
Fully automated discrimination of Alzheimer’s disease using resting-state electroencephalography signals
Introduction
Alzheimer’s disease (AD) is the leading cause of dementia in the elderly population, and its incidence is high among people over 65 years (1,2). Additionally, the prevalence rate of young-onset dementia (for which the overall prevalence is highest for AD) is not insignificant (3). If no mature treatment measures are used, the number of AD patients will be 75 million in 2030 and 131 million in 2050 (4). Presently, there is no cure for AD; however, if patients could be diagnosed at an early stage, proper interventions could slow down the progress of the disease. Thus, quite a few studies have tried to diagnose mild cognitive impairment (MCI), especially amnestic MCI, which has a higher conversion rate to AD (5-7). However, most diagnostic approaches are time consuming and expensive (8-10) and fail to satisfy the needs of timely diagnosis for most potential AD patients. Early identification and diagnosis is a significant challenge in low-resource environments in particular. Thus, there is a great demand for ‘language-free, culturally fair’ low-cost screening tools for AD.
Electroencephalography (EEG) has been the subject of growing interest as an investigational tool for biomarker development in AD (11-14). In practice, event-related tasks, such as memory-related tasks (15,16), decision-making tasks (17), and sensory-perceptual tasks (18), are widely used to examine the effects of AD on specific brain functions. Event-related tasks provide both behavioral and neural data but require multiple repetitions to achieve a stable response and have experiment times that are usually beyond patients’ tolerance (19). Additionally, some event-related tasks are beyond the cognitive capability of elderly subjects, especially those with low education levels. Even the performance of a simple memory task might cause discomfort and anxiety to patients (19).
Conversely, the instructions of resting-state tasks are easy to understand and follow regardless of the education levels of subjects (20). Resting-state protocols do not require external stimuli, and thus they are simple and comfortable for the elderly population and patients. Many resting-state EEG studies have found statistically significant differences among AD, MCI, and healthy controls (21-23).
To assist in applying resting-state EEG signals in AD diagnosis, researchers have explored numerous resting-state EEG properties. The tendency of brain activity to slow down among AD patients has been commonly observed (15,24). Previous studies showed that the power of low-frequency rhythms (e.g., delta and theta) are increased, and the power of high-frequency rhythms (e.g., alpha, beta, and gamma) are decreased in AD patients compared to healthy controls (21,25,26). This shift is proportional to the progression of AD (20). Another common representation in AD patients is the reduced complexity of EEG signals (24). The potential pathophysiological basis for the decreased EEG complexity in AD is not yet clear. The decrease is likely caused by fewer active neurons and the reduction of non-linear connections between them, which leads to simpler dynamics (24,26,27). Other mechanisms, such as the deficiency of neurotransmitters, are also under discussion. It is well accepted that AD leads to changes in neural synchronization (22,28); however, the specific patterns of these changes are still under debate. Some studies have reported that AD patients’ EEG signal synchronization tends to be reduced, while others have shown that it increases (13,22,28-30). Apart from these 3 widely used features, many other resting-state EEG features have been used to detect AD, such as microstate (31,32), epileptiform activity (33), and visibility graphs (34).
With the development of machine learning, classification methods, such as support vector machine (SVM) (35) and random forest (RF) (36), are used to train models to distinguish AD subjects from healthy controls based on resting-state EEG (37). The sensitivity and specificity of such classifiers can be as high as 90% (24,37,38). However, due to the difficulty of acquiring data with evidence-based medical diagnoses, most studies in this area have no more than 50 subjects (7,13,21,24,38-40). This number is acceptable for statistical analyses; however, the machine learning-based classification results are not reliable for generalization with so few samples. In addition, some classification studies have failed to provide necessary details, such as how to choose the training, development, and testing data, which makes the results less persuasive (21,39,40). Additionally, when simply putting features extracted together without feature selection or feature fusion optimization methods, the extremely high-dimensional features compared with the sample number could lead to over-fitting models (41). With the advancements of graphics processing units in computing, deep-learning approaches, such as convolutional neural networks (42,43), recurrent neural networks (44), and discriminative deep probabilistic models (45), have been applied to detect AD and have achieved remarkable accuracy. However, the high requirement of computing resources and the explainability problem of deep-learning models prevent their clinical application. Apart from the theoretical issues, another practical problem facing the clinical application of resting-state EEG in AD screening is the high labor cost and experience dependence in data preprocessing (20). Thus, the full automation of the discrimination of AD is required.
In the current study, based on a resting-state EEG data set comprising 113 normal cognition (NC) subjects, 116 MCI subjects, and 72 AD subjects with evidence-based medical diagnoses, we aimed to explore the development of a fully automated process, including data preprocessing, feature extraction, and machine learning-based classification, to differentiate among the 3 groups.
We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/qims-21-430).
Methods
Subjects and neuropsychological measures
Data collection was carried out at the Department of Neurology and the Department of Memory Clinic of Tongji Hospital. The study was conducted as per the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of Tongji Hospital (No. K-2017-003-XZ-190130). All subjects provided written informed consent after being given a complete description of the study. In total, the EEG data of 301 subjects (comprising 135 males and 166 females) were acquired in this study. Subjects were recruited from the Department of Neurology and the Memory Clinic of Tongji Hospital. Professional clinical neurologists diagnosed all the subjects via neuroimaging tests, including computed tomography and magnetic resonance image, and a comprehensive neuropsychological battery that included the Mini-Mental State Examination (MMSE) (46), Montreal Cognitive Assessment-Basic (MoCA-B) (47), Rey-Osterrieth Complex Figure Test (ROCF) (48), Trail Making Test (TMT) (49), Hopkins Verbal Learning Test (HVLT) (50), Wechsler Memory Scale (WMS) (51), Verbal Fluency Test (VFT) (52), and Boston Naming Test (BNT) (53). Examinations including biochemical blood assessments of folic acid, vitamin B12, thyroid function (free triiodothyronine, free tetraiodothyronine, and thyroid-stimulating hormone), treponema pallidum, and human immunodeficiency virus (HIV) antibodies were conducted to exclude memory loss caused by other reasons. Their demographic and clinical information were also recorded. Further details of subjects’ demographic information and neuropsychological performance are provided in Table 1.
