
Exploratory ensemble interpretable model for predicting local failure in head and neck cancer: the additive benefit of CT and intra-treatment cone-beam computed tomography features
Introduction
Although local control following chemoradiation (CRT) for head and neck squamous cell carcinoma (HNSCC) approaches 60–80% for patients in phase III trials (1,2), those who do experience local failure (LF) often do so rapidly following therapy completion, with two-thirds of failures occurring within the first year. Further, these early failures have been associated with worse overall survival (OS) (3,4). Despite advancements in radiation techniques to spare normal tissue with intensity-modulated radiotherapy (IMRT) and image guidance techniques, definitive CRT still remains toxic with rates of grade 3+ toxicity approaching 80% in modern trials even with supportive care (5). If patients at high risk of LF could be identified during treatment, then response-adapted approaches could be considered to escalate care with the objective of improving local control or halt CRT if deemed futile with the objective of limiting radiation-associated toxicity and proceeding to alternative treatment. Alternatively, if patients at low risk of LF could be identified, then de-escalation approaches (6) could be considered with the objective of reducing toxicity whilst maintaining excellent local control.
Traditionally, prognostication of HNSCC malignancies treated with definitive CRT has been primarily based on TNM stage (7,8) and clinical factors, such as p16 and smoking status (9). However, attempts have been made at improving the prediction of HNSCC outcomes with machine learning techniques correlating imaging characteristics of the disease with clinical endpoints, such as LF. This process of extracting and quantifying numerous features from medical images is termed radiomics (10,11). Prior studies have shown promising results with the correlation of radiomic features of baseline CT (12-17), [18F] fluoro-2-deoxy-D-glucose (FDG) PET/CT (18-21), and MRI images (22) with LF or loco-regional failure (LRF). Of those that were externally validated, Folkert et al. showed that an FDG-PET/CT-based multiparameter logistic regression model achieved an AUC of 0.68 and sensitivity of 67% for the prediction of LF of oropharyngeal primaries (18), Bogowicz et al. showed that a mixed model using CT-based radiomics of both the primary and lymph nodes achieved an AUC of 0.67 for predicting LRF (12), Giraud et al. showed an XGboost model using CT-based radiomics of oropharyngeal primaries to achieve an AUC of 0.68 and sensitivity of 42% at predicting LRF (17), and Legers et al. showed CT-based radiomic risk models of tumor rim sub-volumes to achieve an AUC of 0.63 to 0.65 at predicting LRF (15). While the performance of these models were modest, they were generated using baseline/pre-treatment imaging and did not incorporate additional intra-treatment imaging that could potentially inform the degree of treatment response, as HNSCC primaries and nodes often shrink during radiotherapy (23) with the degree of change thought to correlate with response to treatment. Recently, the use of intra-treatment imaging in HNSCC to quantify treatment response was explored. Leger et al. evaluated intra-treatment CT-based radiomics at week 2 of radiotherapy, showing the model utilizing intra-treatment CT features achieved a higher AUC than those using baseline CT features for the prediction of LRF (C-index of 0.79 vs. 0.65) (24), supporting the idea that intra-treatment CT-based radiomics may further improve the prognostic ability of machine learning models for HNSCC outcomes.
Although CT scans offer superior imaging quality with less noise and less artifact than Cone-Beam CTs (CBCTs), the incorporation of CTs into the adaptive workflow would require the coordination and acquisition of additional scans, resulting in additional strain on department resources and radiation exposure to the patient. In contrast, CBCT imaging is already routinely employed at many radiation oncology centers as part of daily or weekly treatment setup and verification, making it an attractive modality for the evaluation of treatment response during radiotherapy. Therefore, if the addition of CBCT-based radiomics to a machine learning model improved its prognostic ability, such a model could be incorporated into the clinic workflow without requiring the acquisition of additional imaging over what is already performed as part of standard of care treatment. CBCT-based radiomics have previously been evaluated in non-small cell lung cancer (25-27); however, to the best of our knowledge, CBCT-based radiomics have yet to be evaluated in HNSCC malignancies for the prediction of clinical outcomes. In this study, we hypothesize that changes in CBCT-based radiomics by week 4–5 of radiotherapy will provide additive prognostic benefit for the prediction of LF for locally advanced HNSCC malignancies. Here, we trained and validated an interpretable machine learning model, the Explainable Boosting Machine (EBM) classifier within the InterpretML package (28), utilizing a parallel ensemble technique on an internal retrospective case-control cohort of HNSCC malignancies, predicting LF for primary and nodal structures independently following completion of radiotherapy.
Methods
Participants
Participants treated between April 2014 and October 2019 at the University of Texas Southwestern Medical Center (UTSW) were included if they were diagnosed with locally advanced HNSCC (including oropharynx, supraglottic, glottic, or hypopharynx) and completed a full course of conventionally fractionated definitive radiotherapy with daily or weekly cone-beam computed tomography (CBCT) imaging. Most patients received concurrent chemotherapy (see Table S1). Patients were excluded if they received prior induction chemotherapy, had <1 year follow-up without reaching the endpoint of LF, had distant metastases (DM) at presentation, had a prior history of definitive radiation to the head and neck, or had the presence of a separate active malignancy. Resection of the primary was allowed, as long as there was nodal disease that was not excised and available for assessment (primaries and nodes were evaluated separately in this work). Patients were also excluded if they reached death prior to 1 year with ambiguity of the status of their malignancy, in the event that response assessment of their disease was not determinable. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This retrospective study was reviewed and approved through the institutional review board (IRB), with patient consent waived, and was conducted in accordance with the Declaration of Helsinki.
