
Prior image-guided cone-beam computed tomography augmentation from under-sampled projections using a convolutional neural network
Introduction
Accurate target localization is essential to ensure the delivery precision in the radiation therapy, which is especially true for the stereotactic body radiotherapy (SBRT) due to its high fractional dose and tight planning target volume margins. Cone-beam computed tomography (CBCT) has long been developed to provide the volumetric imaging of the up-to-date patient anatomy to minimize the target localization errors. However, the associated X-ray radiation has the potentials to induce cancers (1). As a fractional verification tool, it is more necessary for the CBCT to reduce imaging dose under the principle of “as low as reasonably achievable (ALARA)”. There are two common ways to reduce the CBCT imaging dose. One is to lower the tube current or exposure time of the X-ray source, and the other is to acquire the CBCT in a sparse-view method. In this study, we focused on the second way to reduce the CBCT imaging dose.
The clinical widely-used Feldkamp-Davis-Kress (FDK) algorithm (2) is infeasible to reconstruct high-quality CBCT images from the sparse projections. It suffers from streak artifacts due to the projection under-sampling. In the past years, various algorithms have been proposed to address the ill-conditioning in the under-sampled CT/CBCT reconstruction.
One category is based on the compressed sensing (CS) theory. With a proper sparsity regularization, CS can reconstruct high-quality images from far fewer projections than required by the Nyquist-Shannon sampling theorem. Total variation (TV) has been developed to remove the noises and streak artifacts. Sidky and Pan (3) proposed the adaptive-steepest-descent (ASD)-projection-onto-convex-sets (POCS) to improve the CBCT image quality against the streak artifacts. In the proposed algorithm, TV regularization was utilized to penalize the image gradients in a globally uniform method, inevitably smoothing out the anatomical edges. Later, various edge-preserving algorithms (4-8) have been proposed to alleviate the edge over-smoothing. In these methods, edges were detected either on the intermediate images (4-7) or on the deformed prior images (8), and then a weighted TV regularization was applied during the iterative reconstruction. The edge-preserving performance relies on the edge detection accuracy, which is usually limited by the degraded intermediate image quality.
Another category is based on the prior-image deformation. Previously, we developed an algorithm (9-12) to estimate the onboard CBCT using the prior CT images, deformation models and onboard projections. In this method, the onboard CBCT is considered as a deformation of the prior CT. The problem is formulated as a 3D/2D registration problem. The deformation vector fields (DVFs) between the prior CT and onboard CBCT are solved by minimizing the deformed-CT projection errors meanwhile maintaining reasonable DVF smoothness. This algorithm realized high-quality CBCT images from under-sampled or limited-angle projections. However, the reconstruction process is time-consuming due to the iterative optimization on the DVFs, and the reconstruction images are prone to deformable registration errors, especially in the regions with low contrast or large prior-to-onboard anatomical changes.
In recent years, deep learning, especially the convolutional neural networks (CNNs), has been widely explored in the sparse-view CT/CBCT reconstruction. The CNN models were developed either (1) to integrate into the existing reconstruction algorithms such as iterative reconstruction (8) and filtered back-projection (13), or (2) to work with the existing algorithms as pre-processing or post-processing tools. Lee et al. (14) proposed a deep neural network to synthesize the fully-sampled sinogram for the sparse-view CT reconstruction. Han et al. (15) proposed a CNN model to enhance the sparse-view CT reconstructed using the filtered back-projection algorithm. We previously developed a symmetric residual CNN model (16) to augment the edge information in the CS-based under-sampled CBCT images. These deep learning models realized high image quality of the under-sampled CT/CBCT. However, when the projections are highly under-sampled, the deep learning models need to fill in most missing data, which highly depends on the patient-specific anatomies. The deep learning models trained using a group of patient data did not account for the inter-patient variabilities, and therefore suffered from degraded reconstruction results. To address this problem, patient-specific training strategy (17,18) has been explored to account for the patient-specific anatomies. In these methods, deep learning model was trained for each individual patient using the augmented intra-patient data. The patient-specific information contained in the prior images was encoded in the deep learning model during the training process. Compared to the conventional group-data-trained deep learning models, these patient-specific models achieved considerable improvements in the under-sampled CT/CBCT image quality, especially when the projections are highly under-sampled. However, the patient-specific model is exclusively trained for an individual patient, and has to be retrained for each new patient. Training of the deep learning models is usually time- and computing resource-consuming. Therefore, it is desirable to train a generalized model that utilizes the patient-specific prior information for image augmentation.
