
Artificial intelligence-based bone-enhanced magnetic resonance image—a computed tomography/magnetic resonance image composite image modality in nasopharyngeal carcinoma radiotherapy
Introduction
Southern China has one of the highest rates of nasopharyngeal carcinoma (NPC) in the world (1), for which radiotherapy is a necessary part of the main treatment. Due to the target volume’s proximity to critical structures such as the brainstem, spinal cord, cochlea, mandible, optical nerves, chiasm and parotids, the setting of the radiation field is the source of most locoregional complications and sequelae. The treatment outcomes of radiotherapy in NPC are therefore highly dependent on accurate delineation of the tumor and organs at risk (OARs) which is used to develop optimal treatment plans that conform to the prescribed dose to the target volume and spare the uninvolved critical structures. However, no single modality is capable of an adequate definition of tumor volumes because of its inherent imaging limitations. For example, in computed tomography (CT), tumor invasion of the bone can be clearly seen, but tumor extension into the surrounding soft tissue may not be visible. Emami et al. (2) have shown that the treatment outcome for Stage III and IV NPC with conventional radiotherapy alone is generally poor. Meanwhile, it has become increasingly clear that using multi-imaging modalities and combined chemoradiotherapy has resulted in significant improvements in treatment outcomes for NPC patients.
Many studies have demonstrated that magnetic resonance imaging (MRI) is superior to CT in the staging and follow-up of NPC patients (3-7). MRI provides better tumor definition, especially for NPC and leads to different target definitions than CT. Applying MRI to radiotherapy has great benefits to improve radiation dosimetry by reducing toxicity to OARs and enabling dose escalation to tumor targets to achieve survival gains (8-10). Thus, treatment planning workflow that uses magnetic resonance image-guided radiotherapy (MRIgRT) have already been proposed (11-13). However, MRI lacks bone structure information and cannot be directly utilized for NPC localization. Therefore, MRIgRT is supplemented by CT which provides reliable surrogates for electron density information for dose distribution calculation (14). The usual practice is the conversion of electron density or HU values into synthetic CT (sCT) based on MRI images. To our knowledge, until now, there is no single imaging modality which shows bone structure with the contrast of CT and soft tissue with the contrast of MRI simultaneously.
To address these problems, many kinds of experiments have explored synthesizing CT images from MRI images. These include atlas-based methods (15-17), segmentation-based methods (18-21), voxel-based methods (22,23), and deep learning methods (24-29). The atlas-based method of producing sCT images requires CT-to-MRI registration in which CT and MRI atlas scan pairs correspond anatomically. This method is time consuming, and the atlas of new cases needs to be recalculated. The segmentation-based methods of producing sCT images are done according to the intensity of MRI voxels. This method is simple and saves time. But the accuracy of dose calculation is seriously affected by the accuracy of the segmentation. The voxel-based method focuses on using voxel by voxel mapping, based on the intensity or spatial location of the MRI image, acquired from different MRI pulse sequences. This method is more accurate than the above methods but is more time and labor intensive. Deep learning methods exhibit a superior ability to learn a nonlinear mapping from one image domain to another image domain. Several convolution neural networks have achieved better sCT than the other three traditional methods (25). According to whether the input data sets contain labels, deep learning methods can be separated into two categories, supervised and unsupervised deep learning methods. We investigated the literature and found that most supervised deep learning methods were completed based on a convolutional neural network (CNN) such as U-Net. Among them, Han (24) used a CNN to establish the prediction model of sCT for the first time and realized the end-to-end transformation from MRI to sCT. Most unsupervised learning is based on GAN (mostly CycleGAN). Wolterink et al. (29) used unpaired brain data to train the unsupervised prediction model for transformation from MRI to sCT.
