
THAN: task-driven hierarchical attention network for the diagnosis of mild cognitive impairment and Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is an irreversible and chronic neurodegenerative disease. It has been a leading cause of disability for the elderly over 65 years old (1). It has been reported that more than 33 million people worldwide were suffering from AD in 2018. The number is predicted to increase to 100 million by 2050. The cost of the disease was about 666 billion US dollars in 2018 and it is forecasted to double by 2030 (2). Mild cognitive impairment (MCI) is a prodromal stage of AD. Studies have shown that 20% of patients with MCI could deteriorate into AD within 4 years (3). Although there is no effective way to block the progression of MCI and AD (denoted as MCI/AD), some treatments have been developed to delay their progression. Therefore, it is becoming increasingly essential for the scientific community to develop effective methods to diagnose MCI/AD as early and accurately as possible.
Structural magnetic resonance imaging (sMRI) images are more useful for the early diagnosis of MCI/AD compared with the clinical assessment of cognitive impairment. This is because brain changes induced by AD have been proven to occur 10–15 years before symptom onset (4), and these brain changes can be non-invasively captured by sMRI images (5). Currently, sMRI images are extensively employed for computer-aided MCI/AD diagnosis based on machine learning methods (6-23). Among these methods, the convolutional neural network (CNN)-based methods (9-23) have demonstrated outstanding performance due to their excellent ability to extract task-driven features.
Existing CNN-based MCI/AD classification methods can be divided into four categories. They are regions-of-interest (ROI)-based methods, whole image-based methods, patch-based methods, and attention-based methods. The ROI-based methods first pre-segment disease-related regions according to the domain knowledge of AD experts. After that, different CNNs are designed to extract features from these regions and make final classification (9-11). However, the pre-segmented disease-related regions vary based on different experts and generally cannot cover all the lesion regions. Moreover, these methods require complex pre-processing steps. The whole image-based methods extract features from entire sMRI images directly (12). They require no expert knowledge and fully exploit the global features of an image. However, this type of method cannot accurately extract disease-related features, because the volume of each sMRI image is large while the lesion regions are small, leading to inaccurate classification. Overall, the ROI-based and whole image-based methods cannot automatically extract features from disease-related regions. The patch-based and attention-based methods relieve such a limitation to some extent.
The patch-based methods automatically select disease-related image patches first. After that, they extract features from these image patches and fuse these features to classify patients from normal controls (NCs). For example, image patches were first selected based on distinct anatomical landmarks in a data-driven manner (13-17). After that, the labels of image patches were assigned to the labels of their corresponding sMRI images. Further, in (13), multi-CNNs were trained for feature extraction and classification of these patches, and the majority voting strategy was used for whole image classification. In (14), an improved deep multi-instance learning network was designed to integrate patch-level features for final sMRI image classification. This network avoids the usage of patch-level labels. In (15,16), Liu et al. extended the deep multi-instance learning network on multi-modality data to improve the diagnosis of AD/MCI. Additionally, Lian et al. (17) further strengthened the correlation information of image patches by designing a hierarchical fully convolutional network (FCN) for MCI/AD diagnosis. Moreover, Qiu et al. (18) trained an FCN model to generate a participant-specific disease probability map based on randomly selected 3,000 image patches. They then selected high-risk voxels from the disease probability map, an implicit patch-selection strategy. Further, they trained a multilayer perceptron for AD diagnosis. These patch-based methods do not require expert knowledge. Moreover, they can better extract the visual features [i.e., low-level features such as luminance and edge (19), which are extracted from the shallow layers of CNNs] and semantic features [i.e., high-level features like objects (19), which are extracted from the deep layers of CNNs] of the disease-related image patches for classification. However, most of the methods completely neglect the remaining image patches, leading to dismissing of the global features of sMRI images.