In this study, we proposed a merging-encoder CNN (MeCNN) to realize the prior-image-guided under-sampled CBCT augmentation, which utilized the patient-specific prior information in an inter-patient generalized method. Overall, MeCNN has an encoder-decoder architecture. In the encoder, image features are extracted from both the prior CT images and the under-sampled CBCT images. And the prior image features are merged with the under-sampled image features at multi-scale levels. In the decoder, merged features are used to restore the high-quality CBCT images. Major contributions of this work include: (I) the proposed method can reconstruct high-quality CBCT images using only 36 half-fan projections, substantially reducing the CBCT imaging dose; (II) we proposed a novel merging-encoder CNN to learn a generalized pattern of utilizing patient-specific prior information in the under-sampled image augmentation; (III) the proposed method was trained on the public datasets, and was tested on both the simulated and the clinical CBCTs from our institution. Results demonstrated the effectiveness of the proposed model in augmenting the under-sampled CBCT image quality with the prior-image guidance. Comparison with other state-of-the-art methods confirmed the superiority of the proposed method.
Methods
Problem formulation
Let x∈RI×J×K represent the real-value CBCT images with a dimension of I×J×K voxels reconstructed from the under-sampled projections using FDK (2), y∈RI×J×K represent the corresponding prior CT images, and z∈RI×J×K represent the corresponding ground-truth CBCT images. Then the problem can be formulated as solving a map from the data pair (x,y) to the data z so that
where f is the image augmentation function that can be estimated by a deep learning model.
Network architecture
In this study, we proposed a deep learning model to augment the image quality of the under-sampled CBCT with the prior image guidance. The model takes the data pair consisting of the high-resolution prior CT images and the under-sampled CBCT images as inputs, and generates high-quality augmented CBCT images. The proposed model contains two parts: (I) a pre-alignment network and (II) a merging-encoder CNN (MeCNN). The overall workflow is shown in Figure 1.
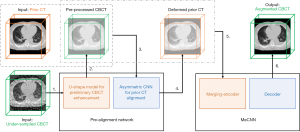
Pre-alignment network
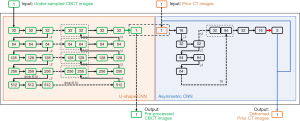
Due to the fixed shape of the convolutional kernels, CNN has limited capabilities of addressing image deformations. To assist the prior-image guidance, a pre-alignment network was designed to preliminarily align the prior CT images with the under-sampled CBCT images. The network takes (I) the prior CT images and (II) the under-sampled CBCT images as inputs, and generates (I) the pre-processed CBCT images with the under-sampling artifacts preliminarily removed and (II) the deformed prior CT images. Specifically, a U-shape CNN (as shown in the orange box in Figure 2) is first used to preliminarily remove the artifacts in the under-sampled CBCT images, yielding the pre-processed CBCT. It is modified based on the classical U-Net (19), which has demonstrated effectiveness in reducing the under-sampling artifacts (15,18). Then an asymmetric CNN (as shown in the blue box in Figure 2) takes the prior CT and the pre-processed CBCT images as inputs and predicts the DVFs between them. At last, a spatial transformation layer generates the deformed prior CT based on the prior CT and the predicted DVFs (20). More details of the pre-alignment network can be found in the Figure 2.
Merging-encoder convolutional neural network (MeCNN)
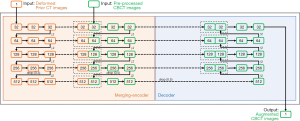
In this study, we proposed a merging-encoder architecture to encode the prior-image features in the under-sampled CBCT augmentation process. The merging-encoder has two branches for the prior CT and the pre-processed CBCT, respectively, to extract and merge image features at multi-scale levels, which can be formulated as Eq. [2]. The merged features are then connected to the decoder at multi-scale levels using shortcuts to yield high-quality CBCT images.
where ICT is the input prior CT image, ICBCT is the input pre-processed CBCT image, Ci is the image features extracted by the i-th convolutional layer, Wi and bi are the kernel weights and bias of the i-th convolutional layer, respectively, L is the convolutional layers in the each encoder branch, K is the convolutional kernel size, ⊗ is the convolutional operation, MaxPool is the maximum pooling operation, Relu is the operation of the rectified linear unit, BN is the batch normalization operation, concat is the feature concatenation in channels. More details of the proposed MeCNN can be found in the Figure 3.
Experiment design
Model training
In the training process, we built a training dataset containing 25 patients with lung cancers. Patient data were enrolled from the public datasets of SPARE (21), 4D-Lung (22) and DIR-LAB (23). For the model validation, we enrolled 3 patients with lung cancers from our institution under an IRB-approved protocol.