The above studies have shown how to generate higher quality cross-modalities images by deep learning methods, but not how to make use of the image features of different image modalities simultaneously. Image fusion allows one to use both CT and MRI information at the same time. Numerous techniques have been proposed in the past decades to deal with medical image fusion. These include average density or pixel-by-pixel selection methods (30-32), wavelet analysis methods (33), weighted averaging to complex multiresolution pyramid method (34), and neural network method (35,36). Previously, synthetic images of single modalities, such as sCT, have been used for radiotherapy, but the fusion of synthetic multimodality images for radiotherapy has not been fully explored. This is a novel concept that opens up a new avenue for the applications of deep learning in MRIgRT.
This paper describes the use of head and neck MRI images to derive sCT images and a method to synthesize a multi-modal image by considering the imaging characteristics between the two modalities. The remaining part of this paper is organized as follows: in Methods, we describe the details of experimental methods; in Results, we evaluate the resulting new modality medical images; in Discussion, the clinical value of the new modal image is discussed in detail; finally, a brief conclusion is given in Conclusions.
Methods
To obtain bone structure information from MRI, we first choose a supervised neural network [U-Net (37)] and an unsupervised generative adversarial network [CycleGAN (38)] to generate the sCT. We then extract the bone structure information and fuse it with the MRI image. The experiment includes four steps as Figure 1: (I) data preprocessing; (II) sCT generation; (III) BeMRI composition; (IV) evaluation.
The study was approved by the Human Research Ethics Committee of the Shenzhen Second People’s Hospital and informed consent was taken from all the patients.
Data preprocessing
Thirty-five patients were retrospectively selected for this study. All patients underwent clinical CT simulation and 1.5 T MRI within the same week at the Shenzhen Second People’s Hospital. The CT images were acquired under the following conditions: a 120 kV tube voltage, a 330 mA current, a 500 ms exposure time, a 0.5×0.5 mm2 in-plane resolution, a 1-mm slice thickness, a 512×512 image size on the SOMATOM Definition Flash (Siemens). T2-weighted MRI images were acquired under the following conditions: a 2,500 ms repetition time, a 123 ms echo time, a 1×1×1 mm3 pixel volume, a 256×256 image size on a 1.5T Avanto scanner (Siemens). The MRI distortion correction was applied by the MRI data acquisition system.
To remove the unnecessary background, we first generated binary head masks through the Otsu threshold method (39). We then used the binary masks with the “AND operation” (i.e., elementwise summation) to exclude the unnecessary background and keep the interior brain structure. We resampled CT images of size 512×512 to 256×256 to match MRI images by bicubic interpolation (40). To align the CT image with the corresponding MRI images, we took the CT as a fixed image. The MRI images were registered to CT ones by rigid registration using the Elastix toolbox (41), and mutual information was taken as the cost function. We randomly selected 28 patients to train neural networks. The other 7 patients were used as test data. On average, each patient’s CT or MRI contains more than 100 2D axial image slices. We rescaled the MRI and CT values of the acquired images to [0, 255], converted this data into a [0, 1] tensor, and normalized each image to [−1, 1]. Instead of image patches, whole 2D images were used to train all models. The axial CT and MRI pairs were put into networks with a size of 256×256 pixels.
sCT generation
U-Net
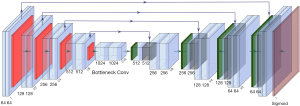
U-Net has achieved great success in medical image processed tasks (24,42). The structure used here is illustrated in Figure 2. It is an end-to-end supervised convolutional neural network, which requires pairs of images as inputs. The network includes an encoder and a decoder (37). The encoder consists of repeated 3*3 convolutions, each with a Batch-Normalization layer (43) and a LeakyReLu layer (44) without input convolution. The max-pooling operation with stride two is used for downsampling. Feature channels are doubled at each downsampling step. The size of the input image was 256×256. The decoder consists of repeated 3×3 convolutions and 4×4 ConvTranspose layers (45), each with a ReLu (46) layer followed by a Batch-Normalization layer. Skip connections are designed to concatenate channels from the encoder to the decoder to use as much information from earlier layers as possible. At the final layer, a sigmoid activation operation is used to map sCT. The output image is kept the same size as the input image, whose size is also 256×256.