Table 1
| Demographics/Performance | NC (n=113) | MCI (n=116) | AD (n=72) | Statistics |
|---|---|---|---|---|
| Age, years | 67.79 (9.88) | 68.17 (10.82) | 73.37 (8.78) | F(2) = 12.02, P<0.001 |
| Gender, male/female | 61/52 | 45/71 | 29/43 | Chi2 = 4.77, P=0.09 |
| Education, years | 10.14 (3.51) | 9.37 (4.55) | 9.12 (4.86) | F(2) = 27.16, P<0.001 |
| MoCA | 23.24 (3.25) | 17.13 (4.32) | 10.90 (5.39) | F(2) = 181.19, P<0.001 |
| MMSE | 27.71 (1.81) | 24.41 (3.40) | 17.72 (6.94) | F(2) = 126.53, P<0.001 |
Values are presented as mean (SD) unless otherwise indicated. NC, normal cognition; MCI, mild cognitive impairment; AD, Alzheimer’s disease; MoCA, Montreal Cognitive Assessment; MMSE, Mini-Mental State Examination.
The subjects were separated into the following 3 groups according to their clinical diagnosis: (I) the NC group (113 subjects); (II) the MCI group (116 subjects); and (III) the AD (72 subjects) group. AD was diagnosed based on the clinical diagnostic criteria for probable AD of the National Institute on Aging and Alzheimer’s Association (54). As there was a lack of in-vivo evidence, such as amyloid positron emission tomography, amyloid/tau cerebrospinal fluid markers, and the presence of autosomal dominant mutations, the subjects in the AD group in the current study could only be referred as probable AD (55). To focus on the etiology of AD, only the amnestic MCI subjects were included in the current study, containing both single-domain and multiple-domain amnestic MCI. According to the general criteria for MCI as defined previously (56,57), subjects were diagnosed with amnestic MCI by neurologists if they met the following specific criteria: (I) the subject and their caregiver complained of memory decline; (II) the subject met the criteria for MMSE (based on the education level, illiterate ≤17, primary school ≤20, or middle school and above ≤24 scores) or MoCA-B (based on the education level, primary school and below ≤19, middle school and high school ≤22, or college ≤24 scores); (III) the subject had a Clinical Dementia Rating Scale (CDR) (58) score of 0.5; (IV) the subject was impaired in the single cognitive domain of memory, or multiple cognitive domains, including the domain of memory, according to the neuropsychological battery tests; (V) the subject had normal daily life function according to the Instrumental Activities of Daily Living (IADL-14) (59) scale. The control group included the subjects who had completed the cognitive tests but were diagnosed as cognitively healthy, which was referred to as NC.
Subjects were excluded from the study if they met any of the following exclusion criteria: (I) were aged below 40 years; (II) had a definite history of stroke; (III) had a definite history of other diseases of the central nervous system, such as infection, demyelinating diseases, and Parkinson’s disease; (IV) had a definite history of mental illness, such as schizophrenia, major depressive disorder; (V) had a serious physical disease; (VI) had an alcohol or drug addiction; (VII) had clinically significant abnormalities in relation to folic acid, vitamin B12, thyroid function, or had positive syphilis or HIV antibodies; (VIII) were unable to complete the neuropsychological tests; (IX) had a Hachinski Ischemic Scale score ≥4; and/or (X) had a Hamilton Rating Scale for Depression (HAMD) score ≥14.
Experiment procedures
All the subjects claimed to have had a sufficient amount of sleep the day before the test. During the resting-state EEG recording, the subjects were sitting in a comfortable upright position and were asked to stay as calm as possible, while keeping their eyes closed for about 5 minutes. According to the 10–20 international system, Electrodes were placed with the reference electrode on the bilateral mastoids. Conductivity was ensured by keeping the impedance of all channels below 10 k Ohm for all the subjects. The EEG signals were acquired at a sampling rate of 1,000 Hz, via a 62-channel [60-channel EEG and 2-channel electrooculogram (EOG)] EEG amplifier (SynAmps2, Neuroscan, USA), armed with Curry 8 data acquisition software. The parameters of the SynAmps2 and Curry 8 were the default settings. In total, 301 subjects’ eye-closed resting-state EEG signals were acquired. The length of the resting-state data was 300±22.1 seconds.
Automated data preprocessing
An automated data preprocessing procedure was developed to remove artifacts, which was done in Matlab with the Fieldtrip (60) and EEGLAB (61) toolbox. First, the raw EEG recordings were filtered to a frequency band between 0.1 and 95 Hz by combining low- and high-pass filters with 95 and 0.1 Hz cut-offs, respectively. The signals were then detrended and downsampled to 500 Hz. The first and last 2 s of the signals were removed to avoid unstable responses. Second, the data of all channels were discarded at the time point when the amplitude of more than 5 channels were larger than 250 μV. In this case, the remaining signals were naturally segmented into several trials because parts of the continuous signals were removed. For each trial, bad channels were identified by a series of statistical indicators, including standard deviation, Hurst exponent, and correlation coefficients. The threshold of each indicator was determined by the median and standard deviation values across all channels. If the indicators were beyond the threshold, the corresponding channel was marked as a bad channel. Before conducting the independent component analysis (ICA), all the bad channels were put aside to extract artificial components, including EOG, electromyography, and electrocardiogram, from the data. After calculating the ICA components, the ADJUST toolbox (62) in EEGLAB was used to identify and remove artificial components. Finally, the marked bad channels were put back and replaced by the interpolation results of the neighboring channels. The Reference Electrode Standardization Technique was then applied to re-reference all the signals (63). The signals were then segmented into epochs of 15 s without overlap.
Feature extraction
The features of the EEG signals include spectral features, complexity, and functional connectivity. This paper used the Band Power Ratio and Continuous Wavelet Transform features as the spectral features. Complexity was used to measure the variability within the EEG signal of each channel, while functional connectivity was used to measure the consistency of EEG signals across channels.
Band power ratio
Power spectrum density was used to present the relative power of the following 6 bands: delta (1–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), low beta (beta1, 12–18.5 Hz), middle beta (beta2, 18.5–21 Hz), and high beta (beta3, 21–30 Hz). To reduce the individual influence of each band, the band power ratio was also calculated, and the following 6 indices were used to evaluate the change of the power of different bands (64):
CWT features
The continuous wavelet transform (CWT) features used in this study were defined as the average magnitude of all CWT coefficients, and were obtained using a selected mother wavelet bump, in that particular frequency band over the entire signal length, which was written as (38):
where fsi is the starting frequency of the ith band, fei is the ending frequency, ts is the starting time, te is the ending time, and Yj,k are the wavelet transform coefficients based on the selected mother wavelet. The frequency bands were the same as those used to calculate band power ratios. The beta1, beta2, and beta3 bands were merged as a beta band.