The included participants who reached LF at either a primary or nodal site of disease were matched with controls based on site of the primary, T stage, N stage, and p16 status. Case-control matching was done in a 1:2 fashion and was performed in SPSS version 26.0 (Armonk, NY: IBM Corp.). Matching was also attempted with 6 variables, adding in smoking status and the chemotherapy agent used; however, less than 60 cases were identified from the pool of controls so these two additional variables were omitted from case-control matching. Baseline characteristics between cases and controls were evaluated for differences with Fisher’s exact tests, Mann-Whitney U tests, and independent samples t-tests for categorical, ordinal, and continuous data, respectively, with a P value of <0.05 set as significant on a 2-tailed test.
Patterns of failure analysis
LF was defined as imaging or pathologic evidence of recurrence within the high dose (70 Gy) volume. If LF was defined by imaging, this was termed positive if these imaging changes were associated with a change in management, including but not limited to surgical resection, neck dissection, re-irradiation, radiation to a distant site, or systemic therapy. If surgery was performed and pathology was negative, then this was not included as a LF event. The dates for LF and DM events were recorded as the date of the imaging scan demonstrating either LF or DM (respectively) that led to a change in management, or when imaging was not present, the date at which a biopsy was performed for confirmation of progression. The date for OS was the date of death from any cause. For all outcomes, the time to event was from the date of completion of radiotherapy to the date of the event. For patterns of failure analysis, baseline scans were compared to post-treatment imaging and pathology reports at time of recurrence (if reached) to determine the primary or nodal structures that met the endpoint of local recurrence.
Actuarial LF and DM rates were estimated with cumulative incidence in R version 4.0.3 (29), with death being the competing risk, using the package “cmprsk” (30,31). Actuarial OS rates were estimated with Kaplan-Meier curves in SPSS.
Imaging parameters and selection
At baseline simulation, all patients were simulated on a Philips 16-slice Brilliance large-bore CT simulator with iodinated IV contrast. Immobilization most commonly included the use of a long IMRT mask and a molded head cushion with or without a bite block depending on the location of the primary.
CBCT imaging was performed either daily or weekly depending on the time period at which they were treated at this facility and the provider. CBCTs were performed on a Varian TrueBeam™ or a Varian VitalBeam™ machine (Varian Medical Systems, Palo Alto, CA) and acquired at 210 degrees of rotation. See supplementary file for imaging parameters (Table S2).
All patients had at least 3 sets of images included for analysis, including: the baseline CT simulation scan (CT1), CBCT prior to the initial fraction (CBCT01), and CBCT prior to fraction 21 (CBCT21). During review of imaging, some CBCTs were noted to be degraded by an artifact appearing to originate primarily along the surfaces of bone and cartilage (see description in the Results section, entitled Reproducibility of Features). When comparing test-retest scans (CBCTa and CBCTb, respectively), this artifact was noted to come and go between scans performed on the same patient prior to the same fraction of radiotherapy often only 2–5 minutes apart. Therefore, patients with CBCT01 or CBCT21 scans affected by this artifact were replaced with a separate repeat scan done prior to the same fraction, preferably, or a CBCT +/− 1 fraction if a repeat scan was not available.
In addition to the above, patients with at least 2 CBCT scans performed prior to the same fraction were selected for test - retest analysis (CBCTa and CBCTb as described above). Scans were not restricted to a specific fraction. If a patient had more than one fraction with test-retest scans, the earliest fraction with repeat scans were chosen.
Segmentation
All initial segmentations on CT1 were contoured by a board-certified radiation oncologist specializing in head and neck radiotherapy. In this study, all primary structures and nodal structures receiving 70 Gy were selected for evaluation. Nodal contours were separated into distinct structures, unless the nodes were abutting each other or matted. Therefore, each patient could have 1 or more separate structures for radiomic assessment.
For CBCT images, all segmentations were deformed from CT1 with rigid + multi-pass deformable image registration in Velocity (Varian Medical Systems, Palo Alto, CA). All deformed contours were manually edited by the physician to verify the inclusion of all affected mucosa if applicable and to exclude any incident bone, air, cartilage, or adipose tissue that may be overlapping the GTV boundaries. This process was repeated for subsequent CBCTs, including CBCT01, CBCT21, CBCTa, and CBCTb.
Of note, segmentations were not modified if there was presence of metal artifacts on the same axial slice as the contour. At study inception, we had assumed that the change in 3D shape or size could potentially be informative for delta radiomics in predicting LF; therefore, we elected to not delete slices affected by metal artifacts during segmentation, as the truncation of the contours would affect the shape features extracted. Overall, 29 primary structures and 19 nodal structures had at least one slice with metal artifact secondary to dental implants.
Radiomic extraction
All features were extracted with PyRadiomics version 3.0.1 (32), which is an Image Biomarker Standardisation Initiative (IBSI) compliant toolbox (33,34). Features were extracted from CT1, CBCT01, CBCT21, CBCTa, and CBCTb, separately, at a fixed bin width of 25 HU, with sitkBSpline interpolation, and resegmentation set to exclude less than −100 Hounsfield Units (HU). The upper limit for resegmentation was set to 7,000 HU to avoid excluding higher HU data. Note that normal non-involved organs, such as bone and cartilage, were manually contoured out prior to radiomic extraction. Normalized weighting, voxel-size resampling, and filters (such as Laplacian of Gaussian or Wavelet filters) were not performed. One hundred and two features were extracted, including 18 first order, 14 shape, 24 gray level co-occurrence matrix (GLCM), 16 gray level run length matrix (GLRLM), 16 gray level size zone matrix (GLSZM), and 14 gray level dependence matrix (GLDM) features. A detailed description of all extracted features can be found in PyRadiomics documentation (pyradiomics.readthedocs.io) and are listed in the supplementary file. To calculate delta radiomic features, or the change in radiomic features over time, features extracted from CBCT01 were subtracted from the same features of CBCT21 to yield the difference. Here, a positive delta feature would be increasing in value from CBCT01 to CBCT21 and a negative feature would be decreasing.