CycleGAN
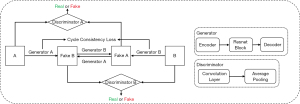
As an unsupervised neural network, CycleGAN can convert the styles of two unregistered images. It alleviates the problem posed by a lack of paired datasets since there is a vast amount of unpaired image data. The structure used here is illustrated in Figure 3. Different from the original GAN, CycleGAN has two generators. The first one was called ‘Generator A’ for transferring input ‘A’ to input ‘B’, and the second one was called ‘Generator B’ for transferring input ‘B’ to input ‘A’. The outputs are called FakeB and FakeA. CycleGAN also has two discriminators, which were designed to identify whether a random input is real training data or fake data synthesized by a generator. The cycle consistency of CycleGAN holds that if input A can be converted to input B through a mapping, then there is also another mapping that converts input B to input A. CycleGAN learns the losses of pair sets of generators and discriminators to improve the quality of the synthetic images and the robustness of the network simultaneously. More details of CycleGAN are given in (38). In this work, we input brain MRI labeled ‘A’ and corresponding brain CT labeled ‘B’.
To compare the quality of the generated image from different networks properly, the parameter settings of the U-Net and CycleGAN model are unified. We set the batch size as one and used Adam (47) to optimize both the generator and discriminator. Four hundred epochs were trained for each model. A fixed learning rate of 2.0×10−4 was applied for the first 200 epochs and linearly reduced to 0 during the training of the remaining 200 epochs. The number of iterations was 1.62×106. For the U-Net model, the paired MRI and CT data are used as input data according to a one-to-one correspondence. For the CycleGAN model, the MRI and CT image data are input randomly.
BeMRI composition
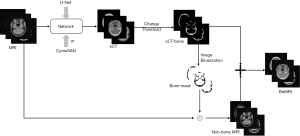
The BeMRI composition based on the sCT is illustrated in Figure 4. First, the ambiguous bone structure in MRI images was extracted from the sCT images. To accurately extract the bone structure information, we used the rescale intercept value and rescale slope value, which was obtained from the ground truth CT header file, to separate the bone and soft tissue according to Eq. [1]. Rescale intercept and rescale slope are DICOM tags that specify the linear transformation from pixels in their stored-on-disk representation to their in-memory representation. In this study, the threshold of 300 Hounsfield Unit (HU) was used to separate the bone and soft tissue. From the CT header file, we knew the Rescale Intercept was −1,024 and the Rescale Slop was 1. We thus chose 1,324 as the pixel value threshold according to Eq. [2] to separate bone and soft tissue. The bone structure extracted from sCT was used to generate sCT-bone masks through image binarization. We then used the bone masks and the “AND operation” to exclude unnecessary bone structure while saving soft tissue in MRI images. Finally, we combined the bone structure extracted from MRI of the eliminated bone structure to generate BeMRI.
We implemented these networks in Pytorch and used a single NVIDIA TITAN X(Pascal) (12GB) GPU for all of the training and testing experiments.
Evaluation
After checking all the BeMRI, the layers without bone structure were deleted, and the remaining results were evaluated. Since the soft tissue area does not change, we only evaluated the results of bone structure. Three metrics were used to compare the ground truth bone and composite bone: MAE according to Eq. [3], PSNR according to Eq. [4] and SSIM according to Eq. [5]. These metrics are widely used in medical image evaluation (28,48,49), and are expressed as follows:
where N is the total number of pixels inside the bone region, i is the index of the aligned pixels inside the bone area. MAX denotes the largest pixel value of each ground truth bone and composite bone images.
SSIM (50) is a metric that can be used to quantify the similarities on the whole image scale, and can be expressed as follows:
where μx and μy are the average pixel values of the bone structure from ground truth CT and sCT, respectively. σx2 and σy2 are the variance of these pixel values for the ground truth CT and sCT, respectively. σxy is the covariance of bone structure from ground truth CT and sCT. c1= (k1L)2, c2= (k2L)2 are two variables used to stabilize the division by the small denominator and L is the dynamic range of the pixel values. k1=0.01, k2=0.03 are set by default.