For each patient, a 4D-CT scan containing 10 respiratory phases was collected. And for each 4D-CT, the end-of-expiration (EOE) phase and the end-of-inspiration (EOI) phase were chosen: one was used as the prior CT images, and the other was used as the ground truth CBCT images, and vice versa. Our previous study (20) has demonstrated that the deformation between the EOI and EOE phases can well represent the motions and anatomy deformations between the prior CT and onboard CBCT. To mimic the clinical positioning mismatch between the plan CT and onboard CBCT in typical modern linac, the prior CT was rotated with a random angle within ±2º along the patient superior-inferior (SI) direction.
To simulate the under-sampled CBCT, half-fan digitally reconstructed radiographs (DRRs) were calculated from the ground truth CBCT images at 36 uniformly distributed angles over 360º. The projection parameters were set to match with the clinical CBCT acquisition settings: the detector was shifted 14.8 cm for the half-fan projection acquisition, the projection matrix was 512×384 with a pixel size of 0.776 mm × 0.776 mm, the source-to-ioscenter distance was 100 cm, and the source-to-detector distance was 150 cm. The under-sampled CBCT images were then reconstructed from DRRs using FDK (2). All the image data were resized to 250×256×96 voxels with a voxel size of 1.5 mm × 1.5 mm × 2.0 mm.
The data pairs containing the under-sampled CBCT image and prior CT images were fed into the proposed network. Model weights were optimized by minimizing the mean-squared-error (MSE) between the augmented CBCT images and the ground truth CBCT images. Optimizer was set to “Adam” (24). Learning rates were set to 0.01 and gradually reduced to 0.001 for the U-shape CNN (in the pre-alignment network) and the MeCNN, and were set to 1×10−4 and gradually reduced to 1×10−5 for the asymmetric CNN (in the pre-alignment network). The proposed model was trained in an end-to-end method. Batch size was 1 due to the memory limitation. The best checkpoint was selected based on the validation errors.
Evaluation using intra-scan simulated CBCT
To evaluate the performance of the proposed model in handling the CBCT under-sampling, 4D-CT data of 11 patients with lung cancers were enrolled in the testing dataset from our institution under an IRB-approved protocol. For each patient, EOE and EOI phase were chosen: one was used as the prior CT, and the other was used as the ground truth CBCT, and vice versa. The simulation of the under-sampled CBCT followed the same process as the training data. The prior CT and the under-sampled CBCT were fed into the trained model. The predicted CBCT was compared against the ground truth CBCT images both qualitatively and quantitatively using metrics of root mean squared error (RMSE), peak signal-to-noise ratio (PSNR), and structure similarity (SSIM) (16). Lower RMSE, higher PSNR, and higher SSIM indicate better image quality. Metrics were calculated within the body, the lung, the bone, and the clinical GTV regions.
To further demonstrate the clinical value of the proposed model, tumor localization accuracy was evaluated using the enhanced CBCT. In this study, only rigid registrations with the 3D translations were performed since only rigid translations are applied for target localization in clinical practice. For each patient, two registration studies were conducted: (I) EOE phase served as the prior CT and EOI phase served as the onboard CBCT, and (II) EOI phase served as the prior CT and EOE phase served as the onboard CBCT. To simulate the initial positioning errors, the volume serving as the prior CT was randomly translated in left-right (LR), anterior-posterior (AP) and superior-inferior (SI) directions. Rigid registrations with only translations were conducted between the onboard CBCT and the prior CT. To avoid the uncertainties of manual registrations, automatic registrations were performed based on the mutual information in the tumor regions using the open-source Elastix (SimpleITK v1.2.4). The tumor localization accuracy is defined as
where xCBCT, yCBCT, and zCBCT are the patient positioning shifts determined by the ground truth CBCT images, and xuCBCT, yuCBCT, and zuCBCT are the shifts determined by the sparse-view CBCT images reconstructed by various methods in the LR, AP and SI directions, respectively.
Evaluation using inter-scan simulated CBCT
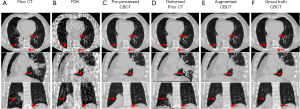
Large anatomy variations can happen from the prior CT to the onboard CBCT, which are hard to be handled by deformation models. To evaluate the performance of the proposed model in handling such large variations, we enrolled a case from the 4D-Lung dataset. For this case, two 4D-CT scans of the patient were selected. The EOE phase of the first 4D-CT scan was used as the prior CT, and the EOE phase of the second 4D-CT scan was used as the ground truth CBCT. As shown in Figure 4, there are large anatomy variations between the two scans. Sparse-view CBCT was simulated from the ground truth CBCT following the same configurations in the training process. The sparse-view CBCT and the prior CT were fed into the trained model, and the augmented CBCT was compared against the ground truth CBCT. In addition, step-by-step results of the proposed method were also compared to demonstrate the effectiveness of the proposed MeCNN. Note that although this case was enrolled from the 4D-Lung dataset, it was not included in the training dataset.