Fused ensemble machine learning pipeline
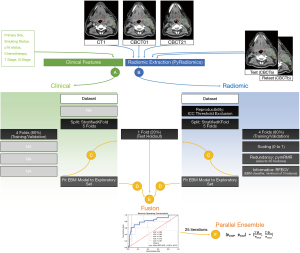
The machine learning pipeline for the fused ensemble model is summarized in Figure 1 and detailed below. For each model, the data was split into 5 folds stratified for the outcome of LF with the sklearn StratifiedKFold function (35). Four folds were used as the exploratory (training/validation) set and one fold was held out (test) for each iteration, with the intent for 5-fold cross-validation. For all steps in radiomic feature selection described subsequently, the exploratory set was used, and the selected features were later applied to the test fold prior to model performance evaluation (the test set was not seen during feature selection). Note clinical features were selected for based on prior knowledge (3,7-9) and did not go through the feature selection process. Clinical features included: primary site of disease, smoking status, p16 status, chemotherapy agent, T stage (AJCC 7), and N stage (AJCC 7).
Feature stability/reproducibility
Repeat radiomic features extracted from CBCTa and CBCTb were compared with Intra-Class Correlation coefficients (ICC) (36) to determine the reproducibility of features. Given artifacts were noted in some CBCTs (as described in the Results section, entitled Reproducibility of Features), a subgroup analysis was performed comparing the ICC values of test-retest scans having the artifact described above with test-retest scans that did not using independent samples t-tests. For subsequent feature selection, a restrictive ICC threshold ≥0.95 was used.
Maximum relevance and minimum redundancy (mRMR)
Selection for features with mRMR (37,38) was performed in python with pymRMR, where features were selected for on the basis of the outcome class (relevance) and then selected against the dependence/similarity to other relevant features (redundancy). For this project, pymRMR was performed with the Mutual Information Difference (MID) scheme with 30 features selected for.
Recursive feature elimination (RFE) with cross-validation
RFE without (39) and with cross-validation (RFECV) (35) have previously been described, where the least important features are iteratively deleted from the model until reaching the combination of features with the highest cross-validation score which are then chosen as the optimal features for the given dataset. For this project, RFECV was adapted for use of the EBM as the classifier. First, the importance scores were calculated for each feature in the model fitted to the exploratory dataset. At each step, the feature with the lowest mean importance score was eliminated from the model, and then 5-fold cross-validation was performed on the exploratory set to determine the mean AUC with the currently included features. This process was repeated until a minimum of 5 features were left in the model. Following completion, the combination of features with the highest mean AUC scores were selected and used in the test set.
EBM model
EBM is a python-based machine learning classifier from InterpretML, termed a “glassbox model”, which is a generalized additive model using methods including bagging, gradient boosting, and automatic interaction detection, producing graphs that not only show model performance but also show details of how the included features contribute to the overall prediction (28). This aids in the overall interpretability of the EBM model (40), given issues may be identified explaining unexpected model predictions during review of specific feature contributions.
EBM models were created for nodal and primary structures separately that predicted LF. Separate models were created for the following sets of features: (I) clinical; (II) radiomic (CT1 + delta features); (III) CT1 radiomic only; (IV) delta radiomic only; and (V) a combined feature model including clinical, radiomic, and delta features in the same EBM model. Subsequently, a (VI) fused model was created by fusing the predicted scores of the separate (I) clinical and (II) radiomic models with the following equation:
Here, prad is the predicted score for the presence of LF from the radiomic model, pclin is the predicted score for the presence of LF from the clinical model, w is the weighting factor, and pF0 and pF1 are the fused predicted scores for the absence and presence of LF, respectively, where the following constraint was met: pF0+ pF1= 1. The weighting factor (w) is defined as:
Here, the AUC was derived from the average of five 5-fold cross-validation iterations of the exploratory (training/validation) sets for each iteration, for the radiomic and clinical models (AUCrad and AUCclin, respectively), where the following constraint was met: w + (1 − w) = 1. Note that AUC values of the test sets were not used to avoid information leakage via the weighting variable at time of fusion.
Following 5-fold cross-validation and calculation of the test predicted scores, the entire above process was re-iterated 25 times, for both the parallel ensemble schema and to reduce the impact of randomness on biasing the reported results.
Ensemble learning methods have previously been employed to reduce variance within datasets and improve accuracy (41,42). Note that the EBM classifier already employs a sequential form of ensemble learning with bagging and gradient boosting in its classification scheme. In this project, we have performed an additional parallel ensemble step where 25 iterations of the above described process are iterated independently with different 5-fold splits, leading to a total of 125 models with 25 used for each separate structure in the test holdout sets. To calculate ensemble scores, these predicted scores were averaged for each structure.
Ensemble scores were plotted on receiver operating characteristic (ROC) and precision-recall (PRC) curves in SPSS for the primary and nodal models. Performance of each model was compared against random chance (where an area under the curve, or AUC, of 0.50 was assumed to be due to random chance) with one-sample t-tests, and performance between models were compared with paired-samples AUC t-tests in SPSS using the ROC analysis function. Significance was set at a P value <0.05 using a 2-tailed test.
Results
Baseline characteristics: patients
Thirty cases (patients who reached LF) were matched with 60 controls and were included in this analysis, with median follow-up times of 420.5 (IQR, 358.5 to 664) and 758.5 (IQR, 617 to 976.5) days, respectively. Of the cases, 23 patients failed in their primary, and 12 patients failed in at least 1 node (with a total of 23 primary and 19 nodal structures reaching the endpoint of LF). Baseline characteristics of the included patients are summarized in Table S1. The site of the primary was significantly different between groups (P=0.021), with relatively more oropharyngeal, glottic, and hypopharyngeal primaries in cases. Furthermore, p16 status significantly differed between groups (P=0.032), being positive in less cases (33.3%) than controls (53.3%), respectively. There was a similar amount of unknown or not applicable p16 cases (33.3% vs. 33.3%). T stage and N stage did not differ significantly between cases and controls (P=0.170 and P=0.493, respectively).