Results
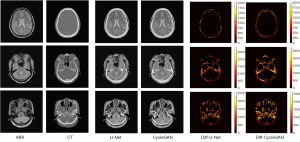
In our experiments, the clinical MRI of 7 patients was acquired to test the proposed models. The details of the BeMRI images for a single patient are shown in Figure 5. The different columns represent different slices in the same image modality, and different rows represent different modalities. Specifically, ground truth CT image, ground truth MRI image, the synthetic BeMRI image from U-Net and the synthetic BeMRI image from CycleGAN are illustrated from top to bottom. To show the boundary of the synthetic bone structure and soft tissue more clearly, we use a red rectangle in each result to magnify the region. It can be seen qualitatively that MRI is more effective in imaging soft tissue, but less effective in imaging bone structure, while the opposite is true for CT. The third column shows that at the boundary between soft tissue and bone structure, MRI cannot sharply identify the boundary while CT can hardly detect the corresponding soft tissue. However, BeMRI can clearly distinguish between soft tissue and bone structure and identify the boundary between them.
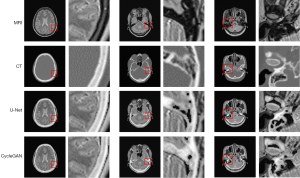
Although it is difficult to distinguish the difference between the BeMRI images generated by U-Net and CycleGAN with the naked eye in Figure 5, it can be seen clearly in the difference map of the normalized pixel value shown in Figure 6. The color bar of the difference map used a gradual color from red to yellow to indicate the difference in pixel value between BeMRI bone structure and ground truth CT. On the whole, the color of the difference map for the result of CycleGAN was lighter than that of U-Net, which indicates the images synthesize by the former are on average noisier than those generated by the latter.
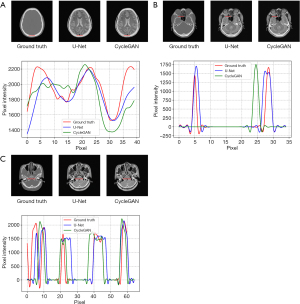
A comparison of 1D profiles passing through three different lines, which were acquired from images generated by the U-Net and CycleGAN models is shown in Figure 7. Since the soft tissue area does not change, we only evaluated the results of the bone structure. Based on these images, the bone structure extracted from sCT by the U-Net model agreed well with the ground truth CT on a pixel-by-pixel scale. For Figure 7A, we chose the parietal part at the same position to evaluate the pixel intensity. Comparing the two deep learning methods with the ground truth, we find they generate similar pixel intensity distributions to the ground truth, which indicated that the U-Net and CycleGAN models can generate reasonable skull structure information for BeMRI. However, as shown in Figure 7B,7C, some details may also be inaccurate compared with the ground truth. In Figure 7B, we found that the details in the ethmoid sinus region were accurately generated using U-Net but inaccurately generated using CycleGAN. Nonetheless, some regions have spurious structures generated by using CycleGAN, which is probably due to its unsupervised learning style. In Figure 7C, we chose a longer horizontal line in the basilar clivus to compare more pixel details of these two methods. It was demonstrated that on a large pixel scale, the image generated by the U-Net model was much better matched with the ground truth image compared to the image generated by the CycleGAN model.
Statistics of the three quantitative metrics over the whole brain image are given in Table 1. The results showed that the U-Net model had smaller MAE and higher SSIM values compared with the CycleGAN model. Otherwise, the P value of MAE between U-Net and CycleGAN is less than 0.05, which means that U-Net model were more accurate than the CycleGAN model used in our experiment.