Complexity
4 indices were adopted to measure the complexity of the EEG signals: Permutation Entropy (P-En), Sample Entropy (S-En), Wavelet Entropy (W-En), and Lempel-Ziv (LZ) complexity. P-En is a dynamic mutation-detection measurement that easily and accurately measures the mutation and amplifies the small change of signals; S-En is used to measure the probability of generating new patterns in signals, and the low value of S-En indicates the high self-similarity of signals; W-En measures the disorder degree of signal energy distribution in different subspaces. Multiscale entropy, which is generated by extending entropy calculation to multiple time scales (65), was also calculated.
Lempel–Ziv (LZ) complexity is another measure used to quantify the complexity of signals, and is usually relevant to the frequency of the same trend in the signal. The EEG signals were converted to binary 0 and 1 before the LZ-complexity calculation. If the value of the current point was larger than the mean value of the total signal, the current point was converted to 1, and otherwise to 0. This step was repeated twice to measure the finer change variation tendency of the signals (66).
Functional connectivity
This paper adopted 2 functional connectivity indices, the correlation coefficient and cross-power spectral density correlation coefficient, to measure amplitude synchronization and spectral synchronization, respectively The correlation coefficient of the 2 signals’ x1 and x2 was calculated as:
where Cov(x1,x2) indicates the covariance of signals x1 and x2, Var(x1) indicates the variance of x1, and Var(x2) indicates the variance of x2.
The cross-power spectral density correlation coefficient was calculated as:
where cpsd(x1,x2) indicates the cross-power spectral density between signals x1 and x2, psd(x1) indicates the power spectral density of x1, and psd(x2) indicates the power spectral density of x2.
Both the complexity and functional connectivity features were calculated based on the signals of 1–30 Hz. All the features were first extracted epoch-wisely and then averaged across epochs within subjects.
Statistical analysis
The analysis of variance (ANOVA) was used to estimate the degree of difference among the NC, MCI, and AD groups for different features. First, the 3 categories of NC, MCI, and AD were analyzed together to identify the regions of the brain with an ANOVA P<0.05. Second, a Tukey-Kramer multiple comparisons (67) was made between every 2 categories, and the electrodes with a P<0.05 were also found.
Classification
The performance of 9 commonly used classifiers, including a linear discriminant analysis (LDA), logistic regression model, naive Bayes classifier, nearest neighbor classifier, SVMs, and ensemble trees [e.g., RF, gradient boosting decision tree (GBDT), XGBoost, and random undersampling (RUS) boosting] were compared on the classification tasks, including AD vs. NC, AD vs. MCI, and MCI vs. NC. The parameters of the models were chosen according to the suggested or default settings in the Classification Learner of MATLAB.
LDA, also known as the Fisher discriminant, assumes that different classes generate data based on Gaussian distributions. The amount of regularization to apply when estimating the covariance matrix of the predictors was set to 0. The logistic regression models the probability of output in terms of input, and can be used to make a classifier, by choosing a cutoff value. It is a common way to make a binary classifier (68). The naive Bayes classifier estimates the parameters of a probability distribution (the kernel smoothing density estimate in our case) using the training data. For any unseen test data, the naive Bayes classifier computes the posterior probability of that sample belonging to each class, and then classifies the test data according to the largest posterior probability. The nearest neighbor classification model categorizes query points based on their distance to points in a training data set. In the present study, Euclidean metrics were used to determine the distance with no weighting. The number of nearest neighbors was set to 10. The SVM with a linear kernel was also recruited, where the scale factor was selected using a heuristic procedure.
Ensemble approaches train many weak classifiers (e.g., trees) and combine their predictions to enhance the performance of a single weak learner. RF and 2 other ensemble tree approaches [GBDT (69) and XGBoost (70)] were conducted. RF builds many trees independently from different bootstrap samples of the training data, while allowing to use of different subsets of variables at the nodes of any tree in the ensemble and then votes their predictions to get a final prediction (36). Additionally, RUS boosting, which is an especially effective approach to classifying imbalanced data, was recruited due to the mild imbalance among groups in the current study. The learning rate was set to 0.1. The number of ensemble learning cycles of the ensemble trees was set to 30. The maximal number of decision splits was set to the number of sample sizes in the training sample.
The samples were divided into a testing set and a training set by 5-fold cross-validation (1 fold for testing, and 4 other folds for training). The classification models were trained on the training set. The recognition accuracy, specificity, sensitivity, and area under the curve (AUC) with 95% confidence intervals were used to evaluate the overall performance of the models using the testing set. The sample sizes were similar among the groups; however, a permutation test was conducted for each classification to determine the empirical chance level accuracy, which is the 95 percentile of empirical performance distribution established by randomly permuting the labels 1,000 times. Permutation tests help identify the over-fit of models and predict that the given model is significantly better than the one built under the same conditions but on random data.
Some features are uninformative, irrelevant or redundant for classification, so reducing the number of features could speed up computation and improve classification performance (71). This paper applied principal component analysis (PCA) to the feature matrix to extract the latent components representing 95% of all variability (72).
The data processing flowchart shows how the raw resting EEG data was processed and how the features were extracted and used for classification (see Figure 1).
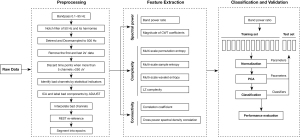
Results
NC could be statistically distinguished from AD but barely from MCI
To include enough information, the spectral, complexity, and functional connectivity features were considered in the current study. The spectral features included 6 kinds of band power ratio and CWT features in 4 frequency bands for each channel, resulting in 600 features. The complexity included 3 kinds of entropy and LZ complexity. For entropy, the multiscale entropy of a scale from 1 to 20 was obtained for every channel, so each entropy indices had 1,200 features. Combining the LZ-complexity value of each channel, there were 3,660 complexity features in total. 2 connectivity matrices were calculated as the correlation coefficient and cross-power spectral density correlation coefficient. As the functional connectivity matrices were symmetric, only the upper triangular matrices were used to avoid redundant information, resulting in 3,540 features. The number of features is shown in Table 2.
