
Ectosome biogenesis and release processes observed by using live-cell dynamic imaging in mammalian glial cells
Introduction
Intercellular communication, essential for organ function, can be mediated by direct cell-to-cell-contact, e.g., tight junction (1), or secreted molecules, e.g., single-molecule-driven signal transduction (2). In the last two decades, a concept “signaling organelle” (3) involves subcellular vesicles and extracellular vehicles (EVs) that are defined as a heterogeneous group of membrane-bound particles containing signaling molecules (4). EVs are released from the cell to deliver signals to target cells (5) for signal transduction cascades as the long- and short-distance intercellular communication mechanism.
As EVs modulate tumor microenvironment behavior via the immune, pro-inflammatory, and inflammatory response (6), they conjure up hope to appear to be promising targets in the diagnosis therapy of diseases, especially cancer. There are two types of EVs, exosomes, and ectosomes. Exosomes are released by the exocytosis of multivesicular bodies (MVBs). Ectosomes are generated by outward budding and pinching off from the plasma membrane, which is generally referred to ubiquitous vesicles as including platelet-derived microvesicles (7), shedding vesicles (8), microparticles (9), and apoptotic bodies (10). The release of EVs is regulated by a variety of stimuli such as ATP (11), protein phosphorylation (12), calmodulin activation, cytoskeleton reorganization (13), and other factors. The release of ectosomes is visible soon as a new type of membrane protrusion resembles a ‘beads-on-a-string’ structure after stimulation and lasts for several minutes, companying an increase in the concentration of free Ca2+ (10) reflecting on a new mechanism of apoptotic body formation in monocytes. Loss of membrane-cytoskeletal adhesion in the formation of ectosomes is involved in plasma membrane phosphatidylinositol 4,5-bisphosphate (PIP2) adhesion energy exchanges between the cytoskeleton and the plasma membrane (14); however, the underlined dynamic process of biogenesis of ectosomes remain largely unknown.
The biogenesis of ectosomes is believed to be assembled by the regulated outward budding of plasma membrane domains (15). Such plasma membrane domain-induced budding in endothelial cells depends on caveolae (16) or CCV (17). Many cells shed their ectosomes (also call microparticles) into circulation. Among them, platelet microparticles are the most abundant ectosomes. These ectosomes are typically characterized by their phosphatidylserine exposure (PS) on their outer membrane, as indicated by annexin V binding (14). Since phosphatidylserine (PS) is exclusively expressed on the plasma membrane’s inner leaflet, the budding of a fragment of the plasma membrane as a source of ectosome biogenesis may not cover the whole situation. The contents of ectosomes include IL-1β and other cytokines that are usually secreted into the extracellular environment or in the lumen of vesicles within the glial cells (18) and also include TNF family, TNF-related apoptosis-inducing ligand, Fas ligand, or CD40 ligand that are released by damaged renal tubular epithelial cells (19). How cells generate ectosomes that contain a secretory form of molecules, just like the intracellular vesicles, is still a puzzle. By stimulating the primary cultured mouse glia cells and recording the dynamic process of ectosome release, we visualized the dynamic process of shedding and investigated the possible mechanism of ectosome release in live-cells.
Methods
This study was approved by the institutional ethics board of the Shenzhen University, in compliance with the national guidelines for the care and use of animals established by the National Natural Science Foundation of China.
Reagents and antibodies
DMEM culture medium, neurobasal medium, and FBS were purchased from Gibco. Colchicine was purchase from Sigma Aldrich. Cy5-conjugated donkey anti-mouse second antibody was purchased from Jackson ImmunoResearch. ER-tracker Blue and Fluo 4 were purchased from Invitrogen. The primary antibodies, mouse monoclonal anti-serca 3, were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-NGF antibody was purchased from Abcam. FITC-annexin V kit was purchased from Beyotime.