Table 1
| Patient | MAE ± SD | SSIM ± SD | PSNR ± SD (dB) | |||||
|---|---|---|---|---|---|---|---|---|
| U-Net | CycleGAN | U-Net | CycleGAN | U-Net | CycleGAN | |||
| 1 | 112.78±22.04 | 123.25±29.34 | 0.93±0.06 | 0.89±0.10 | 24.65±1.97 | 22.97±2.11 | ||
| 2 | 125.43±26.79 | 132.86±31.63 | 0.89±0.10 | 0.87±0.10 | 24.15±1.71 | 22.70±1.73 | ||
| 3 | 131.22±28.99 | 135.27±35.18 | 0.86±0.09 | 0.82±0.12 | 23.61±1.90 | 22.30±1.89 | ||
| 4 | 128.29±27.89 | 131.52±35.47 | 0.88±0.09 | 0.83±0.11 | 23.67±1.80 | 22.34±1.87 | ||
| 5 | 127.70±28.70 | 130.01±37.48 | 0.89±0.07 | 0.83±0.11 | 23.60±1.85 | 22.27±1.98 | ||
| 6 | 126.21±29.31 | 130.37±36.67 | 0.93±0.05 | 0.89±0.08 | 23.67±1.93 | 22.29±1.94 | ||
| 7 | 127.21±31.90 | 129.98±40.04 | 0.86±0.08 | 0.84±0.09 | 23.55±2.04 | 22.30±2.09 | ||
| Average | 125.55±5.48 | 130.47±3.43 | 0.89±0.027 | 0.85±0.028 | 23.84±0.38 | 22.45±0.25 | ||
MAE, mean absolute error; SSIM, structural similarity index; PSNR, peak signal-to-noise ratio; SD, standard deviation.
Discussion
In this study, we proposed a new image modality: a multi-modality composite image, which contains both high contrast soft tissue information and high contrast bone structure. BeMRI experiments do not only simply fuse the images with MRI images and sCT images. Instead, a deep learning method is first used to transform an MRI to into an sCT one which is then embedded in the bone structure part of an MRI image. In this way we are able to make up for the shortcomings of single-modality medical images and acquire multimodal medical image information simultaneously in one image.
In the synthesis of a BeMRI image, supervised and unsupervised deep learning models were compared. Qualitative and quantitative comparison show that U-Net achieved the best results, with the lowest overall average MAE and the highest SSIM. The quality of the synthetic image in some regions and ROI extraction needs to be further studied and improved. First, the standard MRI is mainly aimed at soft tissue imaging, so that errors in the bone structure image can still be propagated into the sCT and BeMRI. Second, we only use the method of threshold segmentation to extract bone structure. The intensity of the synthetic image generated through the deep learning network may not accurately correspond to the CT value. Due to the existence of air in the bone, the intensity of the adjacent points is discontinuous, and the threshold method cannot accurately extract bone structure. Many studies have shown that deep learning can significantly improve the performance of bone segmentation (51,52). Third, we only chose the classic deep learning algorithms without any improvement to achieve the task. U-Net was the first design for biomedical image segmentation, and CycleGAN was first used to synthesize a natural image. However, the quality of the synthetic image can certainly be improved by selecting different networks and algorithms for datasets, such as a recurrent neural network (RNN), attention model (AM), deep residual network (DRN), long/short term memory (LSTM), graph network (GN).
Despite the above drawbacks, we consider that BeMRI has great potential in MRIgRT. Thornton et al. (53) have shown, MRI more accurately segments brain tumors than CT. Meanwhile, Wang et al. (28) has shown that the planning dose based on sCT obtained from MRI is similar to that based on true CT in patients with NPC. A treatment planning that uses the BeMRI workflow in Figure 4 should thus be feasible. There are several clinical benefits of BeMRI in radiotherapy: (I) BeMRI combines the advantages of MRI images with high soft tissue contrast and CT images with high bone contrast; (II) MRI can provide functional evaluation of the treatment outcome of radiotherapy; (III) Through the CT part in BeMRI, the Digitally Reconstructed Radiograph can be generated and used for tumor localization in radiotherapy. In addition, such systematic errors as MRI-CT co-registration which may introduce geometrical uncertainties of ~2 mm for the brain and neck region (11), random errors will also be introduced if the doctors have to alternate between observing two independent image modes. By combining the information from the latter in one image mode, BeMRI thus makes it easier to accurately detect and locate tumors during NPC radiotherapy and minimizes the random error.