The attention-based methods generate attention maps with different weights. By combining an attention map with the feature map from a certain layer, some features can be emphasized and meanwhile the other features are not neglected, either. Jin et al. (20) designed a weighted attention block inserted in 3D ResNet (24) to improve the ability in extracting features of disease-related regions. The weighted attention block was employed on a middle layer of 3D ResNet and the generation of the attention map has no direct constraint. Li et al. (21) proposed an iterative attention focusing strategy for the localization of pathological regions and for the classification between progressive and stable MCI. The iterative attention focusing strategy was utilized on a deep layer of each iterative sub-network. In addition, Lian et al. (22) constructed a dementia attention block to automatically identify subject-specific discriminative locations from whole sMRI images. This attention block was used on a deep layer of the network. Furthermore, Zhang et al. (23) developed a deep cross-modal attention network, which focused on learning the “deep relations” among different brain regions from diffusion tensor imaging and resting-state functional MRI. Overall, the attention-based methods require no expert knowledge, either. However, most attention maps are solely employed on the deep layers of networks for semantic feature extraction. This results in the visual features of disease-related regions not being well extracted and further affects the semantic feature extraction and final disease diagnosis. Even though a few attention-based methods utilize attention maps for visual feature extraction, these attention maps are generated without direct constraints. Thereby, they may be not closely related to disease regions.
To better extract features of disease-related regions and global features, we developed a task-driven hierarchical attention network (THAN) for MCI/AD diagnosis. THAN consists of an information sub-network (IS) and a hierarchical attention sub-network (HAS), as shown in Figure 1. IS can generate a task-driven information map that automatically highlights disease-related regions and their importance for final classification. It contains an information map extractor, a patch classifier, and a mutual-boosting loss function. The information extractor is assisted by the patch classifier to generate an effective information map. The mutual-boosting loss function aims to promote the performance of the information map extractor and the patch classifier. In HAS, a visual attention module and a semantic attention module are devised to successively enhance the extraction of discriminative features.

Methods
The proposed THAN consists of IS and HAS. IS aims to generate an effective information map. HAS makes use of the information map to produce hierarchical attention maps that guide the sub-network to extract discriminative visual and semantic features for final image classification. In this section, we first present IS and then discuss HAS. After that, the dataset and preprocessing steps are introduced. Moreover, evaluation metrics and implementation details are listed.
Information sub-network
IS, inspired by NTS-Net (25), is a mutual-boosting network, as shown in Figure 2. It is composed of two modules and a loss function. One module is an information map extractor and the other is a patch classifier. The goal of the information map extractor is to generate an effective information map with the assistance of the patch classifier. To be specific, the patch classifier first produces informative image patches based on the information map. It then tries to better classify these image patches and to assist the information map extractor to generate an effective information map. Moreover, the effective information map in turn improves the generation of informative image patches and the profound classification of these patches. The two modules boost each other in the above manner. Further, a mutual-boosting loss function is designed to combine the two modules and to promote IS to generate the final effective information map. The details of the two modules and the mutual-boosting loss function are discussed below.

Information map extractor
Given an sMRI image I, an information map M is generated according to the information map extractor F, denoted as
where M∈RL*W*H, L, W, and H represent the length, width, and height of M, respectively.
The structure of F is shown in Figure 2. It is stacked by Conv1, Maxpool, Block1, Block2, Block3, Block4, and Block5. The layers from Conv1 to Block4 aim to extract discriminative features from I. They have the same structure as 3D ResNet18. Block5 is developed to gradually map the feature map with N channels to the information map M with only one channel. It is made up of multiple transition layers. The specific structure of F is displayed in the middle column of Table 1.

Full table
Patch classifier
An image patch in an original image can be downsampled to a value in a feature map through several layers and with certain scales. Conversely, each value in a feature map can be mapped to an image patch in the original image through the same layers yet with reversed scales. The mapping relations are shown in Figure 3. Since M is obtained by applying 32:1 spatial downsampling to I through F, each value sk∈M (k∈{1, 2, ..., L×W×H}) corresponds to an image patch pk in I, where pk is with the size of 32*32*32. We describe sk as the importance value of pk for final classification because of the semantic character of M.
Given a feature map with two channels, it becomes a 2D vector through the global average pooling (GAP) layer. The 2D vector then produces a binary classification result after passed the Softmax layer. In this process, the highest value in the 2D vector determines the classification result. That is, the highest average value of the two channels in the feature map determines the classification result. This implies that the points with higher values in a feature map make more positive contributions to final classification. Based on this theory of deep convolutional neural networks (DCNNs), the points with higher values in a feature map are selected and highlighted for better classification. For M, the points with higher sk make more contributions to final classification. That means that the higher sk is, the more important pk is.