Primary glial cell culture
Primary cortical neurons and glias were prepared as previously described with minor modifications (20). In brief, brains were dissected under the binocular microscope from postnatal day 1 mouse pups [all under standard guidelines with the Institutional Animal Care and Use Committees (IACUC) approval], and the whole brain was used. After collagenase digestion and centrifugations, neuron and glia cells were grown in neurobasal medium with B27 supplement, 100 U/mL penicillin, and 100 µg/mL streptomycin and 5% FBS on polylysine coated cover glass. The cells were used for time-lapse imaging 7 days after the culture.
Live cell imaging
A Zeiss 710 confocal microscope (Carl Zeiss, GERMANY) was used with 20× or 63× water objectives. The cells were scanned under the laser scanning confocal microscope using the Time Series program. The scanning frequency was 1 time/s. The changes in the fluorescence were recorded through continuous scanning. The drug of the intervention was added to cells in 20–30 s during scanning.
Cell labeling
Primary glial cells were labeled with Fluo-4 AM or ER-Tracker™ Blue-White DPX in phenol Red free DMEM for 30 minutes and washed with phenol Red free DMEM and incubated in the same medium before live-cell imaging.
Extracellular vesicles (including ectosome) collection and staining
Primary glial cells were cultured in two-well poly-L-lysine coated cover glasses for 7 days and then washed three times with phenol Red free DMEM before adding fresh phenol Red free DMEM culture medium with or without 30 µM colchicines for 30 minutes. The culture medium was transferred to a 1.5 mL tube for extracellular vesicle preparation. The cells cultured on the cover-glass chamber were fixed with cold methanol for immunofluorescence. For EVs preparation, the collected culture medium was centrifuged at 3,000 rpm for 5 minutes to remove the cells. The supernatant was then transferred to another 1.5 mL tube to centrifuge at 145,000 rpm for 5 minutes to collect the EVs. FITC-conjugated annexin V (1:100) diluted by PBS were used to incubate with the EVs for 30 minutes and washed with PBS by centrifugation, and then anti-NGF mouse monoclonal antibody (1:100) diluted by PBS containing 0.1% triton ×100 were used to incubate with the EVs for 30 minutes and washed with PBS by centrifugation. Cy5-conjugated donkey anti-mouse second antibody was used to labeling NGF signal for 30 minutes and washed with PBS by centrifugation. The precipitate was suspended with 50 µL PBS and dropped on a glass slide for imaging. Immunofluorescence for cells on the cover-glass chamber was performed using an anti-serca 3 mouse monoclonal antibody. The same second antibody, as above, was used for the staining.
Quantitation of annexin V positive and NGF-containing EVs
The total numbers of EVs were counted under the DIC image by granule counting. Annexin V positive and NGF positive EVs were counted, respectively. Co-localized signals were counted for those both of them are positive.
Results
Ectosomes budding from the plasma membrane
The formation of the ectosome has been recognized as outward budding from the plasma membrane. The cytoskeleton’s reorganization can induce the release of ectosomes by the formation of ‘beads-on-a-string’ protrusions (10). Simultaneously, disruption of actin-membrane interactions dissociates the platelet membrane skeleton from its membrane attachment sites, shedding the plasma membrane domain to procoagulant-rich microvesicles (21). To investigate whether the disassembly of a microtubule can promote the ectosome release and whether the process of release can be visualized in glia cells, we labeled the primary cultured glial cells with fluor 4, treated the cells with 20 µM colchicine, and observed the dynamic process using the Zeiss 710 confocal microscope. Dynamic changes in calcium signal were recorded in a time serial program at 1-frame per second. Vesicle-like structures in the cells were observed with increasing calcium-driven signals (Figure 1, Video 1) up to 20 µM of colchicine. The nucleus was labeled with a robust calcium signal in the center of each cell.