Evaluation using clinical CBCT
To further validate its clinical utility, performance of the proposed model was tested on a lung cancer patient using the clinical planning CT and the onboard breath-hold CBCT from our institution. The CBCT projections were acquired by the TrueBeam (Varian Medical Systems, Inc., Palo Alto, CA) using 20 mA and 15 ms and extracted under an IRB-approved protocol. The projection acquisition settings were the same as mentioned in the section of Model training, except that the detector was shifted 16.0 cm for the half-fan projection acquisition. The reference CBCT images were reconstructed by the Varian iTools-Reconstruction (Varian Medical Systems iLab GmbH) using full-sampling acquisition of 894 projections with scatter and intensity correction. To alleviate the image quality degradation resulting from the clinical projection scatters and noises, sparse-view CBCT images were reconstructed from DRRs which were simulated from the iTools-reconstructed reference CBCT images. Specifically, 36 uniformly distributed DRRs over 360º were simulated from the reference CBCT images using the same acquisition settings as the clinical projections. The planning CT and the under-sampled CBCT were fed into the trained model. The predicted CBCT was compared against the reference fully-sampled CBCT images. Due to the limited image quality of the reference CBCT images, only qualitative evaluations were performed. Noted that, once the model was trained on the DRRs, no further re-training or fine-tuning was conducted for the clinical projections.
Comparison with the state-of-the-art methods
The performance of the proposed model was compared to several state-of-the-art methods, including FDK (2), ASD-POCS (3), and 3D U-Net (18). (I) FDK is a CBCT reconstruction algorithm that is widely used in clinical practice. In this study, intensity values of the FDK images were rescaled to the range of the ground truth CBCT using the z normalization. (II) ASD-POCS is an iterative reconstruction algorithm based on the CS theory using the TV regularization. (III) 3D U-Net is modified based on the classical U-Net (19) to adapt to the 3D image data. Details of the model architecture can be referred to in (18). The model takes 3D sparse-view CBCT images as input and generates augmented 3D CBCT images. In the training process, EOI or EOE phase of the 4D-CT scan was used as the ground truth CBCT, and was used to simulate the input sparse-view CBCT. The model was trained and validated using the same datasets and configurations as mentioned in the section of Modeling training.
Statistical analysis
Mann-Whitney U test was conducted using Matlab 2019a to evaluate the differences between metrics of the proposed method and those of other methods mentioned in the previous section.
P<0.05 was considered statistically significant.
Results
Evaluation using intra-scan simulated CBCT
Figure 5 shows the representative slices of the simulated CBCT reconstructed by various algorithms. Due to the projection under-sampling, FDK images suffered from severe streak artifacts, making the structure edges distorted and undistinguishable. The ASD-POCS images well removed the streak artifacts, but they had blurred edges and some small pulmonary structures were smoothed out. The 3D U-Net enhanced the pulmonary structure details. However, as indicated by the arrows in Figure 5, the bone edges were still blurred and even missing, and the soft tissues were not well restored. In comparison, CBCT images reconstructed by the proposed model demonstrate high quality in the pulmonary structures, bones, and soft tissues.
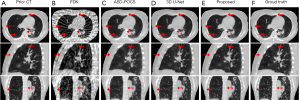
Table 1 shows the quantitative analysis of the CBCT image quality. Metrics were calculated in the body, the lung, the bone, and the clinical GTV regions of all the 11 testing patients. Metrics of the prior CT demonstrated the obvious variations from the prior CT to the onboard CBCT. Compared to the FDK, ASD-POCS, and 3D U-Net, the proposed method had better performance in the under-sampled CBCT augmentation by demonstrating lower RMSE, higher PSNR, and higher SSIM. All the P values were less than 0.05, demonstrating that the improvements made by the proposed method were significant over other methods. The quantitative results agreed with the visual inspection.