K largest importance values are selected from M, denoted as {s1, ..., sk, ..., sK}. Then the corresponding K image patches are obtained from I, denoted as {p1, ..., pk, ..., pK}. Therefore, they are the K most informative patches in I and are most likely related to the category of I. Their labels are hence assigned to the label of I. We utilize one 3D ResNet10 to classify these K patches. The structure of 3D ResNet10 is shown in the last column of Table 1.
Mutual-boosting loss function
The mutual-boosting loss function consists of a patch-level classification loss function and a calibrating loss function.
The patch-level classification loss function is used to improve the classification performance of the K informative patches. It is realized by the widely used classification loss function, i.e., cross-entropy loss,
where yk,n is the one-hot format of the ground truth of the patch pk; ck,n represents the probability that pk belongs to the nth category (n∈{1, 2, …, N}).
In the output of pk, each element (i.e., ck,n) indicates the probability that pk belongs to the corresponding category. Specifically, ck,n is the probability that pk belongs to the nth category. The category that has the highest probability is the true category of pk. Therefore, we define the highest probability as the confidence score of pk being truly classified, which is denoted as ck,
The calibrating loss function is used to keep the consistency between the importance values of informative patches and their confidence scores. It is formalized as follows,
where sk∈M represents the importance value of pk and ck denotes the confidence score of pk. The loss function is the sum of the distances between each pair of sk and ck. It encourages the short distance between sk and ck and penalizes the long distance between them.
The mutual-boosting loss function is the joint of the above two loss functions, that is,
It encourages the better classification of informative patches and the consistency between the importance values and the confidence scores of the informative patches. That means that the patch classifier better classifies the informative patches, which are extracted based on the information map. It meanwhile assists in improving the performance of the information map extractor to produce a more effective information map. The information map in turn boosts the generation of informative patches and the performance of the patch classifier. Therefore, the proposed IS is a mutual-boosting network.
Hierarchical attention sub-network
The task-driven information map M has the semantic information for MCI/AD diagnosis. It implies not only disease-related regions but also their importance for image classification. Therefore, we employ M to generate hierarchical attention maps to construct HAS. HAS is thereby guided to pay more attention to the visual and semantic features of disease-related regions, and also to consider the features of the other regions. Therefore, HAS can well exploit the discriminative features of sMRI images for disease diagnosis.
The framework of HAS is depicted in Figure 4. Two attention modules are devised to focus on the discriminative feature extraction. They are the visual attention module and the semantic attention module. The visual attention module aims to extract discriminative low-level features, i.e., features from the shallow layers of HAS. It is comprised of two attention blocks, i.e., Attention1 and Attention2 in Figure 4. The semantic attention module is used to extract discriminative high-level features, i.e., features from the deep layers of HAS. It consists of another two attention blocks, i.e., Attention3 and Attention4 in Figure 4. Since the size of M is different from that of the feature maps in HAS, M cannot be directly used as an attention map. To solve this, each attention block produces an attention map Mi' based on M and a feature map Fi first. Then Mi' is combined with Fi to enhance the features of the disease-related regions in Fi. Each attention block can be formalized as
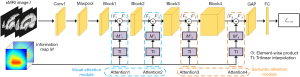
where M and Fi are the input of each attention block; Fi' is the output; TI is the trilinear interpolation function, making Mi' the same size as Fi; ☉ is the element-wise multiplication to highlight important features in Fi. The backbone of HAS is 3D ResNet34. Its structure is shown in the last column in Table 1.
It is worth noting that the proposed HAS is superior to the general spatial attention network. This is because all the attention maps in HAS are generated based on the effective information map and they can correctly highlight the features of disease-related regions, no matter the visual features from the shallow layers or the semantic features from the deep layers. However, the general spatial attention network usually generates attention maps from the network itself. This causes the attention maps generated at shallow layers not closely coinciding with disease-related regions due to gradient vanishing. This further leads to the visual features not being well extracted. The non-discriminative visual features impact the extraction of semantic features, which blocks the good performance of the general spatial attention network.
The cross-entropy loss function is employed in HAS for the final classification of I,
where yn is the one-hot format of the true label of I and cn represents the probability that I, together with M, belongs to the nth category.