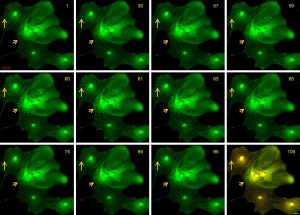
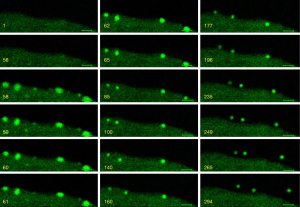
Dot-like structures with strong calcium signals appeared near the cells’ edge (Figure 1, arrows). The calcium signal lasted about 30 seconds, but no ectosome release was observed. The increase of cytosolic free Ca2+ induces the cytoskeleton’s disassembly is believed to respond to the local signal for triggering the EV release (12). The stimulation of colchicine at this concentration seemed not strong enough to trigger the ectosome release. To visualize the budding process and snip-off of ectosome release in the cells, we increased the concentration of colchicine to 30 µM. About a minute post-treatment, many vesicles appeared near the plasma membrane’s edge with increasing calcium signals in the structures (Figure 2, Video 2). All the vesicles with calcium signals occurred at almost the same time and had about the same size. A process including plasma membrane budding, extension, and shedding-off was observed in these cells (Figure 3, Video 3). The whole process lasted about 3 minutes. Vesicles with a more prominent area were found initially, and then the vesicles shrunk into a smaller one (Figure 2, arrows). It took about one minute for the process of shrinking (Figure 2, 58 to 120) and one minute (Figure 2, 120 to 200) for the vesicles shedding from the plasm membrane. The whole process happened the same as those described in other cell types (5).
Ectosomes squeezed out of the plasma membrane
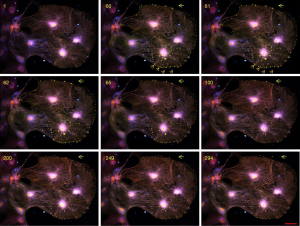
Primary cultured neural cells were mixed with astrocyte-like cells. Since different cell types may have their own way of ectosome release, we further recorded the dynamic changes of neurons or astrocytes based on their morphology. In astrocyte-like cells, a different way of release was observed. Soon after stimulating the cells with 30 µM colchicine, a series of ectosomes pumped out one by one from the same site of the plasma membrane rapidly (Figure 3, Video 4). This forming process was visualized in Video 4. Considering the membrane compensation in such a short time, we thought that it was unlikely to remove a part of the plasma membrane domain consecutively each time at the same position. Most of the membrane that covered the shedding ectosomes might not be derived from the plasma membrane but rather from the intracellular membrane system. To have more shreds of evidence to support this hypothesis, we continued to search for different ways of ectosome release in glia cells. As the outcome, another type of ectosomes that might derive from the endomembrane system was observed (Figure 4). A large number of ectosomes from the whole surface of a cell were released simultaneously after the stimulation of 30 µM colchincine. This type of ectosomes started from a small dot-like structure to a much bigger ball-like shape upon the releasing process (Figure 4, arrows). In the releasing process, ectosomes were squeezed out from the cell surface and became bigger and bigger, and eventually pinched off to release (Figure 4, arrows, and Video 5). The total line-up area of the shedding vesicles might be more extensive than that of the cell’s plasma membrane (Video 5). This process was completely different from the one described as the plasma membrane shedding (Figure 2 and Video 2). The vesicles were more prominent at the beginning and became smaller during release (Figure 3).
Ectosomes might be directly derived from intracellular vesicles
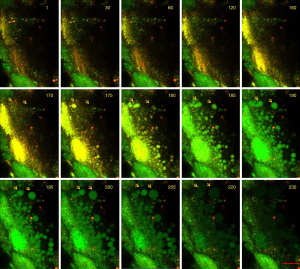
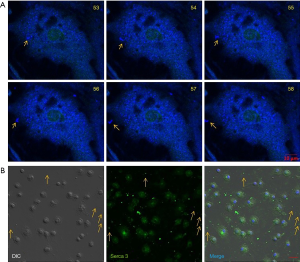
For getting a piece of directed evidence to show that ectosomes might be derived from intracellular vesicles, we labeled the cells with ER-tracker for live-cell imaging. Subcellular vesicles in the cells were very dynamic, even in the absence of extracellular stimulation. When we treated the cell with colchicines, a few of the vesicles labeled with ER-tracker jumped out of cells directly (Figure 5A). A similar phenomenon was also observed in the Fluo 4 labeled cells (Video 6). We further confirmed our findings by fixing the cells with 4% paraformaldehyde after colchicine treatment for ER marker staining. As shown in Figure 5B, Serca 3, a molecule expresses in ER was found both in the cells and in the ectosomes out of the cells. DIC image was used to show the cell boundaries. Some of the signals were located extracellularly.