For the cases, median actuarial time to LF was 135 days, with the actuarial 1 year LF rate being 90% (see cumulative incidence in Figure S1). Of note, one patient in the control group did experience regional failure outside of the high dose (70 Gy) volume at 305 days following therapy completion; however, this was not counted as LF given the criteria for this study. The actuarial 1 year DM rate for the cases was 40%, which was higher than controls (6.7%, P<0.001; see cumulative incidence in Figure S2). Median actuarial time to death was 520 days for cases and not reached for controls, with actuarial 2-year OS rates of 33.0% and 100% for cases and controls, respectively (P<0.001, see Kaplan-Meier curves in Figure S3).
Baseline characteristics: structures
Most patients had ≤5 separate structures contoured for analysis (83.3% cases, 88.3% controls; P=0.519), including both the primaries and nodes. A total of 23 primary and 19 nodal structures reached LF (with 21 primary events and 15 nodal events occurring before year 1), with an associated 66 primary and 194 nodal structures which did not (controls). For primary structures, the primary site (P=0.004) and T stage (P=0.049) differed between cases and controls. The median actuarial time to LF was 173 days for primary cases, with a 1-year actuarial rate of LF for all primary structures being 33.7%. For nodal structures, the p16 status (P=0.001) and T stage (P=0.013) differed between cases and controls, and the median actuarial time to LF for cases was 127 days and 1-year actuarial rate of LF for all nodal structures was 7.0%. See Tables S3,S4 and Figure S4.
Reproducibility of features
Of the 90 included patients, 57 patients had at least two CBCTs performed before the same fraction of radiotherapy and were included in the reproducibility analysis. A total of 56 primary and 144 nodal structures were included. For primary structures, about half were oropharyngeal (17 base of tongue, 9 tonsillar, 2 soft palate, and 3 oropharynx NOS) and the rest were non-oropharyngeal (14 supraglottic, 5 glottic, and 6 hypopharyngeal). For nodal structures, some spanned >1 neck level due to size or if they were matted or abutting adjacent involved nodes; therefore, >1 neck level could have been recorded for each structure. The most common involved level was level II (n=87) followed by levels III (n=51), IV (n=23), VII (n=10), I (n=8), V (n=5), and VI (n=3).
When comparing extracted radiomic features of the test-retest CBCTs (CBCTa and CBCTb), 78.4% and 30.4% features for primaries and 92.2% and 40.2% features for nodes had ICC scores >0.75 and >0.95, respectively (see Figure 2 and Table S5). However, during review of these test-retest images, it was noted that some scans were affected by an artifact that appeared to originate along the surface of bone and cartilage as shown in Figure 3. This artifact was noted to come and go between test-retest scans within minutes of each other of unclear etiology. Twenty patients (35%, 20/57) were noted to have at least one test-retest scan with this artifact. A subcohort analysis was performed comparing the ICC values between patients with and without this artifact, and it was noted to worsen the reproducibility of features, with a mean ICC score for all nodal features of 0.839 vs. 0.900 (presence vs. absence of artifact, P=0.003) and for all primary features of 0.750 vs. 0.792 (presence vs. absence of artifact, P=0.099). The main feature group affected by this artifact appeared to be GLCM features, reaching significance for nodes (mean ICC of 0.788 vs. 0.883, presence vs. absence of artifact, P=0.024) and trended towards significance for primaries (0.681 vs. 0.775, P=0.068) (see Tables 1,2). Therefore, it was decided to avoid this artifact in subsequent radiomic extractions of delta features. For CBCT01 or CBCT21 scans where this artifact was identified, another CBCT done prior to the same fraction was preferentially selected if present or another CBCT ±1 fraction if not.
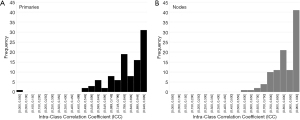
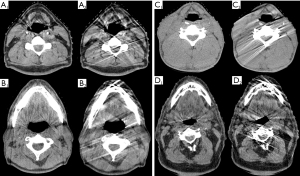
Table 1
| Feature class | (−) No artifact | (+) Artifact present | Difference | P value† | |||
|---|---|---|---|---|---|---|---|
| Mean ICC | St. Dev. | Mean ICC | St. Dev. | ||||