It is noted that since the generation of hierarchical attention maps is directly dependent on M and M is a task-driven information map, we regard the generation of hierarchical attention maps as a task-driven operation. Therefore, we name the proposed network as a THAN. Moreover, even though there are studies about hierarchical attention networks, the specific frameworks of these hierarchical attention networks are different and they are designed based on their own tasks, which are not efficient for other tasks.
Dataset and preprocessing
We utilized the public dataset of Alzheimer’s Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu/) as our experimental data. ADNI was launched in 2003 as a public-private partnership. Its primary goal is to test whether serial magnetic resonance imaging, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. In this work, we downloaded the 1.5T T1-weighted sMRI images of 1,139 subjects at the baseline time point from ADNI, including 396 MCI, 327 AD, and 416 NC. Table 2 shows the demographic and clinical information of the 1,139 subjects, where MMSE represents mini-mental state examination and std represents standard deviation.

Full table
All the sMRI images were pre-processed with a “Minimal” pipeline (26), i.e., skull stripping and affine registration (the other is an “Extensive” pipeline, including skull stripping and non-linear registration). The two pre-processing steps were realized with the tool of FMRIB Software Library 5.0 (https://fsl.fmrib.ox.ac.uk/). Affine registration was used to linearly align the sMRI images with the template of MNI152 (27,28) to remove global linear differences and also to resample the sMRI images into the spatial resolution of 1×1×1 mm3. After this, each sMRI image contains the unnecessary background portion, which has no impact on image classification but increases computation time. Therefore, we removed the background portion of each sMRI image along the minimum vertical external matrix of the brain portion. After this operation, image sizes become different. They are 148±2, 182±2, and 153±3 (in the format of mean ± standard deviation). To keep the same size of these sMRI images, we scaled each sMRI image until the maximum side of each sMRI image is up to 128 by utilizing trilinear interpolation. Additionally, we padded the other two sides of each sMRI image to 128. Thereby, all the sMRI images have the size of 128*128*128. It is noted that we scale images into the size of 128*128*128 because 128 is the multiple of 32 (the size of image patches). Two hundred and fifty-six is not used because images with the size of 256*256*256 are far bigger than that of 128*128*128 and they require more graphics processing unit (GPU) memory and time to train and validate.
Evaluation metrics and implementation details
The proposed THAN is evaluated on four classification tasks: MCI against (vs.) NC, AD vs. NC, AD vs. MCI, and AD vs. MCI vs. NC. In the first two tasks, NC subjects are regarded as negative cases and AD or MCI subjects are regarded as positive ones. For AD vs. MCI, AD subjects are denoted as the positive cases and MCI subjects represent the negative ones. For AD vs. MCI vs. NC, one type of subject is regarded as positive cases and the remaining two types are as negative cases, which is referred to (29). According to (30), this task was conducted three times and the positive cases varied each time. The average of the three times was computed as the final performance of AD vs. MCI vs. NC. Six metrics are assessed. They are accuracy (ACC), specificity (SPE), sensitivity (SEN, also called Recall), precision (PRE), F1-score (F1), and area under the curve (AUC) of receiver operating characteristic.
For every task, we divide all the corresponding sMRI images into the training, validation, and test sets with the ratio of 7:2:1. It is noted that each type of sMRI image in every task also has the same ratio (i.e., 7:2:1) among the training, validation, and test sets, which can avoid data imbalance to some extent. For instance, for the task of MCI vs. NC, the number of sMRI images of MCI subjects and that of NC subjects have the ratio of 7:2:1 in the training, validation and test sets. The training and validation sets are used to train THAN. Specifically, for every task, we first trained IS with 80 epochs. The best trained IS were preserved when the training loss converged to 0 and the validation loss converged to less than 1. After that, an information map was generated for every sMRI image in the training and validation sets using the preserved best-trained IS. Every sMRI image and its information map formed a pair. Furthermore, pairs of sMRI images and information maps were used to train HAS with 80 epochs. The best trained HAS was saved when the ACC of the validation set reached the largest value. The independent test set aims to evaluate the trained THAN, i.e., the saved best-trained IS and HAS. To demonstrate the robustness of THAN, we utilized 3-fold cross-validation for all the table-related experiments. Results are the mean of the 3 experiments. For figure-related experiments, we display the results of the certain one among the 3 times, since we do not find a proper way to fuse the experimental results of the 3 times. Moreover, all the methods, including THAN and the comparison methods, were implemented on PyTorch and they were trained and evaluated on an NVIDIA RTX 2080Ti GPU. During the training process, these methods utilized the optimizer of stochastic gradient descent with the momentum (31) of 0.9 and with the weight decay of 0.0001. The learning rates of these methods were increased to 0.001 using the warmup strategy (32) and then were lowered by a tenth every 30 epochs. Further, the batch sizes of these methods were set to 4. In addition, no data augmentation was used in these methods.