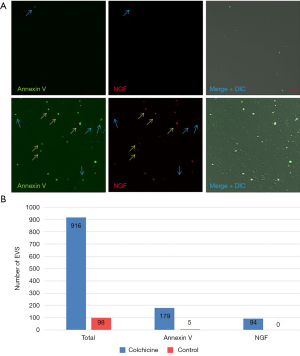
We further prepared ectosomes by collecting culture medium from the cells treated with or without colchicine and labeled them with FITC-conjugated annexin V to show whether the ectosomes have the surface expression of PS like intracellular vesicles. As indicated in Figure 6A, only a few ectosomes were labeled with annexinV (Figure 6A upper panel) when cells were not treated by colchicine. The number of Annexin V-positive ectosomes increased significantly after the treatment of colchicine. Some of the annexin-V ectosomes were NGF positive, which means that the ectosomes had a surface distribution of PS and contained NGF in the vesicle. Both of these signals shared the same characteristic as intracellular vesicles. The total number of ectosomes increased about ten times and annexin-V positive ectosomes increased more than 30 times after colchicine treatment (Figure 6B).
Discussion
The shedding of ectosomes is believed to result in removing small portions of the plasma membrane. To maintain the plasma membrane equilibrium during membrane turnover, cells are required to take measures for compensating the significant reduction of the cell surface by opposite changes in the area such as exosome release to regulate the membrane traffic (5) and to rework their plasma membrane area and surface tension for membrane strength to maintain a given size or shape (22). In some cells, the formation (Figure 1) and the release (Figure 2) of ectosomes appear to be separated into two processes: associated calcium increase was observed in the first step, and membrane shedding happened in the second step. The vesicles were much more prominent at the beginning and then became smaller. A shrinking of the structure into a small one may present a packing process during the biogenesis of ectosomes. The whole process takes minutes to complete. In this case, a small part of the plasma membrane with the cytoplasm components was lost from the cell as the ectosomes release.
In the case of a rapid release (Figure 4, Video 4), it takes less than one minute for the whole process of release. The removal of the membrane happened all at once in such a short time; the opposite process may not be fast enough for the membrane compensation, especially for the situation that ectosomes are generated from the whole surface of the cell at the same time (Figure 4). It appears that the vesicles squeeze from a “pore” on the plasma membrane and become bigger and bigger, keeping a rounded structure from the beginning to their release. The total area of ectosomes may be larger than the surface area of the cell. Therefore, it is not reasonable to say that the plasma membrane shedding generates this type of ectosomes. These observations resonate with a previous report that ectopic overexpression of Caveolin-1 protein in Sf21 insect cells drives the accumulation of hundreds of cytoplasmic vesicles with the same diameter (50–120 nm) as native caveolae seen in mammalian cells [Figure 3E,F of (23)]. The expression of either caveolin α- or β-isoform results in the intracellular accumulation of a homogeneous population of caveolae-sized vesicles with a diameter between 50 and 120 nm (80.3±14.8 nm), which was confirmed as approximately 50–100 nm membranous structures by whole-mount electron microscopy (23).