Table 1
| Metrics | Prior CT | FDK | ASD-POCS | 3D U-Net | Proposed |
|---|---|---|---|---|---|
| Body RMSE | 0.037±0.013 | 0.050±0.012 | 0.051±0.015 | 0.020±0.005 | 0.015±0.003 |
| Body PSNR | 29.76±3.039 | 26.33±1.975 | 28.53±2.232 | 34.42±2.161 | 37.02±1.930 |
| Body SSIM | 0.880±0.047 | 0.613±0.057 | 0.756±0.041 | 0.831±0.020 | 0.960±0.011 |
| Lung RMSE | 0.054±0.015 | 0.041±0.010 | 0.044±0.011 | 0.022±0.004 | 0.019±0.004 |
| Lung PSNR | 26.28±2.480 | 28.08±1.996 | 29.21±1.951 | 33.53±1.703 | 34.51±1.763 |
| Lung SSIM | 0.711±0.072 | 0.667±0.058 | 0.695±0.045 | 0.896±0.021 | 0.914±0.019 |
| Bone RMSE | 0.033±0.012 | 0.048±0.012 | 0.062±0.018 | 0.032±0.007 | 0.021±0.003 |
| Bone PSNR | 30.77±3.384 | 26.61±2.062 | 26.17±2.204 | 30.32±1.734 | 30.88±1.398 |
| Bone SSIM | 0.961±0.020 | 0.872±0.028 | 0.864±0.018 | 0.941±0.016 | 0.976±0.006 |
| GTV RMSE | 0.035±0.018 | 0.041±0.013 | 0.046±0.022 | 0.024±0.006 | 0.018±0.005 |
| GTV PSNR | 31.22±4.236 | 28.23±2.432 | 29.84±3.260 | 33.06±1.994 | 35.53±2.25 |
| GTV SSIM | 0.934±0.044 | 0.870±0.040 | 0.879±0.079 | 0.966±0.014 | 0.979±0.009 |
CBCT, cone-beam computed tomography.
Table 2 shows the tumor localization errors of the sparse-view CBCT reconstructed by various methods. Compared to the FDK, ASD-POCS and 3D U-Net, the proposed method had better tumor localization accuracy. All the P values for the 3D error distance were less than 0.05, indicating that the proposed method significantly improved the CBCT-based IGRT accuracy compared to other methods. Results demonstrated the clinical value of the CBCT quality improvements made by the proposed method.
Table 2
| Directions | FDK | ASD-POCS | 3D U-Net | Proposed |
|---|---|---|---|---|
| Left-right | 1.9±1.2 | 1.0±1.0 | 0.2±0.1 | 0.2±0.1 |
| Anterior-posterior | 1.7±1.7 | 1.1±1.1 | 0.4±0.3 | 0.2±0.1 |
| Superior-inferior | 1.7±1.0 | 0.9±0.8 | 0.5±0.5 | 0.4±0.4 |
| 3D distance | 3.5±1.6 | 2.0±1.5 | 0.8±0.5 | 0.5±0.4 |
CBCT, cone-beam computed tomography.
Evaluation using inter-scan simulated CBCT
Figure 4 shows the step-by-step results of the proposed method. Prior CT (in the first column) demonstrated large anatomy variations from the ground truth CBCT (in the last column). Streaks in the input FDK-based sparse-view CBCT images were well removed by the U-shape CNN (in the third column). But as indicated by the red arrows in the pre-processed CBCT, the soft tissues were not well enhanced, and the bone edges were blurred and even missing. These details were well restored by the asymmetric CNN (in the fourth column) which aligned the prior CT to the pre-processed CBCT in a deformable way. However, the spatial deformation can hardly handle the large anatomy variations. Obvious errors appeared near the tumor regions as indicated by the red arrows in the deformed prior CT images. The proposed MeCNN (in the fifth column) corrected the errors in the deformed prior CT while maintaining the CT-like appearance. The CBCT augmented by the proposed MeCNN showed clear and accurate pulmonary tissues, soft tissues, and bone edges.
Evaluation using clinical CBCT
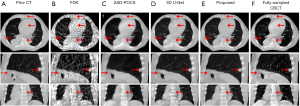
Figure 6 shows the representative slices of the clinical CBCT reconstructed by various algorithms. The FDK and the ASD-POCS showed similar performance to the simulated CBCT evaluation results (in Figure 5). Due to the projection noises, the CBCT images enhanced by the 3D U-Net showed artifacts as demonstrated by red arrows (Figure 6C1). The proposed model overcame the artifacts with the guidance of the prior images, demonstrating CT-like image quality with clear and accurate structures.