Table 2
| Feature category | Features | Dimension |
|---|---|---|
| Spectral | Band power ratio | 6 types * 60 channels = 360 |
| CWT | 4 frequency bands * 60 channels = 240 | |
| Complexity | Multi-scale permutation entropy | 20 scales * 60 channels = 1,200 |
| Multi-scale sample entropy | 20 scales * 60 channels = 1,200 | |
| Multi-scale wavelet entropy | 20 scales * 60 channels = 1,200 | |
| LZ complexity | 60 channels | |
| Functional Connectivity | Correlation coefficient | |
| Cross-power spectral density |
CWT, continuous wavelet transform; LZ, Lempel-Ziv.
An ANOVA was conducted to identify the statistically differential features among AD MCI and NC subjects for the spectral, complexity, and functional connectivity features. For the spectral features, at least 1 channel of all the band power ratio features demonstrated significant differences among the 3 groups (P<0.05 with FDR correction), except for r6. Among the 6 kinds of ratios, r1 and r3 showed the best distinguishable performance in the corrected ANOVA P value, followed by r4 (see Figure 2A). The most distinguishable channels were located in the bilateral temporal lobes and temporoparietal junction areas. However, even for r1 and r3, according to the results of the post-hoc analysis, NC and MCI were not distinguishable for all the channels, while both NC vs. AD and MCI vs. AD were distinguishable (see Figure 2B). The mean value of r1 of AD was greater than that of MCI, while the mean values of MCI were greater than those of NC but there was overlap (see Figure 2C,2D).
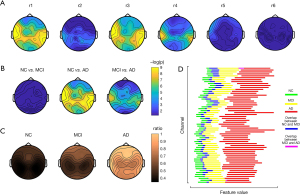
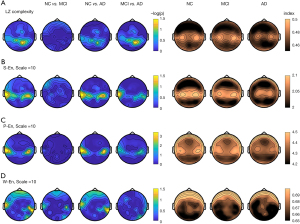
With the complexity features, the LZ complexity of the middle posterior regions indicated significantly different complexity among the 3 groups (P<0.05 with FDR correction), while the bilateral temporal regions of the 3 types of entropy revealed differences among the groups. Both LZ complexity and entropy decreased, as did the increase of cognitive impairment (AD < MCI < NC). According to the post-hoc results, middle posterior LZ complexity distinguished between NC and AD, and MCI and AD, but not MCI and NC (see Figure 3A). Similarly, the bilateral temporal S-EN and P-EN could distinguish between NC and AD, and MCI and AD, but not MCI and NC (see Figure 3B,3C). Conversely, the right temporal W-En distinguished between NC and AD, and NC and MCI, but not MCI and AD (see Figure 3D).
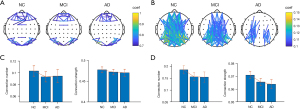
The group-level connectivity networks of the correlation coefficient (see Figure 4A) and cross-power spectral density correlation coefficient (see Figure 4B) in NC, MCI, and AD are shown in Figure 4 (only the nonadjacent connections with the top 10% largest connectivity strength were plotted). The connection number (the left plots in Figure 4C,4D) and the connection strength (the right plots in Figure 4C,4D) in the individual levels were compared among the groups. There were more connections in NC than MCI and AD, and these connections were stronger, but the differences were not statistically significant.
The machine-learning models distinguished among the 3 groups
When separating AD from NC, among all the classifiers, the RF had the best performance in terms of the AUC (80.08%), and RUS Boosting was the best in terms of accuracy (72.43%). The specificity of the RF was as high as 85.84%, but the sensitivity was only 50%. Conversely, the GBDT had a relatively balanced performance (with an AUC of 79.52%, an accuracy of 71.59%, a specificity of 73.21%, and a sensitivity of 70.12%). Table 3 shows the classification performance of all the classifiers. Regardless of the classifier algorithm, all the classifiers had a significantly higher level of accuracy at distinguishing AD from NC (P<0.001, permutation test) than the empirical chance level accuracy.
Table 3
| Classifier | Accuracy (%) | Empirical chance level accuracy (%) | Specificity (%) | Sensitivity (%) | AUC (95% CI) (%) |
|---|---|---|---|---|---|
| LDA | 70.27 | 52.97 | 87.61 | 43.06 | 76.16 (68.34–82.54) |
| Logistic regression | 66.49 | 51.89 | 78.76 | 47.22 | 67.47 (58.84–75.05) |
| Naive bayes | 69.73 | 48.11 | 87.61 | 41.67 | 73.88 (65.91–80.54) |
| SVM | 70.81 | 56.22 | 91.15 | 38.89 | 76.06 (68.05–82.57) |
| Nearest neighbor | 64.32 | 54.05 | 97.35 | 12.50 | 73.97 (65.91–80.69) |
| Random forest | 71.89 | 51.89 | 85.84 | 50.00 | 80.08 (72.64–85.89) |
| XGBoost | 68.75 | 51.56 | 74.89 | 65.96 | 75.86 (74.02–76.96) |
| GBDT | 71.59 | 49.76 | 73.21 | 70.12 | 79.52 (78.92–81.25) |
| RUS boosting | 72.43 | 45.41 | 76.99 | 65.28 | 77.51 (69.72–83.76) |
Empirical chance level accuracy is the 95 percentile of empirical performance distribution established by randomly permuting the labels 1,000 times, representing that the original classification is significant with P<0.05. CI, confidence interval; LDA, linear discriminant analysis; SVM, support vector machine; GBDT, gradient boosting decision tree; RUS, random under sampling.
People with amnestic MCI are more likely to have an AD etiology, and have a higher conversion rate to AD. The machine-learning classifiers were also able to separate AD from amnestic MCI. Among all the classifiers, the RUS boosting achieved the best performance (with an AUC of 70.82%, an accuracy of 69.11%, a specificity of 69.75%, and a sensitivity of 62.42%). Most classifiers, except for the logistic regression classifier (P=0.16, permutation test) had a significantly higher level of accuracy (P<0.001, permutation test) than the empirical chance level accuracy. More details of the classification performance are set out in Table 4.