| Total (all features) | 0.792 | 0.151 | 0.750 | 0.208 | −0.042 | 0.099 | |
| Shape | 0.941 | 0.032 | 0.986 | 0.016 | 0.045 | 0.000 | |
| First order | 0.678 | 0.206 | 0.620 | 0.245 | −0.058 | 0.447 | |
| GLCM | 0.775 | 0.110 | 0.681 | 0.218 | −0.094 | 0.068 | |
| GLRLM | 0.815 | 0.139 | 0.748 | 0.158 | −0.068 | 0.207 | |
| GLSZM | 0.785 | 0.125 | 0.764 | 0.125 | −0.021 | 0.639 | |
| GLDM | 0.798 | 0.128 | 0.782 | 0.185 | −0.016 | 0.016 | |
†ICC values were compared across groups with independent-samples
Table 2
| Feature class | (−) No artifact | (+) Artifact present | Difference | P value† | |||
|---|---|---|---|---|---|---|---|
| Mean ICC | St. Dev. | Mean ICC | St. Dev. | ||||
| Total (all features) | 0.900 | 0.098 | 0.839 | 0.179 | −0.062 | 0.003 | |
| Shape | 0.994 | 0.011 | 0.995 | 0.008 | 0.001 | 0.726 | |
| First order | 0.879 | 0.112 | 0.773 | 0.244 | −0.105 | 0.105 | |
| GLCM | 0.883 | 0.090 | 0.788 | 0.177 | −0.095 | 0.024 | |
| GLRLM | 0.894 | 0.075 | 0.850 | 0.097 | −0.044 | 0.162 | |
| GLSZM | 0.872 | 0.123 | 0.818 | 0.219 | −0.054 | 0.398 | |
| GLDM | 0.904 | 0.090 | 0.864 | 0.099 | −0.041 | 0.408 | |
†ICC values were compared across groups with independent-samples
Models predicting LF of primary structures
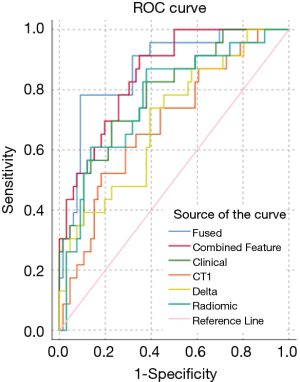
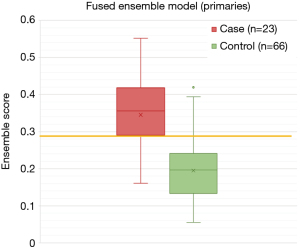
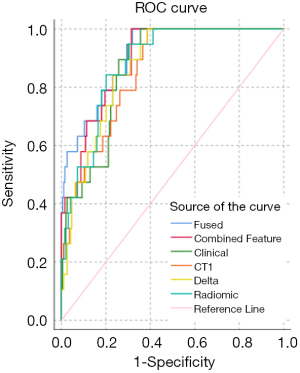
All primary ensemble models were significantly different from random chance (P≤0.004). The fused ensemble model achieved the highest AUC of 0.871 at predicting LF for head and neck primaries in the test cohort, which was numerically higher than the clinical ensemble but did not reach significance (AUC =0.788, P=0.134) and was only marginally higher than the combined feature ensemble (AUC =0.853, P=0.494). However, the fused ensemble model’s AUC was significantly higher than the radiomic ensemble (AUC =0.770, P=0.017), CT1 ensemble (AUC =0.687, P=0.004), and delta ensemble models (AUC =0.696, P=0.002). The maximum Youden J statistic for the fused ensemble model was 0.692 and correlated with a sensitivity of 78.3% and 90.9% for this model. See a summary of the primary models in Table 3, ROC curves in Figure 4, and the distribution of scores of the top performing fused ensemble model in Figure 5.
Table 3
| Model | AUC | 95% CI (lower) | 95% CI (upper) | P value (vs. random chance)† | P value (vs. fused ensemble)‡ | Max Youden J statistic | Predicted score threshold at max J statistic | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|---|
| Fused ensemble | 0.871 | 0.788 | 0.954 | 0.000 | NA | 0.692 | 0.290 | 78.3 | 90.9 |
| Combined feature ensemble | 0.853 | 0.771 | 0.935 | 0.000 | 0.494 | 0.565 | 0.161 | 91.3 | 56.5 |
| Clinical only ensemble | 0.788 | 0.680 | 0.895 | 0.000 | 0.134 | 0.469 | 0.252 | 69.6 | 77.3 |
| Radiomic ensemble (CT1 + Delta) | 0.770 | 0.655 | 0.885 | 0.000 | 0.017 | 0.491 | 0.237 | 87.0 | 49.1 |
| CT1 only ensemble | 0.687 | 0.561 | 0.813 | 0.004 | 0.004 | 0.340 | 0.291 | 52.2 | 81.8 |
| Delta only ensemble | 0.696 | 0.571 | 0.822 | 0.002 | 0.013 | 0.345 | 0.208 | 73.9 | 60.6 |
†ROC curves were compared against random change (AUC =0.5) with one-sample
Models predicting LF of nodal structures
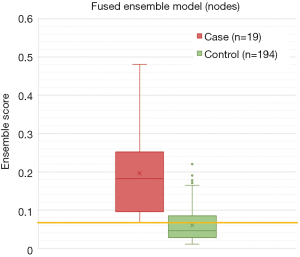
All nodal ensemble models performed relatively well achieving AUCs >0.80 and were significantly different than random chance (P<0.001). The fused ensemble model also achieved the highest AUC of 0.910 at predicting LF for head and neck nodes in the test cohort, which was numerically higher than the clinical ensemble but only trended towards significance (AUC =0.865, P=0.080) and was again only marginally higher than the combined feature ensemble (AUC =0.893, P=0.212). The fused ensemble model was not significantly different from the radiomic ensemble (AUC =0.880, P=0.268) or delta ensemble (AUC =0.867, P=0.150) models, and was only significantly better than the CT1 ensemble (AUC =0.854, P=0.026) model. The maximum Youden J statistic for the fused ensemble model was 0.680 and correlated with a sensitivity of 100.0% and a specificity of 68.0% for this model. See a summary of the nodal models in Table 4, ROC curves in Figure 6, and the distribution of scores of the top performing fused ensemble model in Figure 7.
