region of interest; CNN, convolutional neural network; AUC, area under the receiver operator characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
Table 5
| Abnormal BMD | AUC | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|
| Training | 0.942 | 0.919 | 0.838 | 0.867 | 0.905 | 0.880 |
| 3-fold cross validation | 0.940 | 0.914 | 0.824 | 0.851 | 0.899 | 0.871 |
| Validation | 0.929 | 0.923 | 0.763 | 0.727 | 0.935 | 0.828 |
BMD, bone mineral density; BMFF, bone marrow fat fraction; AUC, area under the receiver operator characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
In terms of the performance in predicting osteoporosis, the model selected seven radiomic features with all three clinical variables, as plotted in the bottom of Figure 5. The t-SNE visualization of the features are shown in the right of Figure 6. It yielded an AUC of 0.926 in the training cohort as shown in the right of Figure 4. For ROC analysis in the temporal validation cohort, this model achieved an AUC of 0.899/0.899 and an accuracy of 0.813/0.844 with manual/CNN segmented ROIs, respectively. In addition, it achieved a sensitivity of 0.778/0.818 and a specificity of 0.667/0.873 with manual/CNN segmented ROIs, respectively. The results are summarized in Table 6. In particular, the model achieved an excellent NPV of 0.957/0.941 for manual segmented ROIs, CNN segmented ROIs and external validation set using CNN segmented ROIs, respectively. This is clinically important for identifying patients without osteoporosis as screening tool. Similarly, a model was also trained using BMFF average and clinical variables only, and the results are shown in Table 7. These results showed AUC was lower in the temporal validation cohort (0.887) compared to using seven radiomic features and three clinical variables. The accuracy was 0.766 which is also lower than the radiomic model results.
Table 6
| Osteoporosis | AUC | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|
| Training | 0.926 | 0.877 | 0.852 | 0.552 | 0.971 | 0.857 |
| 3-fold cross validation | 0.907 | 0.793 | 0.844 | 0.518 | 0.953 | 0.836 |
| Temporal validation | 0.899 | 0.778 | 0.818 | 0.412 | 0.957 | 0.813 |
| Validation uNet | 0.899 | 0.667 | 0.873 | 0.462 | 0.941 | 0.844 |
| External validation uNet | 0.913 | 0.857 | 0.944 | 0.857 | 0.944 | 0.920 |
ROI, region of interest; CNN, convolutional neural network; AUC, area under the receiver operator characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
Table 7
| Osteoporosis | AUC | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|
| Training | 0.914 | 0.959 | 0.754 | 0.446 | 0.989 | 0.789 |
| 3-fold cross validation | 0.909 | 0.875 | 0.776 | 0.449 | 0.970 | 0.793 |
| Validation | 0.887 | 0.889 | 0.745 | 0.364 | 0.976 | 0.766 |
BMFF, bone marrow fat fraction; AUC, area under the receiver operator characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
For the external validation cohort, CNN based ROI was used. The abnormal BMD prediction achieved an AUC and accuracy of 0.688 and 0.68. The osteoporosis prediction model achieved a high AUC of 0.913 and an accuracy and NPV of 0.920 and 0.944.
Discussion
This work is the first attempt to predict ABD and osteoporosis using BMFF map radiomics and achieved promising prediction performance validated in a temporal validation set. When applied on temporal validation cohort, the ABD prediction model achieved a high AUC of 0.929/0925 and NPV of 0.939/0.938 for manual and CNN segmentation respectively. For the osteoporosis prediction, this model achieved an AUC of 0.899/0.899 and an accuracy of 0.813/0.844 for manual/CNN segmented ROIs, respectively. The model also achieved a high NPV of 0.938/0.938 for manual and CNN segmentation, respectively. Our study focused on predicting ABD and osteoporosis by radiomic analysis with fully automated CNN, providing a potential noninvasive biomarker, which can be easily adapted as a clinical screening tool using lumbar spine MRI images with artificial intelligence.