Results
Comparison results with two advanced attention modules
The proposed THAN was compared with two advanced attention modules. One is the squeeze-and-excitation (SE) module (33). It aims to pay attention to the relationships among channels of a feature map. The other is the convolutional block attention module (CBAM) (34). It is an extension of SE and utilizes both channel-wise and spatial-wise attention on a feature map. We kept the backbone of HAS unchanged and rewrote the SE and CBAM modules from 2D to 3D, respectively. We then replaced the visual and semantic attention modules (four attention blocks totally) in HAS with four 3D SE and CBAM modules, respectively. The two new networks are denoted as ResNet + SE and ResNet + CBAM. The comparison results of ResNet, ResNet + SE, ResNet + CBAM, and THAN are summarized in Table 3.

Full table
We can observe that (I) for MCI vs. NC, THAN gains the best values in terms of the five metrics among the four networks, and its SEN is just 0.7% lower than the SEN of ResNet + CBAM. Further, compared with ResNet (i.e., the baseline network), THAN and ResNet + CBAM achieve better values on the six metrics, and ResNet + SE obtains larger values on five metrics except the SEN value, which is the same with ResNet’s SEN. (II) For AD vs. NC, THAN achieves the best values among the four networks in terms of the six metrics. However, both ResNet + SE and ResNet + CBAM are all behind ResNet for the six metrics. (III) For AD vs. MCI, THAN achieves the best performance for the five metrics except SEN among the four networks, with SEN being 0.1% behind ResNet + SE’s. ResNet + CBAM is superior to ResNet except SPE and PRE, yet ResNet + SE does not have obvious advantages than ResNet. (IV) For AD vs. MCI vs. NC, THAN attains the best performance among the four networks in terms of the six metrics. The performance of ResNet + SE and ResNet + CBAM is similar and is better than that of ResNet. These results indicate that, overall, THAN is better than the two state-of-the-art attention modules and the baseline network for the four tasks; However, SE and CBAM cannot always promote the performance of the four tasks. This can be explained by the following reasons: since the SE and CBAM modules used in shallow layers do not have direct constraints, they are not well trained due to gradient vanishing. This causes the result that the visual features extracted using the SE and CBAM modules are not well extracted, which further degrades the extraction of semantic features and the performance of image classification. However, for THAN, the visual and semantic attention modules generate attention maps based on an effective information map, highlighting disease-related regions. Therefore, the attention maps in both shallow and deep layers can emphasize disease-related features. This benefits discriminative visual and semantic feature extraction and further facilitates image classification.
Comparison results with state-of-the-art methods
Regarding the four kinds of methods mentioned in Introduction, since their codes are not public, we realized one method in each kind and compared them with THAN. Specifically, for the attention-based methods, the approach in (20) was realized because the other attention-based methods are for different tasks (21,22) or use multi-modality data (23). The only whole image-based method (12) was realized and compared. For the patch-based methods, since the approach in (18) is the most recent study, it was compared. For the ROI-based method, we just realized the method in (9). The four comparison methods were conducted on our dataset and using the same 3-fold cross-validation. Their mean comparison results are summarized in Table 4.

Full table
It can be seen that (I) for MCI vs. NC, THAN outperforms these comparison methods in terms of ACC, SPE, PRE, F1, and AUC, and it just 0.2% falls behind the patch-based method of (18) and achieves excellent performance compared with the ROI-based methods. (II) For AD vs. NC, AD vs. MCI, and AD vs. MCI vs. NC, THAN achieves the most excellent performance when compared with these comparison methods. These results imply that THAN improves the performance of MCI/AD diagnosis and achieve better results when compared with the above methods.