Table 4
| Model | AUC | 95% CI (lower) | 95% CI (upper) | P value (vs. random chance)† | P value (vs. fused ensemble)‡ | Max Youden J statistic | Predicted score threshold at max J statistic | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|---|
| Fused ensemble | 0.910 | 0.853 | 0.967 | 0.000 | NA | 0.680 | 0.066 | 100.0 | 68.0 |
| Combined feature ensemble | 0.893 | 0.819 | 0.941 | 0.000 | 0.212 | 0.686 | 0.046 | 100.0 | 68.6 |
| Clinical only ensemble | 0.865 | 0.802 | 0.929 | 0.000 | 0.080 | 0.648 | 0.061 | 94.7 | 70.1 |
| Radiomic ensemble (CT1 + Delta) | 0.880 | 0.819 | 0.941 | 0.000 | 0.268 | 0.643 | 0.060 | 94.7 | 69.6 |
| CT1 only ensemble | 0.854 | 0.784 | 0.924 | 0.000 | 0.026 | 0.613 | 0.056 | 100.0 | 61.3 |
| Delta only ensemble | 0.867 | 0.803 | 0.930 | 0.000 | 0.150 | 0.613 | 0.054 | 100.0 | 61.3 |
†ROC curves were compared against random change (AUC =0.5) with one-sample
Discussion
In this exploratory study, we demonstrated that a novel approach of analyzing discrete primary and nodal HNSCC structures within the same patient separately yielded high discriminatory ability at predicting LF in a fused ensemble model developed from a single institutional cohort predominantly consisting of early LF (≤12 months). Here, the thought was that radiographic changes within one structure may not be indicative of response in another; therefore, we sought to evaluate these independently. The highest AUC values were achieved with the fused models using an interpretable machine learning algorithm (EBM) and a parallel ensemble design for both the primaries (AUC =0.871) and nodes (AUC =0.910), incorporating both clinical and radiomic (baseline CT1 and delta CBCT) features as input. However, when compared to the clinical only model, the fused model’s performance only trended towards improvement and did not reach statistical significance for both primary (P=0.134) and nodal (P=0.080) clinical comparisons. Although we could not prove that radiomic findings provided additive prognostic benefit over traditional clinical features in an EBM model statistically, we view these results as promising given the limited sample size may have inhibited our ability to detect a difference secondary to inadequate power and given the absolute AUC improvement was quite large for both the primary (0.083) and nodal (0.045) fused/clinical comparisons.
The most frequently included delta features were shape features, with change in maximum 3D diameter and change in sphericity being the most commonly included radiomic features in primary (69.6%) and nodal (80.8%) models, respectively (see Tables S6,S7). The inclusion of these features are intuitive in that volume reduction of a tumor is often equated to response, and prior small retrospective experiences have supported that shrinkage of the tumor over the course of or following radiotherapy is associated with better local outcomes (43,44). In contrast, change in texture features were not commonly incorporated in EBM models, occurring less than ~20% of the time due to exclusion during feature selection. The seemingly weaker informativeness of delta texture features may be due to the (I) inherent artifacts of CBCT scans and metal artifacts from dental implants that could interfere with accurate quantification of texture features and (II) the inherent heterogeneity of both primary and nodal structures in this dataset making broad changes in texture features over time difficult to interpret. Prior retrospective CT studies evaluating the evolution of morphology of nodes following CRT have shown that baseline hypodensity/fat sub-volume component, volume/size, and HU standard deviation were associated with response/regression following therapy (45). It is unclear if delta texture analysis would be improved if subgroups were created based on baseline imaging features, such as presence/absence of a necrotic center, single/matted nodes, etc.; however, the size of this cohort precluded meaningful subgroup analyses.
To our knowledge, this is the first published experience using CBCT-based delta radiomics for head and neck malignancies in machine learning, as prior experiments incorporating CBCT-based delta radiomic features have primarily focused on non-small cell lung cancer (NSCLC) (27,46-49). Initial findings have been disparate, with one study showing stratification of OS based on delta features (48) and two studies failing to demonstrate an additive prognostic benefit for CBCT-based radiomics when added to planning CT features for OS and LRR (47) or PFS (49). This discrepancy could be due to differences in datasets, acquisition parameters of scans, segmentation and deformable image registration, feature selection strategies, and data analysis. Of these, the study by van Timmeren et al. included the largest dataset from four separate institutions, correlating CBCT-based delta radiomic features, planning CT, and clinical features with LRR and OS. After their model was trained on data from the first institution and validated on the other three, the additive prognostic benefit of CBCT-based delta radiomic features could not be confirmed (47), which the authors noted differences in image quality (varying treatment machines), segmentation, and deformable image registration between centers may have affected the ability for these models to be validated across institutions. Indeed, it is well known that CBCT-based radiomic features vary substantially based on the scanner and acquisition parameters (25), which may limit the generalizability of a model developed at a single center; however, external validation remains to be tested for our proposed models.
The primary model was limited in that it included multiple sites of disease in the head and neck that are known to behave differently. This heterogeneity of morphology at baseline could decrease the potentially informative associations of various features, given it is possible that different disease sites may exhibit different delta CBCT-based radiomics over time when responding to or not responding to treatment. If this were the case, different sites of disease would be better suited to separate models. However, the limited size of the cohort limited meaningful subgroup analyses in this study and did not allow us to test this hypothesis. Other published models of LF or LRF in the head and neck region have tended to be more restrictive with the primary sites included, e.g., oropharyngeal (17-19), laryngeal/hypopharyngeal (20), hypopharyngeal (22). Still, our model compared favorably to previously published models predicting LF or LRF of HNSCC (as discussed in the introduction). Additionally, our analysis is limited by a lack of comparison of delta radiomic features to other known imaging modalities, such as PET/CT (18-21), that have been shown to be informative for LF prediction of HNSCC. Therefore, we cannot comment on the superiority of this radiomic method in comparison to other radiomic methods that use features from other modalities.
From a clinical standpoint, this model is limited from its retrospective nature and small sample size. Although case-control matching was performed to reduce variances in baseline characteristics between cohorts, this selection method may introduce bias in that the controls may not be representative of the normal population (here, “normal population” references patients with HNSCC that undergo definitive radiotherapy with or without chemotherapy and achieve local control). In addition, the small sample size (n=90) of the dataset used may not adequately reflect the extent of imaging variations of both baseline CT and intra-treatment CBCT scans over the course of radiotherapy. Therefore, further validation of this model in larger prospective cohorts is warranted.