Furthermore, we performed external validation using data acquired from another site with a scanner from a different vendor (GE vs. Philips in the training and temporal validation cohorts). Although the prediction accuracy was found to be moderate for abnormal BMD at 0.68, the prediction accuracy for osteoporosis was found to be excellent at 0.92. As shown in Table 2, the scanning protocol for the external validation cohort was vastly different from the protocol used for training/validation/testing cohort. The voxel size is 1.5×1.7 mm2 for Philips machine and 2.0×2.0 mm2 for GE machine. The slice thickness is 3 mm for the Philips machine and 8mm for the GE machine. The difference in image voxel size will impact feature extracted (22). It is expected that the performance of the pipeline will be worse on external validation cohort. The accuracy of 0.68 for abnormal BME prediction is moderate considering the difference in the scanning protocol. The accuracy of 0.92 for osteoporosis prediction is actually higher than the accuracy of the temporal validation set. This high accuracy is encouraging, but this could be due to the small sample size of the external validation set (7 osteoporosis out of 25 subjects). Future study with multi-site data with large sample size is needed to confirm the performance.
Radiomics is a relatively new technique that can provide potential biomarkers for clinical outcomes by image feature extraction and analysis (27). As shown in Figure 7, the patterns of GLCM texture maps of osteoporosis and normal subjects are different. An increased bone marrow heterogeneity is observed in osteoporosis patients. Moreover, according to Figure 5, nine and seven radiomic features were selected by the model for ABD and osteoporosis prediction, respectively. Among these selected features, the median value and two features from GLCM matrix of the fraction map were selected by both models, while the mean value of the fraction map (also known as BMFF average) was excluded by both models. One possible reason is that the intensity values within ROIs do not necessarily follow normal distribution, suggesting the potential of the median value to better describe the first order statistics of the ROIs.
The abnormal BMD and osteoporosis prediction performance generated by CNN segmentation in our study were similar to that by manual segmentation. This model achieved an AUC of 0.929/0.925 and an accuracy of 0.859/0.844 with manual/CNN segmented ROIs, respectively. Furthermore, the mean Dice coefficient was 0.912±0.062 between U-Net ROIs and the manual ROIs in the validation cohort. The capability of automatic segmentation of vertebrae in fat fraction map in our work was similar with previous studies that used T1-weighted or T2-weighted images for spine segmentation (28,29). Another similar result was from the study of automatic vertebral body segmentation that based on deep learning of Dixon images with an AUC of 0.92 and a mean Dice coefficient of 0.849±0.091 (10). These parameter values suggest that solely using BMFF average and demographic characteristics is not as effective as using radiomic models. This is not surprising since radiomics is utilizing texture features not captured by a simple BMFF average. The machine learning radiomic approach is rapidly gaining popularity in radiology. It allows for the exploitation of patterns in imaging data and in medical records for a more accurate and precise quantification, diagnosis and prognosis.
QCT could be performed on any CT scanner with the use of calibration phantom and dedicated analysis software to obtain volumetric BMD (30). In our study we have used Mindways QCT pro (Model 3) with a synchronously calibrated phantom in local hospital and used Mindways CliniQCT (Model 4) with an asynchronously calibrated phantom in external hospital to obtain lumbar BMD. Both of these QCT systems have good accuracy assessment for volumetric trabecular BMD in the spine and good short-term precision and it has been demonstrated in previous studies (31-33). This is especially meaningful to verify the performance of the BMFF map Radiomics model for diagnosis of osteoporosis and ABD as QCT is the reference in both local and external data set.