Table 4
| Classifier | Accuracy (%) | Empirical chance level Accuracy (%) | Specificity (%) | Sensitivity (%) | AUC (95% CI) (%) |
|---|---|---|---|---|---|
| LDA | 64.92 | 54.26 | 87.39 | 27.78 | 67.30 (58.78–74.81) |
| Logistic regression | 60.73 | 63.72 | 78.99 | 30.56 | 56.14 (47.23–64.67) |
| Naive bayes | 68.06 | 49.47 | 80.67 | 47.22 | 69.05 (60.65–76.35) |
| SVM | 64.92 | 57.45 | 89.92 | 23.61 | 67.09 (58.68–74.53) |
| Nearest neighbor | 64.40 | 55.32 | 93.28 | 16.67 | 63.46 (54.93–71.22) |
| Random forest | 67.02 | 52.66 | 84.03 | 38.89 | 69.05 (60.58–76.42) |
| XGBoost | 61.38 | 52.42 | 62.67 | 60.16 | 66.01 (64.22–67.69) |
| GBDT | 61.43 | 51.62 | 63.30 | 60.29 | 67.85 (66.19–69.11) |
| RUS boosting | 69.11 | 56.54 | 69.75 | 62.42 | 70.82 (62.69–77.81) |
Empirical chance level accuracy is the 95 percentile of empirical performance distribution established by randomly permuting the labels 1,000 times, representing that the original classification is significant with P<0.05. CI, confidence interval; LDA, linear discriminant analysis; SVM, support vector machine; GBDT, gradient boosting decision tree; RUS, random under sampling.
With differentiating MCI from NC cases, only the SVM, nearest neighbor, and ensemble trees (RF, XGBoost, GBDT, and RUS boosting) had a significantly higher level of accuracy (P<0.05, permutation test) than the empirical chance level accuracy. Among them, the nearest neighbor achieved the highest AUC of 63.95%, but had a relatively low sensitivity. The RUS boosting had a balanced performance in terms of AUC of 60.21%, an accuracy of 59.91%, a specificity of 62.83%, and a sensitivity of 57.14%. More details of the classification performance are listed in Table 5.
Table 5
| Classifier | Accuracy (%) | Empirical chance level Accuracy (%) | Specificity (%) | Sensitivity (%) | AUC (95% CI) (%) |
|---|---|---|---|---|---|
| LDA | 53.02 | 56.90 | 50.44 | 55.46 | 55.34 (47.86–62.57) |
| Logistic regression | 43.53 | 56.47 | 39.82 | 47.06 | 58.54 (51.07–65.64) |
| Naive bayes | 53.88 | 56.90 | 69.03 | 39.50 | 58.92 (51.45–66.00) |
| SVM | 56.03 | 55.33 | 57.52 | 54.62 | 56.85 (49.35–64.04) |
| Nearest neighbor | 58.62 | 56.03 | 77.88 | 40.34 | 63.95 (56.50–70.80) |
| Random forest | 59.48 | 56.47 | 68.14 | 51.26 | 61.20 (53.71–68.19) |
| XGBoost | 54.04 | 45.13 | 57.60 | 51.30 | 56.21 (54.86–58.60) |
| GBDT | 54.38 | 44.76 | 57.04 | 52.87 | 57.31 (54.94–60.32) |
| RUS boosting | 59.91 | 56.03 | 62.83 | 57.14 | 60.21 (52.67–67.31) |
Empirical chance level accuracy is the 95 percentile of empirical performance distribution established by randomly permuting the labels 1000 times. Larger than empirical chance level accuracy represents that the original classification is significant with P<0.05. CI, confidence interval; LDA, linear discriminant analysis; SVM, support vector machine; GBDT, gradient boosting decision tree; RUS, random under sampling.
The distinguishable power of demographic information was also examined to ensure that the classification performance was not due to the potential influence of age, gender, or education level. For example, in differentiating AD from NC, the accuracy of the RF classifier using demographic information was 71.35%, and the accuracy using both demographic information and resting-state EEG features was 74.59%. In differentiating AD from MCI, the accuracy using demographic information was 61.78%, and the accuracy using both demographic information and resting-state EEG features was 66.49%. In differentiating MCI from NC, the accuracy using demographic information was 57.33%, and the accuracy using both demographic information and resting-state EEG features was 59.05%. For all the classifiers in all the conditions, a classifier that used demographic information always performed worse than a classifier that used both demographic information and resting-state EEG features or resting EEG features alone.
Discussion
In the current study, a process was developed to implement the automated assistant diagnosis of AD using resting-state EEG signals. By combining the statistical analysis and classification methods, the results confirmed the potential of distinguishing among NC, amnestic, MCI, and probable AD subjects on a large data set. In line with previous studies, the statistical analysis results demonstrated the tendency to slow brain activity and reduced complexity with the progression of AD (15,24). Among them, entropy and spectral power features were better able to differentiate NC from AD, LZ complexity was better able to differentiate MCI from AD. At the same time, only the right temporal W-En showed statistical separability between NC and MCI (see Figures 2,3). Concerning the connectivity, our results provide evidence (see Figure 4) that the connectivity became weaker with the progression of AD (15,22,28). More and more complex features have been developed and explored in the emerging literature; however, our results showed that the plain spectral features are good candidates for filling the gap between clinical use and empirical studies. To assist clinicians to use EEG more efficiently to distinguish among patients in different states, we compared the EEG features among the NC, MCI, and AD groups more intuitively by showing the detailed distribution range of Band Power Ratio theta/(alpha + beta1) of the 3 groups with 95% confidence intervals (see Figure 2D). The values of the AD subjects were higher than those of the NC and MCI subjects in every channel; however, there was an overlap between the NC and MCI subjects. The red bars in Figure 1D can be accepted as an EEG biomarker of AD distribution with high confidence, and be used by clinicians to identify AD patients.
The classification results also supported the clinical diagnosis of AD via EEG signals. The AUC of AD vs. NC on the testing set was 80.08%, and the AUC of AD vs. MCI was 70.82%. It was also easier to classify AD vs. NC than AD vs. MCI in terms of classification accuracy. Notably, the AD group in the current study was only probable AD; thus, some patients may have been misdiagnosed, which should be noted when interpreting the results. Compared to the results of distinguishing AD from the other 2 groups, the performance of distinguishing NC from MCI was less satisfactory, but still surpassed the empirical chance level. Previous studies (22,40,73) have shown that MCI and NC groups can be distinguished using similar features. This may be because of the variable physiological states of MCI subjects in different studies, as the MCI state is easily influenced by many factors, such as non-pathological factors, even though the labels are evidence-based medical diagnoses. In our dataset, the MCI group comprised amnestic MCI patients, single-domain or multiple-domain, while the NC group comprised outpatients of the Department of Neurology with a subjective complaint of memory decline. Those who were diagnosed as NC were taken as the control group. In some studies, this kind of subject was described as having subjective cognitive decline (SCD) based on the results of SCD-Q9 screenings or other SCD questionnaires. Unfortunately, we did not have these data; thus, the NC group included the subjects who were diagnosed as cognitively healthy. Previous studies have shown significant differences in EEG markers between SCD and healthy controls (74,75). The limited differences between MCI and controls in this study may partly be due to this.