In another aspect, several reports have shown that the contents of ectosomes usually include IL-2 (18) and other cytokines (19). These molecules have the signal peptides in their N-terminal so that they may be left in the lumen of the endoplasmic reticulum (ER) after translation but not in the cytoplasm. Usually, they are secreted into the extracellular environment by exocytosis or in the lumen of vesicles within the cells. The extracellular vesicles that contain the same components as the vesicles in the cell gave us a hint that the source of ectosomes may be derived directly from intracellular vesicles. The intracellular vesicles maintain their integrity after getting out of the cell without fusion with the plasma membrane and without releasing their contents.
Furthermore, a negatively charged phospholipid PS is found almost exclusively on the plasma membrane inner leaflet in healthy cells. But many ectosomes that are recognized as shedding from the plasma membrane have a characteristic of surface expression of PS in host cell plasma membrane-derived vesicles as the immune mechanism against pathogen infection Trypanosoma cruzi (24). Although the surface expression of PS on ectosomes may relate to TAT-5 phospholipid flippase activity and lipid phosphatidylethanolamine and associated PI3Kinase VPS-34, Beclin1 homolog BEC-1, DnaJ protein RME-8, and the uncharacterized Dopey homolog PAD-1 (25), it cannot explain why these ectosomes have both the surface expression of PS and cytokines as their contents (24).
Intracellular vesicles are usually in a dynamic state. We can even observe the dynamic vesicle directly jump out of the cell for release (Figure 5A and Video 6). The finding of this type of vesicle release may mean that cytokines are also released in two different ways. One is secreted from cells by exocytosis, and the other is released in membrane-bounded vesicles. Whether the crucial cytokines and hormones like insulin have two forms of state in the bloodstream remain to be proved.
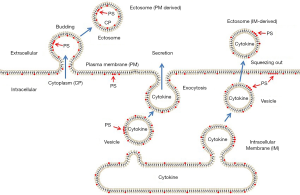
Our data support the hypothesis that intracellular membranes or vesicles should be at least part of the membrane source of these ectosomes. According to the source of membranes (Figure 7), ectosomes can be divided into plasma membrane-derived ectosomes and intracellular membrane-derived ectosomes. Plasma membrane-derived ectosomes are slow in the generation and release. This vesicle’s size was more prominent at the beginning and much smaller after release (Figure 2). Intracellular membrane-derived vesicles are fast in the process of generation and release. Surface expression of PS and cytokines as their contents may act as the markers for intracellular membrane-derived ectosomes. If the model is correct, colchicine treatment should increase the proportion of Annexin-V positive EVs, which biochemically isolated EVs might be isolated from the culture medium and quantified. Such evidence for the presence of extracellular vesicles maintaining an intracellular orientation might provide the transition phase of the process; however, it seems that the quantified algorithm of such dynamic process of EVs-traffic has not yet fruitfully matured for dynamic transitions; however, the imaging of phase transitions in living cells (26) might be relevant. In fact, some slow-motion video-imaging could show that the puncta observed “extracellularly” are indeed outside of the cells and not belonging to some protrusions. It is unclear why such releasing response is related to the sizing of EVs. Tracking patients’ EV-releasing-responses to therapeutics over time might shed light on this issue and has implications for predicting disease severity and adjusting the therapeutic dosage and drug delivery timing.