Evaluation of the IS
We first show the selection of the hyper-parameter K in IS. After that, we visualize the information map generated by IS to show its effectiveness in highlighting disease-related regions.
The only hyper-parameter K in IS (i.e., the number of informative patches) was investigated based on the classification performance of THAN. We first varied K from 3 to 10 for each task and kept the other settings unchanged. We found that the performance of the four tasks increases first and then degrades. With this trend, we then observed the results when K is 20 and 30, and found that the performance of the four tasks continues falling. The trend that the larger number of patches does not have better performance is due to the following reasons: the selected informative patches are highly related to disease lesions and their corresponding regions in original sMRI images are assigned higher weights for image classification. However, not all the image patches contain lesions. With the increase of K, some of the selected informative patches do not contain disease lesions while they are assigned higher weights for image classification. The incorrect weights on these incorrect informative patches lead to the falling of classification performance. Therefore, there is the trend that the larger number of patches does not have better performance. All the results are shown in Table 5. We can see that (I) for MCI vs. NC, the four metrics of ACC, SPE, PRE, and F1 are the best when K =6; SEN and AUC achieve the highest points when K is 20 and 7, respectively. According to the overall performance, we set K =6 for MCI vs. NC. (II) For AD vs. NC, ACC, SPE, PRE, and F1 achieve the best points when K =4. The other two metrics are the highest when K =5. Therefore, we set K =4 for AD vs. NC. (III) For AD vs. MCI, ACC, SEN, and F1 obtain the largest values when K =6, SPE is the best when K =5, and PRE and AUC are the highest when K =8. Therefore, we set K =6 for this task. (IV) For AD vs. MCI vs. NC, the values of ACC, SEN, and F1 are the largest when K =6, and SPE and AUC values are the best when K =5. According to the overall performance, we set K =6 for this task. Even though the optimal values of K for the four tasks are different, they are determined after the network of each task is well trained. That means we do not need to consider K in the test process.

Full table
To display the effectiveness of IS in localizing disease-related regions, we visualized the generated information maps of an MCI subject in MCI vs. NC and an AD subject in AD vs. NC, respectively. Specifically, we first transformed the generated 3D information map of a subject into the same size as the input sMRI image using Eq. [6]. We then selected the 40th, 50th, 60th, and 70th 2D maps from the coronal, sagittal, and transverse views of the transformed 3D information map, respectively. After that, we combined 12 maps with the 2D slices from the input sMRI image to produce the visualized 2D information maps. The results are displayed in Figure 5. The color indicates the importance of regions for classification. The redder the color is, the more important the whole region is for AD or MCI diagnosis. It can be seen that (I) for both AD and MCI subjects, the redder regions contain hippocampi and ventricles. This indicates that hippocampus and ventricle play important role in image classification, which is in accordance with the conclusion that hippocampus atrophy and ventricle enlargement are the biomarkers of MCI and AD (35). This observation implies that IS can effectively highlight disease-related regions. Nevertheless, even though hippocampi and ventricles are of importance for disease diagnosis, experts still observe other regions of images and make final diagnosis considering the features of the entire images in clinical trials. Moreover, since the pathogenesis of AD has not been discovered and there may be latent lesion regions related to MCI and AD, it is necessary to consider the features of the remaining regions except hippocampi and ventricles. The contributions of the features of other regions for image classification are decided based on the weights of these attention maps. (II) The redder regions of the MCI subject are larger than that of the AD subject. This may be because the MCI and NC subjects are harder to distinguish than the AD and NC subjects, and IS requires to focus on more regions to generate more discriminative features for MCI diagnosis. It is noted that some redder regions are in the background. This is caused by the mapping relations between image patches and the values of an information map. Figure 3 displays the mapping relations. According to the mapping relations, a value in the information map corresponds to a 32*32*32 image patch in the original image. Therefore, some image patches, mapped from some values in the information map, contain both brain tissue and background. Further, for coarsely visualizing the importance of image patches, a value in the information map can be mapped into a 32*32*32 patch with values and these values on the patch are realized through the trilinear interpolation to the value in the information map. It is noted that the patch with values and the image patch forementioned are two different concepts. Due to the trilinear interpolation, when the forementioned image patch, containing both brain tissue and background, corresponds to a higher value in the information map, it will be combined with a patch with higher values, which indicate redder color. Therefore, the background portions in these image patches are also with redder color. This coarse visualization is a common phenomenon in current methods, such as in (36) and (37). However, this phenomenon does not affect the performance of THAN. Since information maps are used to generate attention maps in different layers of HAS, after multiple convolution layers, HAS can recognize the useless background portions.