Runtime
The proposed MeCNN model was implemented with the Keras (v2.2.4) framework using TensorFlow (v1.11.0) backend. The model training and evaluations were performed on a computer equipped with a GPU of NVIDIA Quadro RTX 8000 (48GB memory), a CPU of Intel Xeon, and 128GB memory. The entire workflow shown in Figure 1 took about 3.3 seconds to predict the augmented CBCT volumes (dimensions: 256×256×96, voxel size: 1.5 mm × 1.5 mm × 2.0 mm).
Discussion
CBCT has been widely used in clinics as a fractional verification tool to ensure the patient positioning accuracy in the radiation therapy. Considering the potential cancer-inducing risks associated with the X-ray radiation, it is desirable to reduce the CBCT imaging dose while maintaining the high image quality. In this study, we proposed a method to enhance the quality of the CBCT images reconstructed from highly under-sampled projections. It takes advantage of the deep learning-based algorithms to remove the under-sampling artifacts and enhance structure edges. Meanwhile, it utilizes the patient-specific prior information via a novel merging-encoder to account for the inter-patient variabilities, addressing the severe ill-conditioning in the highly under-sampled CBCT reconstruction.
Although trained on EOI and EOE phase data, the proposed model was not trained to predict deformations or organ/tumor locations in the CBCT. Instead, the model was trained to learn from the corresponding anatomical structures in the high-quality prior CT images to augment those in the CBCT. When there is deformation from CT to CBCT, the model needs to learn to register the corresponding structures under deformation before image augmentation. In this sense, simulating deformations from prior image to current image is important to train the model. However, this deformation does not need to match exactly with a patient’s CT-CBCT deformation since the model is trained to learn the fundamental rules of image registration and augmentation regardless of any specific deformation. In our training process, we mimic the deformation from prior to current images using the EOI and EOE phases from a 4D-CT scan, which represents the largest deformation in a breathing cycle. In addition, data augmentation was performed in the training process to simulate the positioning errors.
As demonstrated by the results, our proposed MeCNN method realized high-quality CBCT images with accurate structures from only 36 half-fan projections. The patient-specific information-guided augmentation pattern learned by the proposed model is applicable to all patients without the need for retraining. Results in this study preliminarily demonstrated that the proposed model is robust against the detector shift in the half-fan acquisition (training shift: 14.8 cm, clinical testing shift: 16.0 cm). Whether the model is robust against other changes in the acquisition configurations requires further evaluation, which is warranted in the future studies.
The FDK algorithm (2) reconstructed the CBCT images by back-projecting the projections along the ray directions, suffering from severe streak artifacts when the projections were under-sampled. The compressed sensing theory-based algorithm (3) introduced the TV regularization to penalize the image gradients while maintaining the projection fidelity during the iterative reconstruction, effectively suppressing the streak artifacts and noises. However, the TV regularization item also penalized the structure edges globally, which blurred the edges and even smoothed out small structures in the under-sampled images.
Conventional deep learning-based algorithms (15,16) trained the model based on large numbers of samples, mapping the under-sampled images to their fully-sampled counterparts. These models aimed to learn a common pattern to restore volumetric information from the under-sampled images. However, medical images contain a lot of patient-specific structure details. When the projections are highly under-sampled, these details in the input under-sampled images can be indistinguishable from artifacts, and can hardly be accurately restored with a common pattern learned from group data. As shown in the results (Figures 5,6), the 3D U-Net that employed the conventional group-based training strategy had limited performance in enhancing the soft tissues, bone edges, and small structures, since these details were mixed with the streak artifacts in the input under-sampled images. And these errors can hardly be corrected by further enhancement using additional group-trained models without any patient-specific constraint. Another concern for the deep learning models is the robustness against the variations from the training dataset (25,26). The input images are the only volumetric information source for the models to enhance the image quality. Artifact or noise variations in the input images can cause the models to misbehave in unexpected ways. As demonstrated by the results of the clinical CBCT evaluation (in Figure 6), the input under-sampled images were reconstructed from the clinical projections, which had different scatters, noises, and streak artifacts from the training data. The restoring pattern learned by the 3D U-Net from the training DRRs cannot handle such variations. Thus, the CBCT images enhanced by the 3D U-Net showed unexpected artifacts.