Additionally, different acquisition conditions and processing methods could also affect the classification results. Relatively speaking, more reliable and generalizable conclusions could be drawn from results based on a larger scale of samples, such as the current study. Conversely, the timely diagnosis of MCI patients is also necessary to ensure they receive intervention and treatment to slow down the conversion to AD and better overall disease management. Thus, further investigation still needs to be conducted of the EEG features that better distinguish NC from MCI to diagnose MCI subjects.
Demonstrating the separability among the 3 categories could potentially be useful to achieve the early diagnosis of AD, but it is not sufficient. A longitudinal study (rather than a cross-sectional study) should be conducted to reveal the neural indicator of disease progressions. Ideally, separating MCI subjects with and without an AD etiology based on follow-ups is a straightforward approach for developing clinically useful tools to aid with early diagnosis. There are some other directions worthy of further study. The automated preprocessing methods are labor-saving, especially for large-scale data. Our study developed an automated method to preprocess the raw EEG data, but the results of the automated preprocessing were not quantitatively verified because of the limited available specifically labeled data. Additionally, more EEG data are required when using deep learning to train a more effective model.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (2018YFC1314700), the National Science Foundation of China (81671307), Database Project of Tongji Hospital of Tongji University (TJ(DB)2102), the Postdoctoral Foundation of Anhui Province, China (2019B289), the Shanghai Sailing Program, China (20YF1442000), and the Hospital Program and Qihang Program of Shanghai Mental Health Center (2020-QH-01, 2020-YJ01).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/qims-21-430
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-430). YC, SW, and XL are employees, and ZL has served as a paid consultant in the iFLYTEK Research division of iFLYTEK CO., LTD. YD is a former employee of iFLYTEK CO., LTD. ZL, SW, and XL own iFLYTEK stock. YC has a related pending patent. These authors’ employment and remuneration do not depend on the outcomes or publication of this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Tongji Hospital (No. K-2017-003-XZ-190130). All subjects provided written informed consent after being given a full description of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brookmeyer R, Evans DA, Hebert L, Langa KM, Heeringa SG, Plassman BL, Kukull WA. National estimates of the prevalence of Alzheimer's disease in the United States. Alzheimers Dement 2011;7:61-73. [Crossref] [PubMed]
- Fratiglioni L, Grut M, Forsell Y, Viitanen M, Grafström M, Holmén K, Ericsson K, Bäckman L, Ahlbom A, Winblad B. Prevalence of Alzheimer's disease and other dementias in an elderly urban population: relationship with age, sex, and education. Neurology 1991;41:1886-92. [Crossref] [PubMed]
- Hendriks S, Peetoom K, Bakker C, van der Flier WM, Papma JM, Koopmans R, et al. Global Prevalence of Young-Onset Dementia: A Systematic Review and Meta-analysis. JAMA Neurol 2021; Epub ahead of print. [Crossref] [PubMed]
- Winblad B, Amouyel P, Andrieu S, Ballard C, Brayne C, Brodaty H, et al. Defeating Alzheimer's disease and other dementias: a priority for European science and society. Lancet Neurol 2016;15:455-532. [Crossref] [PubMed]
- Silveira M, Marques J. Boosting Alzheimer Disease Diagnosis Using PET Images. 2010 20th International Conference on Pattern Recognition, 2010:2556-9.
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR Jr. Alzheimer's disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage 2008;39:1186-97. [Crossref] [PubMed]
- Jeong J, Gore JC, Peterson BS. Mutual information analysis of the EEG in patients with Alzheimer's disease. Clin Neurophysiol 2001;112:827-35. [Crossref] [PubMed]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature 2009;461:916-22. [Crossref] [PubMed]
- Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, Perrin RJ, Bales KR, Soares H, Fagan AM, Holtzman DM. Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer's disease diagnosis and prognosis. PLoS One 2011;6:e18850. [Crossref] [PubMed]
- Kim S, Lee Y, Jeon CY, Kim K, Jeon Y, Jin YB, Oh S, Lee C. Quantitative magnetic susceptibility assessed by 7T magnetic resonance imaging in Alzheimer's disease caused by streptozotocin administration. Quant Imaging Med Surg 2020;10:789-97. [Crossref] [PubMed]
- Pritchard WS, Duke DW, Coburn KL, Moore NC, Tucker KA, Jann MW, Hostetler RM. EEG-based, neural-net predictive classification of Alzheimer's disease versus control subjects is augmented by non-linear EEG measures. Electroencephalogr Clin Neurophysiol 1994;91:118-30. [Crossref] [PubMed]
- Suk HI, Lee SW, Shen DAlzheimer’s Disease Neuroimaging Initiative. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct 2015;220:841-59. [Crossref] [PubMed]
- Adler G, Brassen S, Jajcevic A. EEG coherence in Alzheimer's dementia. J Neural Transm (Vienna) 2003;110:1051-8. [Crossref] [PubMed]
- van der Hiele K, Vein AA, Reijntjes RH, Westendorp RG, Bollen EL, van Buchem MA, van Dijk JG, Middelkoop HA. EEG correlates in the spectrum of cognitive decline. Clin Neurophysiol 2007;118:1931-9. [Crossref] [PubMed]
- Hogan MJ, Swanwick GR, Kaiser J, Rowan M, Lawlor B. Memory-related EEG power and coherence reductions in mild Alzheimer's disease. Int J Psychophysiol 2003;49:147-63. [Crossref] [PubMed]