Ablation study of the HAS
To validate the effectiveness of the combination of the visual and semantic attention modules in boosting the discriminative feature extraction for disease diagnosis, we conducted the ablation study on different combinations of the two attention modules with the backbone of HAS. The results are illustrated in Table 6.

Full table
For MCI vs. NC, we find that (I) the backbone + visual attention and the backbone + semantic attention are superior to the backbone in terms of ACC, SEN, F1, and AUC. (II) The backbone + visual attention module + semantic attention module (i.e., THAN) achieves the best performance referring to the six metrics when compared with the backbone and the backbone + semantic attention, and it obtains the highest values referring to the five matrices except SEN when compared with the backbone + visual attention. These results imply that both the visual attention module and the semantic attention module facilitate the extraction of discriminative features and further promote the performance of MCI vs. NC to some extent. Moreover, the performance of MCI vs. NC is best improved when the two attention modules are combined.
For AD vs. NC, the following results can be acquired. (I) Both the backbone + visual attention module and the backbone + semantic attention module are a little better than the backbone overall. (II) THAN achieves the best performance among the four methods. These results imply that both the visual attention module and the semantic attention module boost the performance of AD vs. NC. Furthermore, the performance of AD vs. NC is improved to the highest point when the two attention blocks are employed together.
For AD vs. MCI, we can see that (I) the backbone + visual attention module is better than the backbone in terms of ACC, SPE, PRE, F1, and AUC; the backbone + semantic attention module is superior to the backbone for ACC, SPE, PRE, and F1. (II) THAN achieves the best performance for all the matrices among the four methods. These results imply that the performance of AD vs. MCI is promoted by the visual attention module and the semantic attention module, respectively. Moreover, it is best improved when the two attention modules are combined.
For AD vs. MCI vs. NC, we observe that (I) the backbone + visual attention module is better than the backbone in terms of the six metrics; the backbone + semantic attention module is superior to the backbone for ACC, SPE, PRE, and F1. (II) THAN achieves the best performance for all the metrics when compared with the four methods. These results indicate that AD vs. MCI vs. NC is improved by the two attention modules, respectively, and it is improved to the best point when the two attention modules are combined.
Overall, the performance of the four tasks is promoted by the visual attention block and the semantic attention block, respectively, and it is highly improved by combining the visual and semantic attention modules into the backbone. This demonstrates that HAS can effectively extract both discriminative visual and semantic features for disease diagnosis.
Discussion
We first investigate the execution time of THAN. Moreover, we visualize the features extracted using THAN. Finally, we compare THAN with state-of-the-art methods.
Execution time of THAN
To discuss the efficiency of THAN, we described the execution time of THAN on the test sets for the four tasks, as shown in Table 7. The mean time indicates the average execution time of an sMRI image in a task. The max time indicates the maximum execution time of an sMRI image in a task. The min time indicates the minimum execution time of an sMRI image in a task. It can be seen that the mean time, the min time and even the max time of every sMRI image in the four tasks are far less than 1 s. This demonstrates that the proposed THAN is very efficient, even though it is made up of two sub-networks.

Full table
Visualization of the original images and their features extracted using THAN
To further demonstrate the ability of THAN in extracting discriminative features, t-distributed stochastic neighbor embedding (t-SNE) (38) was utilized to visualize the original images of the test set and the features of the test set extracted using THAN. t-SNE is a technique to visualize high-dimensional data by giving each datapoint a location in a 2D or 3D map (38). To be specific, it projects features from a high dimension to a low dimension and meanwhile attempts to preserve the local structures of these features. Since the dimensions of high-dimensional features have no specific meanings, the two dimensions of the projected 2D features also have no detailed meanings. Furthermore, since the 2D map is simpler than the 3D map, it is chosen to visualize each datapoint. It is noted that t-SNE cannot demonstrate the classification performance of THAN, which is determined by both the features extracted and the classifier used.