X-ray and CT are the most commonly used imaging modalities to study bone health in machine learning according to literature. Rastegar et al. developed predictive models to classify patients with osteoporosis, osteopenia and normal bone density using radiomics by lumbar and pelvis radiography. The AUC range from 0.50–0.78 (9). Valentinitsch et al. demonstrated an automated pipeline for opportunistic osteoporosis screening using 3D texture features and regional BMD by CT images. The AUC of identifying patients with osteoporotic vertebral fractures is 0.88 (34). Recently, a variety of imaging techniques have been used to predict osteoporosis and BMD beyond conventional X-ray. Scanlan et al. and Vogl et al. used bone acoustics to infer bone health (35,36). Kim et al. and Hwang et al. used dental radiographs to predict jaw osteonecrosis and BMD, respectively (37,38). Studies using machine learning based on MRI images to predict BMD and fracture is few. Ferizi et al. and Deniz et al. used MRI to help with the diagnosis and segmentation of the images, respectively (39,40). Ferizi et al. compared the performance of different machine learning classifiers to predict osteoporotic bone fracture by MRI data and found that the RUS-boosted trees, the logistic regression and the linear discriminant are best and the sensitivity and specificity of the best model are 0.62 and 0.67, respectively (39). Compared with these studies, deep learning using the lumbar fat fraction map by mDixon sequence in our work has great performance for predicting ABD and osteoporosis and is free from exposure of ionizing radiation. Lumbar spine MR is routinely performed in patients with lower back pain to detect degenerative disc disease. mDixon Quant is an efficient and fast quantitative method that can be easily added into the routine protocol. Incorporating mDixon allows prediction for ABD and osteoporosis with automatic segmentation and BMFF map radiomics without additional radiation exposure. Thus, physicians can get information of not only discs, nerves and muscles, but also bone density for the patients with lower back pain through one MRI scan. In addition, mDixon Quant can be used as screening tool. Physicians can use the information generated from mDixon Quant to decide necessary further work-up which may involve DXA or QCT (8). During the study, we choose the midsagittal plane of the scan as it is the most conventional position of the lumbar spine. It can provide all the lumbar vertebral information in one slice. It is easier to obtain in routine scans, and the midsagittal plane also reduced the bias of analyzing different vertebrae in different images.
Our work is the first attempt using radiomics to predict osteoporosis with BMFF map, we also further validated the finding using independent external validation set. There is a competition between adipogenesis and osteogenesis in bone marrow (41). Previous work has demonstrated an inverse correlation between vertebral BMFF and BMD after being adjusted for age, sex and BMI and shown that BMFF has a good performance to predict osteoporosis and ABD (8). In this work, the abnormal BMD and osteoporosis prediction performance from radiomic model are better than model generated using only BMFF with clinical information except positive predictive value (PPV) in the same patient cohort as shown in Tables 4-7. In comparison to using BMFF averages only, radiomics brings additional and complementary information of the fat fraction map. It evaluates bone marrow fat texture features while BMFF is a quantitative indicator of bone marrow fat. The relationship of BMFF and BMD is not linear (8). BMFF is only an indicator of bone marrow fat content while radiomics contain more feature information of the fat fraction map, such as bone marrow heterogeneity. This will provide more comprehensive assessment of bone marrow fat and may contain certain unknown histologic features. Furthermore, age, sex and BMD has been demonstrated correlated with BMD. These factors were included in our radiomics model to improve the accuracy.
There are still some limitations in this work. First, as this cohort was recruited from the general population for screening osteoporosis and ABD, the subject number is unbalanced between osteoporosis, osteopenia and normal bone density group. This imbalance may cause bias during prediction temporal validation and might contribute to the low PPV in the validation set. Secondly, although QCT is more accurate than DXA for measurement of BMD, DXA is the gold standard for osteoporosis screening in clinics. Thirdly, fracture risk assessment is also very important in clinical. Further study is needed to pursue this. Furthermore, radiomic features were extracted from 2-dimensional (2D) ROIs. 3D radiomic analysis has the potential benefit of better capturing the heterogeneity of the full volume lumbar ROIs, however it suffers from more partial volume effect due to the fact that slice thickness is much larger than the in-plane resolution. As a result, 3D ROIs might lead to larger variation and we decided to use 2D ROIs in this study. Moreover, although the osteoporosis prediction performance of this prospective study is encouraging, the external validation data set sample size is relatively small and only from one external institute. Further multi-center study is needed to establish this as a screening tool.