- Karrasch M, Laine M, Rinne JO, Rapinoja P, Sinervä E, Krause CM. Brain oscillatory responses to an auditory-verbal working memory task in mild cognitive impairment and Alzheimer's disease. Int J Psychophysiol 2006;59:168-78. [Crossref] [PubMed]
- Wojcik GM, Masiak J, Kawiak A, Kwasniewicz L, Schneider P, Postepski F, Gajos-Balinska A. Analysis of Decision-Making Process Using Methods of Quantitative Electroencephalography and Machine Learning Tools. Front Neuroinform 2019;13:73. [Crossref] [PubMed]
- Caravaglios G, Costanzo E, Palermo F, Muscoso EG. Decreased amplitude of auditory event-related delta responses in Alzheimer's disease. Int J Psychophysiol 2008;70:23-32. [Crossref] [PubMed]
- Kim JS, Lee SH, Park G, Kim S, Bae SM, Kim DW, Im CH. Clinical implications of quantitative electroencephalography and current source density in patients with Alzheimer's disease. Brain Topogr 2012;25:461-74. [Crossref] [PubMed]
- Cassani R, Estarellas M, San-Martin R, Fraga FJ, Falk TH. Systematic Review on Resting-State EEG for Alzheimer's Disease Diagnosis and Progression Assessment. Dis Markers 2018;2018:5174815. [Crossref] [PubMed]
- Bennys K, Rondouin G, Vergnes C, Touchon J. Diagnostic value of quantitative EEG in Alzheimer's disease. Neurophysiol Clin 2001;31:153-60. [Crossref] [PubMed]
- Koenig T, Prichep L, Dierks T, Hubl D, Wahlund LO, John ER, Jelic V. Decreased EEG synchronization in Alzheimer's disease and mild cognitive impairment. Neurobiol Aging 2005;26:165-71. [Crossref] [PubMed]
- Huang C, Wahlund L, Dierks T, Julin P, Winblad B, Jelic V. Discrimination of Alzheimer's disease and mild cognitive impairment by equivalent EEG sources: a cross-sectional and longitudinal study. Clin Neurophysiol 2000;111:1961-7. [Crossref] [PubMed]
- Dauwels J, Srinivasan K, Ramasubba Reddy M, Musha T, Vialatte FB, Latchoumane C, Jeong J, Cichocki A. Slowing and Loss of Complexity in Alzheimer's EEG: Two Sides of the Same Coin? Int J Alzheimers Dis 2011;2011:539621. [Crossref] [PubMed]
- Dauwels J, Vialatte F, Cichocki A. Diagnosis of Alzheimer's disease from EEG signals: where are we standing? Curr Alzheimer Res 2010;7:487-505. [Crossref] [PubMed]
- Jeong J. EEG dynamics in patients with Alzheimer's disease. Clin Neurophysiol 2004;115:1490-505. [Crossref] [PubMed]
- Jeong J, Kim SY, Han SH. Non-linear dynamical analysis of the EEG in Alzheimer's disease with optimal embedding dimension. Electroencephalogr Clin Neurophysiol 1998;106:220-8. [Crossref] [PubMed]
- Babiloni C, Lizio R, Marzano N, Capotosto P, Soricelli A, Triggiani AI, Cordone S, Gesualdo L, Del Percio C. Brain neural synchronization and functional coupling in Alzheimer's disease as revealed by resting state EEG rhythms. Int J Psychophysiol 2016;103:88-102. [Crossref] [PubMed]
- Besthorn C, Förstl H, Geiger-Kabisch C, Sattel H, Gasser T, Schreiter-Gasser U. EEG coherence in Alzheimer disease. Electroencephalogr Clin Neurophysiol 1994;90:242-5. [Crossref] [PubMed]
- Dauwels J, Vialatte FB, Cichocki A. On the early diagnosis of Alzheimer’s disease from EEG signals: a mini-review. Advances in Cognitive Neurodynamics 2011;709-16. [Crossref]
- Tait L, Tamagnini F, Stothart G, Barvas E, Monaldini C, Frusciante R, Volpini M, Guttmann S, Coulthard E, Brown JT, Kazanina N, Goodfellow M. EEG microstate complexity for aiding early diagnosis of Alzheimer's disease. Sci Rep 2020;10:17627. [Crossref] [PubMed]
- Smailovic U, Koenig T, Laukka EJ, Kalpouzos G, Andersson T, Winblad B, Jelic V. EEG time signature in Alzheimer’s disease: Functional brain networks falling apart. Neuroimage Clin 2019;24:102046. [Crossref] [PubMed]
- Vossel KA, Tartaglia MC, Nygaard HB, Zeman AZ, Miller BL. Epileptic activity in Alzheimer's disease: causes and clinical relevance. Lancet Neurol 2017;16:311-22. [Crossref] [PubMed]
- Ahmadlou M, Adeli H, Adeli A. New diagnostic EEG markers of the Alzheimer's disease using visibility graph. J Neural Transm (Vienna) 2010;117:1099-109. [Crossref] [PubMed]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273-97. [Crossref]
- Breiman L. Random Forests. Mach Learn 2001;45:5-32. [Crossref]
- Lehmann C, Koenig T, Jelic V, Prichep L, John RE, Wahlund LO, Dodge Y, Dierks T. Application and comparison of classification algorithms for recognition of Alzheimer's disease in electrical brain activity (EEG). J Neurosci Methods 2007;161:342-50. [Crossref] [PubMed]
- Durongbhan P, Zhao Y, Chen L, Zis P, De Marco M, Unwin ZC, Venneri A, He X, Li S, Zhao Y, Blackburn DJ, Sarrigiannis PG. A Dementia Classification Framework Using Frequency and Time-Frequency Features Based on EEG Signals. IEEE Trans Neural Syst Rehabil Eng 2019;27:826-35. [Crossref] [PubMed]
- Abásolo D, Hornero R, Espino P, Alvarez D, Poza J. Entropy analysis of the EEG background activity in Alzheimer's disease patients. Physiol Meas 2006;27:241-53. [Crossref] [PubMed]
- Tzimourta KD, Giannakeas N, Tzallas AT, Astrakas LG, Afrantou T, Ioannidis P, Grigoriadis N, Angelidis P, Tsalikakis DG, Tsipouras MG. EEG Window Length Evaluation for the Detection of Alzheimer's Disease over Different Brain Regions. Brain Sci 2019;9:81. [Crossref] [PubMed]
- Riley RD, Ensor J, Snell KIE, Harrell FE Jr, Martin GP, Reitsma JB, Moons KGM, Collins G, van Smeden M. Calculating the sample size required for developing a clinical prediction model. BMJ 2020;368:m441. [Crossref] [PubMed]
- Lopez-Martin M, Nevado A, Carro B. Detection of early stages of Alzheimer's disease based on MEG activity with a randomized convolutional neural network. Artif Intell Med 2020;107:101924. [Crossref] [PubMed]