Conclusions
In this work, we proposed a new medical image modality-CT/MRI composite, BeMRI, by using deep learning networks to derive sCT images from MRI images and fuse them to form what we call BeMRI images. Prospects for future work include using more advanced deep learning networks and real clinical data with tumor lesions. This work is expected to find useful applications in NPC MRIgRT.
Acknowledgments
Funding: This work is supported in part by grants from the National Key Research and Develop Program of China (2016YFC0105102), the Leading Talent of Special Support Project in Guangdong (2016TX03R139), Shenzhen matching project (GJHS20170314155751703), the Science Foundation of Guangdong (2020B1111140001, 2017B020229002, 2015B020233011), CAS Key Laboratory of Health Informatics, Shenzhen Institutes of Advanced Technology, the National Natural Science Foundation of China (U20A20373, 61871374, 61901463), the Sanming Project of Shenzhen (SZSM201612041), and the Shenzhen Science and Technology Program of China grant (JCYJ20200109115420720).
Footnote
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-20-1239). The special issue “Artificial Intelligence for Image-guided Radiation Therapy” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Human Research Ethics Committee of the Shenzhen Second People’s Hospital and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol 2002;13:1007-15. [Crossref] [PubMed]
- Emami B, Sethi A, Petruzzelli GJ. Influence of MRI on target volume delineation and IMRT planning in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 2003;57:481-8. [Crossref] [PubMed]
- Ng SH, Chong VF, Ko SF, Mukherji SK. Magnetic resonance imaging of nasopharyngeal carcinoma. Top Magn Reson Imaging 1999;10:290-303. [Crossref] [PubMed]
- Nishioka T, Shirato H, Kagei K, Abe S, Hashimoto S, Ohmori K, Yamazaki A, Fukuda S, Miyasaka K. Skull-base invasion of nasopharyngeal carcinoma: magnetic resonance imaging findings and therapeutic implications. Int J Radiat Oncol Biol Phys 2000;47:395-400. [Crossref] [PubMed]
- Sakata K, Hareyama M, Tamakawa M, Oouchi A, Sido M, Nagakura H, Akiba H, Koito K, Himi T, Asakura K. Prognostic factors of nasopharynx tumors investigated by MR imaging and the value of MR imaging in the newly published TNM staging. Int J Radiat Oncol Biol Phys 1999;43:273-8. [Crossref] [PubMed]
- Olmi P, Fallai C, Colagrande S, Giannardi G. Staging and follow-up of nasopharyngeal carcinoma: magnetic resonance imaging versus computerized tomography. Int J Radiat Oncol Biol Phys 1995;32:795-800. [Crossref] [PubMed]
- Curran WJ, Hackney DB, Blitzer PH, Bilaniuk L. The value of magnetic resonance imaging in treatment planning of nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 1986;12:2189-96. [Crossref] [PubMed]
- Farjam R, Tyagi N, Deasy JO, Hunt MA. Dosimetric evaluation of an atlas-based synthetic CT generation approach for MR-only radiotherapy of pelvis anatomy. J Appl Clin Med Phys 2019;20:101-9. [Crossref] [PubMed]
- Korhonen J, Kapanen M, Keyriläinen J, Seppälä T, Tenhunen M. A dual model HU conversion from MRI intensity values within and outside of bone segment for MRI-based radiotherapy treatment planning of prostate cancer. Med Phys 2014;41:011704 [Crossref] [PubMed]
- Xiang L, Wang Q, Nie D, Zhang L, Jin X, Qiao Y, Shen D. Deep embedding convolutional neural network for synthesizing CT image from T1-Weighted MR image. Med Image Anal 2018;47:31-44. [Crossref] [PubMed]
- Nyholm T, Nyberg M, Karlsson MG, Karlsson M. Systematisation of spatial uncertainties for comparison between a MR and a CT-based radiotherapy workflow for prostate treatments. Radiat Oncol 2009;4:54. [Crossref] [PubMed]
- Ulin K, Urie MM, Cherlow JM. Results of a multi-institutional benchmark test for cranial CT/MR image registration. Int J Radiat Oncol Biol Phys 2010;77:1584-9. [Crossref] [PubMed]
- Nakazawa H, Mori Y, Komori M, Shibamoto Y, Tsugawa T, Kobayashi T, Hashizume C. Validation of accuracy in image co-registration with computed tomography and magnetic resonance imaging in Gamma Knife radiosurgery. J Radiat Res 2014;55:924-33. [Crossref] [PubMed]
- McGee KP, Hu Y, Tryggestad E, Brinkmann D, Witte B, Welker K, Panda A, Haddock M, Bernstein MA. MRI in radiation oncology: Underserved needs. Magn Reson Med 2016;75:11-4. [Crossref] [PubMed]
- Kops ER, Herzog H. Alternative methods for attenuation correction for PET images in MR-PET scanners. IEEE Nucl Sci Symp Conf Rec 2007;6:4327-30.