In conclusion, this is the first study to demonstrate the feasibility of using radiomic analysis and fully automatic segmentation of fat fraction map to predict ABD and osteoporosis. It provides potential noninvasive biomarkers using a chemical shift-based water-fat separation sequence without ionizing radiation. This information would be valuable to the patients. This approach is suitable for ABD and osteoporosis prediction as a clinical screening tool. Further studies are expected to improve the prediction model and validate the findings using multicenter data. The model and the deep learning segmentation algorithm will be made publicly available on GitHub upon publication (link will be provided upon publication).
Acknowledgments
The authors would like to thank Juin-Wan Zhou for proofreading this manuscript.
Funding: This study received financial support from the National Natural Science Foundation of China (No. 81801653 and No. 82102018).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/qims-21-587
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-587). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of The Third Affiliated Hospital of Southern Medical University and The Fifth Affiliated Hospital of Sun Yat-sen University (No. 201711008 and NCT04647279), and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu J, Curtis EM, Cooper C, Harvey NC. State of the art in osteoporosis risk assessment and treatment. J Endocrinol Invest 2019;42:1149-64. [Crossref] [PubMed]
- Aparisi Gómez MP, Ayuso Benavent C, Simoni P, Aparisi F, Guglielmi G, Bazzocchi A. Fat and bone: the multiperspective analysis of a close relationship. Quant Imaging Med Surg 2020;10:1614-35. [Crossref] [PubMed]
- Ergen FB, Gulal G, Yildiz AE, Celik A, Karakaya J, Aydingoz U. Fat fraction estimation of the vertebrae in females using the T2*-IDEAL technique in detection of reduced bone mineralization level: comparison with bone mineral densitometry. J Comput Assist Tomogr 2014;38:320-4. [Crossref] [PubMed]
- Gokalp G, Mutlu FS, Yazici Z, Yildirim N. Evaluation of vertebral bone marrow fat content by chemical-shift MRI in osteoporosis. Skeletal Radiol 2011;40:577-85. [Crossref] [PubMed]
- Kühn JP, Hernando D, Meffert PJ, Reeder S, Hosten N, Laqua R, Steveling A, Ender S, Schröder H, Pillich DT. Proton-density fat fraction and simultaneous R2* estimation as an MRI tool for assessment of osteoporosis. Eur Radiol 2013;23:3432-9. [Crossref] [PubMed]
- Yoo HJ, Hong SH, Kim DH, Choi JY, Chae HD, Jeong BM, Ahn JM, Kang HS. Measurement of fat content in vertebral marrow using a modified dixon sequence to differentiate benign from malignant processes. J Magn Reson Imaging 2017;45:1534-44. [Crossref] [PubMed]
- Ma J. Dixon techniques for water and fat imaging. J Magn Reson Imaging 2008;28:543-58. [Crossref] [PubMed]
- Zhao Y, Huang M, Ding J, Zhang X, Spuhler K, Hu S, Li M, Fan W, Chen L, Zhang X, Li S, Zhou Q, Huang C. Prediction of Abnormal Bone Density and Osteoporosis From Lumbar Spine MR Using Modified Dixon Quant in 257 Subjects With Quantitative Computed Tomography as Reference. J Magn Reson Imaging 2019;49:390-9. [Crossref] [PubMed]
- Rastegar S, Vaziri M, Qasempour Y, Akhash MR, Abdalvand N, Shiri I, Abdollahi H, Zaidi H. Radiomics for classification of bone mineral loss: A machine learning study. Diagn Interv Imaging 2020;101:599-610. [Crossref] [PubMed]
- Zhou J, Damasceno PF, Chachad R, Cheung JR, Ballatori A, Lotz JC, Lazar AA, Link TM, Fields AJ, Krug R. Automatic Vertebral Body Segmentation Based on Deep Learning of Dixon Images for Bone Marrow Fat Fraction Quantification. Front Endocrinol (Lausanne) 2020;11:612. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. editors. U-Net: Convolutional Networks for Biomedical Image Segmentation 2015; Cham: Springer International Publishing.
- Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1-73. [Crossref] [PubMed]
- Dobbin KK, Simon RM. Optimally splitting cases for training and testing high dimensional classifiers. BMC Med Genomics 2011;4:31. [Crossref] [PubMed]
- Li N, Li XM, Xu L, Sun WJ, Cheng XG, Tian W. Comparison of QCT and DXA: Osteoporosis Detection Rates in Postmenopausal Women. Int J Endocrinol 2013;2013:895474. [Crossref] [PubMed]
- Adams JE. Quantitative computed tomography. Eur J Radiol 2009;71:415-24. [Crossref] [PubMed]
- Engelke K, Adams JE, Armbrecht G, Augat P, Bogado CE, Bouxsein ML, Felsenberg D, Ito M, Prevrhal S, Hans DB, Lewiecki EM. Clinical use of quantitative computed tomography and peripheral quantitative computed tomography in the management of osteoporosis in adults: the 2007 ISCD Official Positions. J Clin Densitom 2008;11:123-62. [Crossref] [PubMed]
- ACR-SPR-SSR practice parameter for the performance of quantitative computed tomography (QCT) bone densitometry The American College of Radiology; 2013.
- Ward RJ, Roberts CC, Bencardino JT, Arnold E, Baccei SJ, Cassidy RC, Chang EY, Fox MG, Greenspan BS, Gyftopoulos S, Hochman MG, Mintz DN, Newman JS, Reitman C, Rosenberg ZS, Shah NA, Small KM, Weissman BN. ACR Appropriateness Criteria® Osteoporosis and Bone Mineral Density. J Am Coll Radiol 2017;14:S189-202. [Crossref] [PubMed]
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980; 2014.
- Glorot X, Bengio Y. editors. Understanding the difficulty of training deep feedforward neural networks. Proceedings of the thirteenth international conference on artificial intelligence and statistics; 2010.
- Ding J, Stopeck AT, Gao Y, Marron MT, Wertheim BC, Altbach MI, Galons JP, Roe DJ, Wang F, Maskarinec G, Thomson CA, Thompson PA, Huang C. Reproducible automated breast density measure with no ionizing radiation using fat-water decomposition MRI. J Magn Reson Imaging 2018;48:971-81. [Crossref] [PubMed]
- Cattell R, Chen S, Huang C. Robustness of radiomic features in magnetic resonance imaging: review and a phantom study. Vis Comput Ind Biomed Art 2019;2:19. [Crossref] [PubMed]
- Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. [Crossref] [PubMed]
- Liao C, Li S, Luo Z. Gene Selection for Cancer Classification using Wilcoxon Rank Sum Test and Support Vector Machine. 2006 International Conference on Computational Intelligence and Security; 2006, 3-6 Nov. 2006.