- Huggins CJ, Escudero J, Parra MA, Scally B, Anghinah R, Vitória Lacerda De Araújo A, Basile LF, Abasolo D. Deep learning of resting-state electroencephalogram signals for three-class classification of Alzheimer's disease, mild cognitive impairment and healthy ageing. J Neural Eng 2021; [Crossref] [PubMed]
- Petrosian AA, Prokhorov DV, Lajara-Nanson W, Schiffer RB. Recurrent neural network-based approach for early recognition of Alzheimer's disease in EEG. Clin Neurophysiol 2001;112:1378-87. [Crossref] [PubMed]
- Bi X, Wang H. Early Alzheimer's disease diagnosis based on EEG spectral images using deep learning. Neural Netw 2019;114:119-35. [Crossref] [PubMed]
- Folstein MF, Robins LN, Helzer JE. The Mini-Mental State Examination. Arch Gen Psychiatry 1983;40:812. [Crossref] [PubMed]
- Julayanont P, Tangwongchai S, Hemrungrojn S, Tunvirachaisakul C, Phanthumchinda K, Hongsawat J, Suwichanarakul P, Thanasirorat S, Nasreddine ZS. The Montreal Cognitive Assessment-Basic: A Screening Tool for Mild Cognitive Impairment in Illiterate and Low-Educated Elderly Adults. J Am Geriatr Soc 2015;63:2550-4. [Crossref] [PubMed]
- Caffarra P, Vezzadini G, Dieci F, Zonato F, Venneri A. Rey-Osterrieth complex figure: normative values in an Italian population sample. Neurol Sci 2002;22:443-7. [Crossref] [PubMed]
- Tombaugh TN. Trail Making Test A and B: normative data stratified by age and education. Arch Clin Neuropsychol 2004;19:203-14. [Crossref] [PubMed]
- Brandt J. The hopkins verbal learning test: Development of a new memory test with six equivalent forms. Clin Neuropsychol 1991;5:125-42. [Crossref]
- Prigatano GP. Wechsler Memory Scale: a selective review of the literature. J Clin Psychol 1978;34:816-32. [Crossref] [PubMed]
- Parks RW, Loewenstein DA, Dodrill KL, Barker WW, Yoshii F, Chang JY, Emran A, Apicella A, Sheramata WA, Duara R. Cerebral metabolic effects of a verbal fluency test: a PET scan study. J Clin Exp Neuropsychol 1988;10:565-75. [Crossref] [PubMed]
- Mack WJ, Freed DM, Williams BW, Henderson VW. Boston Naming Test: shortened versions for use in Alzheimer's disease. J Gerontol 1992;47:154-8. [Crossref] [PubMed]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:263-9. [Crossref] [PubMed]
- Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol 2014;13:614-29. [Crossref] [PubMed]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med 2004;256:183-94. [Crossref] [PubMed]
- Artero S, Petersen R, Touchon J, Ritchie K. Revised criteria for mild cognitive impairment: validation within a longitudinal population study. Dement Geriatr Cogn Disord 2006;22:465-70. [Crossref] [PubMed]
- Morris JC. The Clinical Dementia Rating (CDR). Neurology 1993;43:2412-2412-a.
- Lawton MP, Brody EM. Assessment of older people: self-maintaining and instrumental activities of daily living. Gerontologist 1969;9:179-86. [Crossref] [PubMed]
- Oostenveld R, Fries P, Maris E, Schoffelen JM. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci 2011;2011:156869. [Crossref] [PubMed]
- Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods 2004;134:9-21. [Crossref] [PubMed]
- Mognon A, Jovicich J, Bruzzone L, Buiatti M. ADJUST: An automatic EEG artifact detector based on the joint use of spatial and temporal features. Psychophysiology 2011;48:229-40. [Crossref] [PubMed]
- Zhai Y, Yao D. A study on the reference electrode standardization technique for a realistic head model. Comput Methods Programs Biomed 2004;76:229-38. [Crossref] [PubMed]
- Cui D, Liu J, Bian Z, Li Q, Wang L, Li X. Cortical source multivariate EEG synchronization analysis on amnestic mild cognitive impairment in type 2 diabetes. ScientificWorldJournal 2014;2014:523216. [Crossref] [PubMed]
- Park JH, Kim S, Kim CH, Cichocki A, Kim K. Multiscale entropy analysis of EEG from patients under different pathological conditions. Fractals 2007;15:399-404. [Crossref]
- Abásolo D, Hornero R, Gómez C, García M, López M. Analysis of EEG background activity in Alzheimer's disease patients with Lempel-Ziv complexity and central tendency measure. Med Eng Phys 2006;28:315-22. [Crossref] [PubMed]
- Hayter A. A proof of the conjecture that the Tukey-Kramer multiple comparisons procedure is conservative. Ann Stat 1984;12:61-75. [Crossref]
- Walker SH, Duncan DB. Estimation of the probability of an event as a function of several independent variables. Biometrika 1967;54:167-79. [Crossref] [PubMed]
- Friedman JH. Greedy Function Approximation: A Gradient Boosting Machine. Ann Stat 2001;29:1189-232. [Crossref]
- Chen T, Guestrin C. XGBoost. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY, USA: ACM 2016:785-94.
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR Jr, Barch DM, Petersen SE, Schlaggar BL. Prediction of individual brain maturity using fMRI. Science 2010;329:1358-61. [Crossref] [PubMed]
- Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst 1987;2:37-52. [Crossref]
- Musaeus CS, Engedal K, Høgh P, Jelic V, Mørup M, Naik M, Oeksengaard AR, Snaedal J, Wahlund LO, Waldemar G, Andersen BB. Oscillatory connectivity as a diagnostic marker of dementia due to Alzheimer's disease. Clin Neurophysiol 2019;130:1889-99. [Crossref] [PubMed]
- Alexander DM, Arns MW, Paul RH, Rowe DL, Cooper N, Esser AH, Fallahpour K, Stephan BC, Heesen E, Breteler R, Williams LM, Gordon E. EEG markers for cognitive decline in elderly subjects with subjective memory complaints. J Integr Neurosci 2006;5:49-74. [Crossref] [PubMed]
- López-Sanz D, Bruña R, Garcés P, Camara C, Serrano N, Rodríguez-Rojo IC, Delgado ML, Montenegro M, López-Higes R, Yus M, Maestú F. Alpha band disruption in the AD-continuum starts in the Subjective Cognitive Decline stage: a MEG study. Sci Rep 2016;6:37685. [Crossref] [PubMed]