Our data nonetheless provide new insights into the process of ectosome release and a reasonable explanation of the different sources of ectosomes and their contents. These observations have implications for human diseases, diagnostics, and drug delivery (27). E.g., epithelial-mesenchymal transition (EMT) is closely associated with the development of drug resistance to a combination of simvastatin and paclitaxel for cancer, as simvastatin can disrupt lipid rafts (cholesterol-rich domains) and suppress integrin-β3 and focal adhesion formation, suggesting that inhibiting FAK signaling pathway re-sensitizes the drug-resistant cancer cells to paclitaxel (28). Simvastatin re-polarizes tumor-associated macrophages (TAM), promoting M2-to-M1 phenotype switch via lipid-rafts-cholesterol-associated LXR/ABCA1 regulation. The TAM-repolarization increases TNF-α but attenuates TGF-β, which, in turn, remodels the tumor microenvironment and suppresses EMT. Thus, such an engineer-designer EV formulation can enhance treatment efficacy. Upon that single-cell transcriptomes reveal the mechanism for a breast cancer prognostic gene panel (29), we can combine the GPS imaging systems (30) to track down subclonal migration and growth (31) with therapeutic EV delivery to control cancer subclonal progression (32) with subclone-specific therapeutics (33). These implications remain to be elucidated clinically.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant number 31371358, 2014) and by CHOC Children’s Foundation, CHOC-UCI Joint Research Awards (grant number 16004004, 2016).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-20-1015). SCL serves as an unpaid editorial board member of Quantitative Imaging in Medicine and Surgery. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional ethics board of the Shenzhen University, in compliance with the national guidelines for the care and use of animals established by the National Natural Science Foundation of China.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dudek KA, Dion-Albert L, Lebel M, LeClair K, Labrecque S, Tuck E, Ferrer Perez C, Golden SA, Tamminga C, Turecki G, Mechawar N, Russo SJ, Menard C. Molecular adaptations of the blood-brain barrier promote stress resilience vs. depression. Proc Natl Acad Sci U S A 2020;117:3326-36. [Crossref] [PubMed]
- Li S, Couet J, Lisanti MP. Src tyrosine kinases, Galpha subunits, and H-Ras share a common membrane-anchored scaffolding protein, caveolin. Caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. J Biol Chem 1996;271:29182-90. [Crossref] [PubMed]
- Lisanti M, Li S, Tang Z, Song K, Kubler E, Scherer P. Caveolin, an integral membrane protein of caveolae membranes in vivo: Implications for signal transduction. In: Limbird LE (ed). Alpha2 Adrenergic Receptors: Structure, function and therapeutic implications. New York: Harwood Academic Publishers, 1997:63-76.
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, Wiita AP, Debnath J. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 2020;22:187-99. [Crossref] [PubMed]
- Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 2015;25:364-72. [Crossref] [PubMed]
- Bruno S, Chiabotto G, Favaro E, Deregibus MC, Camussi G. Role of extracellular vesicles in stem cell biology. Am J Physiol Cell Physiol 2019;317:C303-13. [Crossref] [PubMed]
- Holme PA, Solum NO, Brosstad F, Røger M, Abdelnoor M. Demonstration of platelet-derived microvesicles in blood from patients with activated coagulation and fibrinolysis using a filtration technique and western blotting. Thromb Haemost 1994;72:666-71. [Crossref] [PubMed]
- van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28. [Crossref] [PubMed]
- Burger D, Schock S, Thompson CS, Montezano AC, Hakim AM, Touyz RM. Microparticles: biomarkers and beyond. Clin Sci (Lond) 2013;124:423-41. [Crossref] [PubMed]
- Atkin-Smith GK, Tixeira R, Paone S, Mathivanan S, Collins C, Liem M, Goodall KJ, Ravichandran KS, Hulett MD, Poon IK. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun 2015;6:7439. [Crossref] [PubMed]
- Wang J, Pendurthi UR, Rao LVM. Sphingomyelin encrypts tissue factor: ATP-induced activation of A-SMase leads to tissue factor decryption and microvesicle shedding. Blood Adv 2017;1:849-62. [Crossref] [PubMed]