- Liu C, Ding J, Spuhler K, Gao Y, Serrano Sosa M, Moriarty M, Hussain S, He X, Liang C, Huang C. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging 2019;49:131-40. [Crossref] [PubMed]
- Šimundić AM. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009;19:203-11. [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- Rak M, Steffen J, Meyer A, Hansen C, Tönnies KD. Combining convolutional neural networks and star convex cuts for fast whole spine vertebra segmentation in MRI. Comput Methods Programs Biomed 2019;177:47-56. [Crossref] [PubMed]
- Gaweł D, Główka P, Kotwicki T, Nowak M. Automatic Spine Tissue Segmentation from MRI Data Based on Cascade of Boosted Classifiers and Active Appearance Model. Biomed Res Int 2018;2018:7952946. [Crossref] [PubMed]
- Cheng X, Yuan H, Cheng J, Weng X, Xu H, Gao J, Huang M, Wáng YXJ, Wu Y, Xu W, Liu L, Liu H, Huang C, Jin Z, Tian WBone and Joint Group of Chinese Society of Radiology, Chinese Medical Association (CMA), Musculoskeletal Radiology Society of Chinese Medical Doctors Association, Osteoporosis Group of Chinese Orthopedic Association, Bone Density Group of Chinese Society of Imaging Technology, CMA. Chinese expert consensus on the diagnosis of osteoporosis by imaging and bone mineral density. Quant Imaging Med Surg 2020;10:2066-77. [Crossref] [PubMed]
- Wang L, Su Y, Wang Q, Duanmu Y, Yang M, Yi C, Cheng X. Validation of asynchronous quantitative bone densitometry of the spine: Accuracy, short-term reproducibility, and a comparison with conventional quantitative computed tomography. Sci Rep 2017;7:6284. [Crossref] [PubMed]
- Brown JK, Timm W, Bodeen G, Chason A, Perry M, Vernacchia F, DeJournett R. Asynchronously Calibrated Quantitative Bone Densitometry. J Clin Densitom 2017;20:216-25. [Crossref] [PubMed]
- Link TM, Lang TF. Axial QCT: clinical applications and new developments. J Clin Densitom 2014;17:438-48. [Crossref] [PubMed]
- Valentinitsch A, Trebeschi S, Kaesmacher J, Lorenz C, Löffler MT, Zimmer C, Baum T, Kirschke JS. Opportunistic osteoporosis screening in multi-detector CT images via local classification of textures. Osteoporos Int 2019;30:1275-85. [Crossref] [PubMed]
- Scanlan J, Li FF, Umnova O, Rakoczy G, Lövey N, Scanlan P. Detection of Osteoporosis from Percussion Responses Using an Electronic Stethoscope and Machine Learning. Bioengineering (Basel) 2018;5:107. [Crossref] [PubMed]
- Vogl F, Friesenbichler B, Hüsken L, Kramers-de Quervain IA, Taylor WR. Can low-frequency guided waves at the tibia paired with machine learning differentiate between healthy and osteopenic/osteoporotic subjects? A pilot study. Ultrasonics 2019;94:109-16. [Crossref] [PubMed]
- Kim DW, Kim H, Nam W, Kim HJ, Cha IH. Machine learning to predict the occurrence of bisphosphonate-related osteonecrosis of the jaw associated with dental extraction: A preliminary report. Bone 2018;116:207-14. [Crossref] [PubMed]
- Hwang JJ, Lee JH, Han SS, Kim YH, Jeong HG, Choi YJ, Park W. Strut analysis for osteoporosis detection model using dental panoramic radiography. Dentomaxillofac Radiol 2017;46:20170006. [Crossref] [PubMed]
- Ferizi U, Besser H, Hysi P, Jacobs J, Rajapakse CS, Chen C, Saha PK, Honig S, Chang G. Artificial Intelligence Applied to Osteoporosis: A Performance Comparison of Machine Learning Algorithms in Predicting Fragility Fractures From MRI Data. J Magn Reson Imaging 2019;49:1029-38. [Crossref] [PubMed]
- Deniz CM, Xiang S, Hallyburton RS, Welbeck A, Babb JS, Honig S, Cho K, Chang G. Segmentation of the Proximal Femur from MR Images using Deep Convolutional Neural Networks. Sci Rep 2018;8:16485. [Crossref] [PubMed]
- Wang D, Haile A, Jones LC. Dexamethasone-induced lipolysis increases the adverse effect of adipocytes on osteoblasts using cells derived from human mesenchymal stem cells. Bone 2013;53:520-30. [Crossref] [PubMed]