- Hofmann M, Steinke F, Scheel V, Charpiat G, Farquhar J, Aschoff P, Brady M, Schölkopf B, Pichler BJ. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. J Nucl Med 2008;49:1875-83. [Crossref] [PubMed]
- Dowling JA, Lambert J, Parker J, Salvado O, Fripp J, Capp A, Wratten C, Denham JW, Greer PB. An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int J Radiat Oncol Biol Phys 2012;83:e5-11. [Crossref] [PubMed]
- Zaidi H, Montandon ML, Slosman DO. Magnetic resonance imaging‐guided attenuation and scatter corrections in three‐dimensional brain positron emission tomography. Med Phys 2003;30:937-48. [Crossref] [PubMed]
- Lee YK, Bollet M, Charles-Edwards G, Flower MA, Leach MO, McNair H, Moore E, Rowbottom C, Webb S. Radiotherapy treatment planning of prostate cancer using magnetic resonance imaging alone. Radiother Oncol 2003;66:203-16. [Crossref] [PubMed]
- Berker Y, Franke J, Salomon A, Palmowski M, Donker HC, Temur Y, Mottaghy FM, Kuhl C, Izquierdo-Garcia D, Fayad ZA, Kiessling F, Schulz V. MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echotime/Dixon MRI sequence. J Nucl Med 2012;53:796-804. [Crossref] [PubMed]
- Hsu SH, Cao Y, Huang K, Feng M, Balter JM. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Phys Med Biol 2013;58:8419. [Crossref] [PubMed]
- Edmund JM, Kjer HM, Van Leemput K, Hansen RH, Andersen JA, Andreasen D. A voxel-based investigation for MRI-only radiotherapy of the brain using ultra short echo times. Phys Med Biol 2014;59:7501-19. [Crossref] [PubMed]
- Kim J, Glide-Hurst C, Doemer A, Wen N, Movsas B, Chetty IJ. Implementation of a novel algorithm for generating synthetic CT images from magnetic resonance imaging data sets for prostate cancer radiation therapy. Int J Radiat Oncol Biol Phys 2015;91:39-47. [Crossref] [PubMed]
- Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. [Crossref] [PubMed]
- Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, Shen D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. Med Image Comput Comput Assist Interv 2017;10435:417-25.
- Jin CB, Kim H, Liu M, Jung W, Joo S, Park E, Ahn YS, Han IH, Lee JI, Cui X. Deep CT to MR Synthesis Using Paired and Unpaired Data. Sensors (Basel) 2019;19:2361. [Crossref] [PubMed]
- Dinkla AM, Wolterink JM, Maspero M, Savenije MHF, Verhoeff JJC, Seravalli E, Išgum I, Seevinck PR, van den Berg CAT. MR-Only Brain Radiation Therapy: Dosimetric Evaluation of Synthetic CTs Generated by a Dilated Convolutional Neural Network. Int J Radiat Oncol Biol Phys 2018;102:801-12. [Crossref] [PubMed]
- Wang Y, Liu C, Zhang X, Deng W, Synthetic CT. Generation Based on T2 Weighted MRI of Nasopharyngeal Carcinoma (NPC) Using a Deep Convolutional Neural Network (DCNN). Front Oncol 2019;9:1333. [Crossref] [PubMed]
- Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT Synthesis Using Unpaired Data. In: Tsaftaris S, Gooya A, Frangi A, Prince J. editors. Simulation and Synthesis in Medical Imaging. SASHIMI 2017. Lecture Notes in Computer Science, vol 10557. Springer, Cham 2017:14-23.