Conclusions
Overall, the fused ensemble model incorporating both clinical and radiomic (CT1 and delta CBCT) features achieved the highest discriminatory ability for predicting LF of primaries (AUC =0.871) and for nodes (AUC =0.910). Although the fused ensemble model did not reach significance when compared to the clinical only models, this may have been due to a lack of power from the limited sample size of this study, and we find the trend towards better performance promising. If these exploratory models were shown in a larger retrospective and/or in a separate prospective cohort to stratify LF accurately and to be superior to the clinical only model, this could aid in the early decision for response-adapted approaches with an imaging modality that is already incorporated in the routine course of radiotherapy for HNSCC malignancies. The proposed fused ensemble EBM models are worthy of further investigation in larger prospective cohorts.
Acknowledgments
Funding: This research is partially supported by a seed grant from the Department of Radiation Oncology at UT Southwestern Medical Center.
Footnote
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-274). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. MRF reports non-financial support from Varian, Inc., outside of the submitted work. JW reports that this work was funded in part by a UT Southwestern seed grant. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was reviewed and approved through the institutional review board (IRB), with patient consent waived, and was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nguyen-Tan PF, Zhang Q, Ang KK, Weber RS, Rosenthal DI, Soulieres D, Kim H, Silverman C, Raben A, Galloway TJ, Fortin A, Gore E, Westra WH, Chung CH, Jordan RC, Gillison ML, List M, Le QT. Randomized phase III trial to test accelerated versus standard fractionation in combination with concurrent cisplatin for head and neck carcinomas in the Radiation Therapy Oncology Group 0129 trial: long-term report of efficacy and toxicity. J Clin Oncol 2014;32:3858-66. [Crossref] [PubMed]
- Ang KK, Zhang Q, Rosenthal DI, Nguyen-Tan PF, Sherman EJ, Weber RS, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol 2014;32:2940-50. [Crossref] [PubMed]
- Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, Soulieres D, Trotti A, Avizonis V, Ridge JA, Harris J, Le QT, Gillison M. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol 2014;32:3365-73. [Crossref] [PubMed]
- Grønhøj C, Jakobsen KK, Jensen DH, Rasmussen J, Andersen E, Friborg J, von Buchwald C. Pattern of and survival following loco-regional and distant recurrence in patients with HPV+ and HPV- oropharyngeal squamous cell carcinoma: A population-based study. Oral Oncol 2018;83:127-33. [Crossref] [PubMed]
- Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 2019;393:40-50. [Crossref] [PubMed]
- Strohl MP, Wai KC, Ha PK. De-intensification strategies in HPV-related oropharyngeal squamous cell carcinoma-a narrative review. Ann Transl Med 2020;8:1601. [Crossref] [PubMed]
- O'Sullivan B, Huang SH, Su J, Garden AS, Sturgis EM, Dahlstrom K, et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol 2016;17:440-51. [Crossref] [PubMed]
- Amin MB, Edge SB, American Joint Committee on C. AJCC cancer staging manual 2017.
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363:24-35. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging 2020;11:91. [Crossref] [PubMed]
- Bogowicz M, Tanadini-Lang S, Guckenberger M, Riesterer O. Combined CT radiomics of primary tumor and metastatic lymph nodes improves prediction of loco-regional control in head and neck cancer. Sci Rep 2019;9:15198. [Crossref] [PubMed]
- Keek S, Sanduleanu S, Wesseling F, de Roest R, van den Brekel M, van der Heijden M, et al. Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri)tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS One 2020;15:e0232639 [Crossref] [PubMed]
- Keek S, Sanduleanu S, Wesseling F. Correction: Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri)tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS One 2020;15:e0237048 [Crossref] [PubMed]
- Leger S, Zwanenburg A, Leger K, Lohaus F, Linge A, Schreiber A, et al. Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC. Cancers (Basel) 2020;12:3047. [Crossref] [PubMed]
- Welch ML, McIntosh C, McNiven A, Huang SH, Zhang BB, Wee L, Traverso A, O'Sullivan B, Hoebers F, Dekker A, Jaffray DA. User-controlled pipelines for feature integration and head and neck radiation therapy outcome predictions. Phys Med 2020;70:145-52. [Crossref] [PubMed]
- Giraud P, Giraud P, Nicolas E, Boisselier P, Alfonsi M, Rives M, Bardet E, Calugaru V, Noel G, Chajon E, Pommier P, Morelle M, Perrier L, Liem X, Burgun A, Bibault JE. Interpretable Machine Learning Model for Locoregional Relapse Prediction in Oropharyngeal Cancers. Cancers (Basel) 2020;13:57. [Crossref] [PubMed]
- Folkert MR, Setton J, Apte AP, Grkovski M, Young RJ, Schöder H, Thorstad WL, Lee NY, Deasy JO, Oh JH. Predictive modeling of outcomes following definitive chemoradiotherapy for oropharyngeal cancer based on FDG-PET image characteristics. Phys Med Biol 2017;62:5327-43. [Crossref] [PubMed]
- Haider SP, Sharaf K, Zeevi T, Baumeister P, Reichel C, Forghani R, Kann BH, Petukhova A, Judson BL, Prasad ML, Liu C, Burtness B, Mahajan A, Payabvash S. Prediction of post-radiotherapy locoregional progression in HPV-associated oropharyngeal squamous cell carcinoma using machine-learning analysis of baseline PET/CT radiomics. Transl Oncol 2021;14:100906 [Crossref] [PubMed]