- Wiedmer T, Sims PJ. Participation of protein kinases in complement C5b-9-induced shedding of platelet plasma membrane vesicles. Blood 1991;78:2880-6. [Crossref] [PubMed]
- Flaumenhaft R, Dilks JR, Richardson J, Alden E, Patel-Hett SR, Battinelli E, Klement GL, Sola-Visner M, Italiano JE Jr. Megakaryocyte-derived microparticles: direct visualization and distinction from platelet-derived microparticles. Blood 2009;113:1112-21. [Crossref] [PubMed]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 2000;100:221-8. [Crossref] [PubMed]
- Juan T, Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol 2018;74:66-77. [Crossref] [PubMed]
- Andrews AM, Rizzo V. Microparticle-Induced Activation of the Vascular Endothelium Requires Caveolin-1/Caveolae. PLoS One 2016;11:e0149272 [Crossref] [PubMed]
- Li S, Lisanti M, Puszkin S. Purification and molecular characterization of NP185, a neuronal-specific and synapse-enriched clathrin assembly polypeptide. Bioquim Patol Clin 1998;62:5-17. [PubMed]
- Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, Matteoli M, Verderio C. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J 2009;28:1043-54. [Crossref] [PubMed]
- Feigerlová E, Battaglia-Hsu SF, Hauet T, Guéant JL. Extracellular vesicles as immune mediators in response to kidney injury. Am J Physiol Renal Physiol 2018;314:F9-F21. [Crossref] [PubMed]
- Su Q, Cai Q, Gerwin C, Smith CL, Sheng ZH. Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat Cell Biol 2004;6:941-53. [Crossref] [PubMed]
- Fox JE, Austin CD, Boyles JK, Steffen PK. Role of the membrane skeleton in preventing the shedding of procoagulant-rich microvesicles from the platelet plasma membrane. J Cell Biol 1990;111:483-93. [Crossref] [PubMed]
- Morris CE, Homann U. Cell surface area regulation and membrane tension. J Membr Biol 2001;179:79-102. [Crossref] [PubMed]
- Li S, Song KS, Koh SS, Kikuchi A, Lisanti MP. Baculovirus-based expression of mammalian caveolin in Sf21 insect cells. A model system for the biochemical and morphological study of caveolae biogenesis. J Biol Chem 1996;271:28647-54. [Crossref] [PubMed]
- Cestari I, Ansa-Addo E, Deolindo P, Inal JM, Ramirez MI. Trypanosoma cruzi immune evasion mediated by host cell-derived microvesicles. J Immunol 2012;188:1942-52. [Crossref] [PubMed]
- Beer KB, Rivas-Castillo J, Kuhn K, Fazeli G, Karmann B, Nance JF, Stigloher C, Wehman AM. Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc Natl Acad Sci U S A 2018;115:E1127-36. [Crossref] [PubMed]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 2017;168:159-171.e14. [Crossref] [PubMed]
- Jia HR, Zhu YX, Xu KF, Liu X, Wu FG. Plasma membrane-anchorable photosensitizing nanomicelles for lipid raft-responsive and light-controllable intracellular drug delivery. J Control Release 2018;286:103-13. [Crossref] [PubMed]
- Jin H, He Y, Zhao P, Hu Y, Tao J, Chen J, Huang Y. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin β3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics 2019;9:265-78. [Crossref] [PubMed]
- Li SC, Stucky A, Chen X, Kabeer MH, Loudon WG, Plant AS, Torno L, Nangia CS, Cai J, Zhang G, Zhong JF. Single-cell transcriptomes reveal the mechanism for a breast cancer prognostic gene panel. Oncotarget 2018;9:33290-301. [Crossref] [PubMed]
- Miao Y, Chen Z, Li SC. Functional endoscopy techniques for tracking stem cell fate. Quant Imaging Med Surg 2019;9:510-20. [Crossref] [PubMed]
- Li SC, Tachiki LM, Luo J, Dethlefs BA, Chen Z, Loudon WG. A biological global positioning system: considerations for tracking stem cell behaviors in the whole body. Stem Cell Rev Rep 2010;6:317-33. [Crossref] [PubMed]
- Li SC, Lee KL, Luo J. Control dominating subclones for managing cancer progression and posttreatment recurrence by subclonal switchboard signal: implication for new therapies. Stem Cells Dev 2012;21:503-6. [Crossref] [PubMed]
- Lee LX, Li SC. Hunting down the dominating subclone of cancer stem cells as a potential new therapeutic target in multiple myeloma: An artificial intelligence perspective. World J Stem Cells 2020;12:706-20. [Crossref] [PubMed]