The strategy that adopts the intra-patient data to account for the inter-patient variabilities has demonstrated effectiveness in enhancing the image quality of the highly under-sampled images (17,18). The patient prior information helps the models to accurately recover the patient-specific details. However, these methods adopted the patient-specific training strategy by training models on the intra-patient data. The model weights were optimized to fit the specific patient data. Consequently, the trained models were exclusive for individual patients and cannot be generalized among different patients. When a new patient came, a new model was required to be trained using the patient prior data, consuming a lot of time and computing resources. In comparison, our proposed method extracted the patient-specific features from the prior CT images, which were merged with the under-sampled CBCT features to yield CT-like high-quality CBCT images. The proposed model took the prior images together with the under-sampled CBCT images as inputs. Instead of learning the patient-specific structures (17,18), the proposed method learned a generalized pattern to utilize the prior information for the onboard image enhancement. As a result, once trained, the proposed model can be applied to different patients without any further re-training or finetuning. Results demonstrated that the proposed merging-encoder architecture is effective in restoring the patient-specific structure details from the highly under-sampled CBCT projections. Besides, improvements in the tumor localization accuracy demonstrated the clinical value of the proposed method. In addition, with the guidance of the patient-specific prior images, the proposed model preliminarily showed robustness against the input image quality variations (in Figure 6). The structure texture features extracted from the high-quality prior CT were used to complement the anatomy geometry features extracted from the under-sampled CBCT. Thus, the predicted CBCT images had CT-like high quality with clear and accurate structures.
The proposed MeCNN has some limitations. Firstly, in the merging-encoder, convolutional layers were used to extract and merge image features. Due to the fixed shapes of the convolutional kernels, it has limited capabilities in handling the image deformations. To address this problem, we designed a pre-alignment network to alleviate the structure mismatches between the prior and the onboard images. Nevertheless, the DVFs were imperfect due to the limited quality of the pre-processed CBCT. The residual deformation errors, especially those in the low contrast regions, can have adverse effects on the augmented CBCT. In the future, advanced convolution operations, such as the deformable convolutions (27), can be used to handle the image deformations in a more robust method. Secondly, as shown in the second row of Figure 4, although the structure errors in the deformed prior CT were well corrected by the proposed MeCNN, there are still some voxels having inaccurate intensity. In the training process, the data were augmented mainly accounting for the deformations and positioning errors between the prior CT and onboard CBCT. Large anatomy variations were not considered. As such, obvious deformation errors in the deformed prior CT could introduce some inaccurate intensities to the MeCNN-enhanced CBCT. Advanced data augmentation techniques, such as random structure shading in the prior CT, can be explored in future studies to further improve its robustness. Thirdly, scatters and noises in the clinical projections can severely degrade the input under-sampled CBCT image quality, and consequently degrade the model’s augmentation performance. As demonstrated by the results, the proposed MeCNN showed robustness against the residual noises in the projections. But additional scatter correction techniques are still required for the proposed method, avoiding the input image being too corrupted to augment. In the future studies, our proposed model can be incorporated with more advanced projection scatter and noise correction techniques, such as the deep learning-based projection correction, to address the clinical cases in an end-to-end way.
Conclusions
The proposed model demonstrated the effectiveness in augmenting the image quality of the under-sampled CBCT with the prior image guidance. It can generate high-quality CBCT images using only about 4% of the clinical fully-sampled projections (36 projections out of about 900 projections), considerably reducing the imaging dose and improving the clinical utility of CBCT.
Acknowledgments
Funding: This work was supported by the National Institutes of Health under Grant No. R01-CA184173 and R01-EB028324.
Footnote
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-114). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. This work was supported by the National Institutes of Health under Grant No. R01-CA184173 and R01-EB028324. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patient data included in this study are anonymized. Patient data are acquired from the public SPARE (https://image-x.sydney.edu.au/spare-challenge/), DIR-LAB (