- Zhong J, Frood R, Brown P, Nelstrop H, Prestwich R, McDermott G, Currie S, Vaidyanathan S, Scarsbrook AF. Machine learning-based FDG PET-CT radiomics for outcome prediction in larynx and hypopharynx squamous cell carcinoma. Clin Radiol 2021;76:78.e9-78.e17. [Crossref] [PubMed]
- Wang K, Zhou Z, Wang R, Chen L, Zhang Q, Sher D, Wang J. A multi-objective radiomics model for the prediction of locoregional recurrence in head and neck squamous cell cancer. Med Phys 2020;47:5392-400. [Crossref] [PubMed]
- Hsu CY, Lin SM, Ming Tsang N, Juan YH, Wang CW, Wang WC, Kuo SH. Magnetic resonance imaging-derived radiomic signature predicts locoregional failure after organ preservation therapy in patients with hypopharyngeal squamous cell carcinoma. Clin Transl Radiat Oncol 2020;25:1-9. [Crossref] [PubMed]
- Morgan HE, Sher DJ. Adaptive radiotherapy for head and neck cancer. Cancers Head Neck 2020;5:1. [Crossref] [PubMed]
- Leger S, Zwanenburg A, Pilz K, Zschaeck S, Zöphel K, Kotzerke J, Schreiber A, Zips D, Krause M, Baumann M, Troost EGC, Richter C, Löck S. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother Oncol 2019;130:10-7. [Crossref] [PubMed]
- Fave X, Mackin D, Yang J, Zhang J, Fried D, Balter P, Followill D, Gomez D, Jones AK, Stingo F, Fontenot J, Court L. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys 2015;42:6784-97. [Crossref] [PubMed]
- van Timmeren JE, Carvalho S, Leijenaar RTH, Troost EGC, van Elmpt W, de Ruysscher D, Muratet JP, Denis F, Schimek-Jasch T, Nestle U, Jochems A, Woodruff HC, Oberije C, Lambin P. Challenges and caveats of a multi-center retrospective radiomics study: an example of early treatment response assessment for NSCLC patients using FDG-PET/CT radiomics. PLoS One 2019;14:e0217536 [Crossref] [PubMed]
- van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Oberije C, Monshouwer R, Bussink J, Brink C, Hansen O, Lambin P. Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother Oncol 2017;123:363-9. [Crossref] [PubMed]
- Nori H, Jenkins S, Koch P, Caruana R. InterpretML: A Unified Framework for Machine Learning Interpretability. Preprint, 2019.
- R-Core-Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2020.
- Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transplant 2007;40:381-7. [Crossref] [PubMed]
- Gray B. cmprsk: Subdistribution Analysis of Competing Risks. . R package version 2.2-10.2020.
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Zwanenburg A, Leger S, Vallieres M, Lock S. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003.
- Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. [Crossref] [PubMed]
- Pedregosa F, Varoquax G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Duchesnay MPE. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 2011;2825-30.
- Salarian A. Intraclass Correlation Coefficient (ICC). MATLAB Central File Exchange.
- Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 2005;27:1226-38. [Crossref] [PubMed]
- Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005;3:185-205. [Crossref] [PubMed]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning 2002;389-422. [Crossref]
- Caruana R, Lou Y, Gehrke J, Koch P, Sturm M, Elhadad N. Intelligible Models for HealthCare: Predicting Pneumonia Risk and Hospital 30-day Readmission. KDD '15: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2015:1721-30.
- Dietterich TG. Ensemble Methods in Machine Learning. In: Kittler J, Roli F, editors. Multiple Classifier Systems, LCCS-1857: Springer, 2000:1-15.
- Moreno F, Inesta J, Leon P, Mico L. Comparison of Classifier Fusion Methods for Classification in Pattern Recognition Tasks. SSPR /SPR 2006: Structural, Syntactic, and Statistical Pattern Recognition; 2006:705-13.
- Hou J, Guerrero M, Suntharalingam M, D'Souza WD. Response assessment in locally advanced head and neck cancer based on RECIST and volume measurements using cone beam CT images. Technol Cancer Res Treat 2015;14:19-27. [Crossref] [PubMed]
- Nevens D, Vantomme O, Laenen A, Hermans R, Nuyts S. The prognostic value of location and size change of pathological lymph nodes evaluated on CT-scan following radiotherapy in head and neck cancer. Cancer Imaging 2017;17:8. [Crossref] [PubMed]
- Tang C, Fuller CD, Garden AS, Awan MJ, Colen RR, Morrison WH, Frank SJ, Beadle BM, Phan J, Sturgis EM, Zafereo ME, Weber RS, Rosenthal DI, Gunn GB. Characteristics and kinetics of cervical lymph node regression after radiation therapy for human papillomavirus-associated oropharyngeal carcinoma: quantitative image analysis of post-radiotherapy response. Oral Oncol 2015;51:195-201. [Crossref] [PubMed]
- van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P. Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncol 2017;56:1537-43. [Crossref] [PubMed]
- van Timmeren JE, van Elmpt W, Leijenaar RTH, Reymen B, Monshouwer R, Bussink J, Paelinck L, Bogaert E, De Wagter C, Elhaseen E, Lievens Y, Hansen O, Brink C, Lambin P. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: Evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother Oncol 2019;136:78-85. [Crossref] [PubMed]
- Shi L, Rong Y, Daly M, Dyer B, Benedict S, Qiu J, Yamamoto T. Cone-beam computed tomography-based delta-radiomics for early response assessment in radiotherapy for locally advanced lung cancer. Phys Med Biol 2020;65:015009 [Crossref] [PubMed]
- Qin Q, Shi A, Zhang R, Wen Q, Niu T, Chen J, Qiu Q, Wan Y, Sun X, Xing L. Cone-beam CT radiomics features might improve the prediction of lung toxicity after SBRT in stage I NSCLC patients. Thorac Cancer 2020;11:964-72. [Crossref] [PubMed]