- Aguilar M, Garrett A L. Biologically based sensor fusion for medical imaging. Proc SPIE Int Soc Opt Eng 2001;4385:149-58.
- Jiang H, Tian Y. Fuzzy image fusion based on modified Self-Generating Neural Network. Expert Syst Appl 2011;38:8515-23. [Crossref]
- Yang B, Li S. Pixel-level image fusion with simultaneous orthogonal matching pursuit. Information Fusion 2012;13:10-9. [Crossref]
- Seal A, Bhattacharjee D, Nasipuri M, Rodríguez-Esparragón D, Menasalvas E, Gonzalo-Martin C. PET-CT image fusion using random forest and à-trous wavelet transform. Int J Numer Method Biomed Eng 2018; [Crossref] [PubMed]
- Liu Z, Tsukada K, Hanasaki K, Ho YK. Image fusion by using steerable pyramid. Pattern Recognit Lett 2001;22:929-39. [Crossref]
- He C, Lang F, Li H. Medical Image Registration using Cascaded Pulse Coupled Neural Networks. Information Technology Journal 2011;10:1733-9. [Crossref]
- Iscan Z, Yüksel A, Dokur Z, Korürek M, Ölmez T. Medical image segmentation with transform and moment based features and incremental supervised neural network. Digital Signal Processing 2009;19:890-901. [Crossref]
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Med Image Comput Comput Assist Interv 2015:234-41.
- Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. Proceedings of the IEEE International Conference on Computer Vision 2017:2223-32.
- Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979;9:62-6. [Crossref]
- Prashanth HS, Shashidhara HL, KN BM. Image Scaling Comparison Using Universal Image Quality Index. International Conference on Advances in Computing, Control, and Telecommunication Technologies 2009:859-63.
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Lin G, Liu F, Milan A, Shen C, Reid I. RefineNet: Multi-Path Refinement Networks for Dense Prediction. IEEE Trans Pattern Anal Mach Intell 2020;42:1228-42. [PubMed]
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015.
- Xu B, Wang N, Chen T, Li M. Empirical evaluation of rectified activations in convolutional network. arXiv preprint arXiv:1505.00853, 2015.
- Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434, 2015.
- Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th International Conference on Machine Learning 2010:807-14
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- Zhao C, Carass A, Lee J, Jog A, Prince JL. A supervoxel based random forest synthesis framework for bidirectional MR/CT synthesis. Simul Synth Med Imaging 2017;10557:33-40. [Crossref] [PubMed]
- Ben-Cohen A, Klang E, Raskin SP, Soffer S, Ben-Haim S, Konen E, Amitai MM, Greenspan H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng Appl Artif Intell 2019;78:186-94. [Crossref]
- Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13:600-12. [Crossref] [PubMed]
- Noguchi S, Nishio M, Yakami M, Nakagomi K, Togashi K. Bone segmentation on whole-body CT using convolutional neural network with novel data augmentation techniques. Comput Biol Med 2020;121:103767 [Crossref] [PubMed]
- Zhang J, Liu M, Wang L, Chen S, Yuan P, Li J, Shen SG, Tang Z, Chen KC, Xia JJ, Shen D. Context-guided fully convolutional networks for joint craniomaxillofacial bone segmentation and landmark digitization. Med Image Anal 2020;60:101621 [Crossref] [PubMed]
- Thornton AF Jr, Sandler HM, Ten Haken RK, McShan DL, Fraass BA, La Vigne ML, Yanke BR. The clinical utility of magnetic resonance imaging in 3-dimensional treatment planning of brain neoplasms. Int J Radiat Oncol Biol Phys 1992;24:767-75. [Crossref] [PubMed]