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brenner DJ, Hall EJ. Computed tomography--an increasing source of radiation exposure. N Engl J Med 2007;357:2277-84. [Crossref] [PubMed]
- Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm. JOSA A 1984;1:612-9. [Crossref]
- Sidky EY, Pan X. Image reconstruction in circular cone-beam computed tomography by constrained, total-variation minimization. Phys Med Biol 2008;53:4777-807. [Crossref] [PubMed]
- Cai A, Wang L, Zhang H, Yan B, Li L, Xi X, Li J. Edge guided image reconstruction in linear scan CT by weighted alternating direction TV minimization. J Xray Sci Technol 2014;22:335-49. [Crossref] [PubMed]
- Tian Z, Jia X, Yuan K, Pan T, Jiang SB. Low-dose CT reconstruction via edge-preserving total variation regularization. Phys Med Biol 2011;56:5949-67. [Crossref] [PubMed]
- Liu Y, Ma J, Fan Y, Liang Z. Adaptive-weighted total variation minimization for sparse data toward low-dose x-ray computed tomography image reconstruction. Phys Med Biol 2012;57:7923-56. [Crossref] [PubMed]
- Lohvithee M, Biguri A, Soleimani M. Parameter selection in limited data cone-beam CT reconstruction using edge-preserving total variation algorithms. Phys Med Biol 2017;62:9295-321. [Crossref] [PubMed]
- Chen Y, Yin FF, Zhang Y, Zhang Y, Ren L. Low dose CBCT reconstruction via prior contour based total variation (PCTV) regularization: a feasibility study. Phys Med Biol 2018;63:085014 [Crossref] [PubMed]
- Zhang Y, Yin FF, Segars WP, Ren L. A technique for estimating 4D-CBCT using prior knowledge and limited-angle projections. Med Phys 2013;40:121701 [Crossref] [PubMed]
- Zhang Y, Yin FF, Pan T, Vergalasova I, Ren L. Preliminary clinical evaluation of a 4D-CBCT estimation technique using prior information and limited-angle projections. Radiother Oncol 2015;115:22-9. [Crossref] [PubMed]
- Harris W, Zhang Y, Yin FF, Ren L. Estimating 4D-CBCT from prior information and extremely limited angle projections using structural PCA and weighted free-form deformation for lung radiotherapy. Med Phys 2017;44:1089-104. [Crossref] [PubMed]
- Zhang Y, Yin FF, Zhang Y, Ren L. Reducing scan angle using adaptive prior knowledge for a limited-angle intrafraction verification (LIVE) system for conformal arc radiotherapy. Phys Med Biol 2017;62:3859-82. [Crossref] [PubMed]
- Wurfl T, Hoffmann M, Christlein V, Breininger K, Huang Y, Unberath M, Maier AK. Deep Learning Computed Tomography: Learning Projection-Domain Weights From Image Domain in Limited Angle Problems. IEEE Trans Med Imaging 2018;37:1454-63. [Crossref] [PubMed]
- Lee H, Lee J, Kim H, Cho B, Cho S. Deep-neural-network-based sinogram synthesis for sparse-view CT image reconstruction. IEEE Transactions on Radiation and Plasma Medical Sciences 2018;3:109-19. [Crossref]
- Han YS, Yoo J, Ye JC. Deep residual learning for compressed sensing CT reconstruction via persistent homology analysis. arXiv preprint arXiv:161106391. 2016.
- Jiang Z, Chen Y, Zhang Y, Ge Y, Yin FF, Ren L. Augmentation of CBCT Reconstructed From Under-Sampled Projections Using Deep Learning. IEEE Trans Med Imaging 2019;38:2705-15. [Crossref] [PubMed]
- Shen L, Zhao W, Xing L. Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning. Nat Biomed Eng 2019;3:880-8. [Crossref] [PubMed]
- Jiang Z, Yin FF, Ge Y, Ren L. Enhancing digital tomosynthesis (DTS) for lung radiotherapy guidance using patient-specific deep learning model. Phys Med Biol 2021;66:035009 [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; Springer, 2015.
- Jiang Z, Yin FF, Ge Y, Ren L. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys Med Biol 2020;65:015011 [Crossref] [PubMed]
- Shieh CC, Gonzalez Y, Li B, Jia X, Rit S, Mory C, Riblett M, Hugo G, Zhang Y, Jiang Z, Liu X, Ren L, Keall P. SPARE: Sparse-view reconstruction challenge for 4D cone-beam CT from a 1-min scan. Med Phys 2019;46:3799-811. [Crossref] [PubMed]
- Hugo GD, Weiss E, Sleeman WC, Balik S, Keall PJ, Lu J, Williamson JF. A longitudinal four-dimensional computed tomography and cone beam computed tomography dataset for image-guided radiation therapy research in lung cancer. Med Phys 2017;44:762-71. [Crossref] [PubMed]
- Castillo E, Castillo R, Martinez J, Shenoy M, Guerrero T. Four-dimensional deformable image registration using trajectory modeling. Phys Med Biol 2010;55:305-27. [Crossref] [PubMed]
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
- Papernot N, McDaniel P, Goodfellow I. Transferability in machine learning: from phenomena to black-box attacks using adversarial samples. arXiv preprint arXiv:160507277. 2016.
- Eykholt K, Evtimov I, Fernandes E, Li B, Rahmati A, Xiao C, Prakash A, Kohno T, Song D. Robust physical-world attacks on deep learning visual classification. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018.
- Dai J, Qi H, Xiong Y, Li Y, Zhang G, Hu H, Wei Y. Deformable convolutional networks. Proceedings of the IEEE international conference on computer vision, 2017.