Specifically, each original image was reshaped into a one-dimensional feature vector first. T-SNE was then utilized to visualize these original images. For the features extracted using THAN, the trained IS was taken to produce information maps for the sMRI images. After that, the first to the GAP layers of the trained HAS were used to produce a 512-dimensional feature vector for each sMRI image. Finally, t-SNE was employed to visualize these 512-dimensional feature vectors of the test set. Results are shown in Figure 6. A red point represents the original image or its feature vector of an MCI subject, a yellow point the original image or its feature vector of an NC subject, and a blue point the original image or its feature vector of an AD subject.

We can find that (I) compared with original images, the features extracted using THAN has the ability to differentiate different categories. This indicates that the proposed THAN can learn discriminative features from sMRI images. (II) The two kinds of features in AD vs. NC are best distinguished compared with those in MCI vs. NC, AD vs. MCI, and AD vs. MCI vs. NC. This result matches the result in Table 6, that is, ACC of AD vs. NC is the best, followed by MCI vs. NC, by AD vs. MCI, and then by AD vs. MCI vs. NC. This indicates that AD vs. NC is the easiest task in the four tasks, and MCI vs. NC, AD vs. MCI, and AD vs. MCI vs. NC require more exploration, especially the task of AD vs. MCI vs. NC. Motivated by the observation, in the future, we will pay more attention to investigate the recognition between MCI and NC, between AD and MCI, and among AD, MCI, and AD.
Acknowledgments
Funding: This work was supported in part by the National Natural Science Foundation of China (grant No. 61901237), Natural Science Foundation of Zhejiang Province (grant No. LQ20F020013), Ningbo Natural Science Foundation (grant No. 2019A610102, 2019A610103, 2018A610057), and Open Progam of the State Key Lab of CAD&CG Zhejiang University (grant No. A1915). Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions such as AbbVie, Alzheimer’s Association, and Araclon Biotech. A complete listing of ADNI investigators can be found online. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-21-91). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jia L, Quan M, Fu Y, Zhao T, Li Y, Wei C, Tang Y, Qin Q, Wang F, Qiao Y, Shi S, Wang Y, Du Y, Zhang J, Zhang J, Luo B, Qu Q, Zhou C, Gauthier S, Jia J. Dementia in China: epidemiology, clinical management, and research advances. Lancet Neurol 2020;19:81-92. [Crossref] [PubMed]
- Patterson C. World Alzheimer report 2018. Available online: https://www.alz.co.uk/research/world-report-2018
- Roberts R, Knopman DS. Classification and epidemiology of MCI. Clin Geriatr Med 2013;29:753-72. [Crossref] [PubMed]
- Reiman EM, Jagust WJ. Brain imaging in the study of Alzheimer's disease. Neuroimage 2012;61:505-16. [Crossref] [PubMed]
- Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 2010;6:67-77. [Crossref] [PubMed]
- Brand L, Nichols K, Wang H, Shen L, Huang H. Joint multi-modal longitudinal regression and classification for alzheimer’s disease prediction. IEEE Trans Med Imaging 2020;39:1845-55. [Crossref] [PubMed]
- Suk HI, Lee SW, Shen D. Deep ensemble learning of sparse regression models for brain disease diagnosis. Med Image Anal 2017;37:101-13. [Crossref] [PubMed]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR Jr, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimer's disease. Brain 2008;131:681-9. [Crossref] [PubMed]
- Aderghal K, Benois-Pineau J, Afdel K. Classification of sMRI for Alzheimer's disease Diagnosis with CNN: Single Siamese Networks with 2D+? Approach and Fusion on ADNI. ICMR 2017: Proceedings of the 2017 ACM on International Conference on Multimedia Retrieval; June 6-9; Bucharest, Romania. New York: ACM, 2017:494-8.
- Cui R, Liu M. Hippocampus Analysis by Combination of 3-D DenseNet and Shapes for Alzheimer's Disease Diagnosis. IEEE J Biomed Health Inform 2019;23:2099-107. [Crossref] [PubMed]
- Xia Z, Yue G, Xu Y, Feng C, Yang M, Wang T, Lei B. A Novel End-to-End Hybrid Network for Alzheimer's Disease Detection Using 3D CNN and 3D CLSTM. ISBI 2020: 2020 IEEE 17th International Symposium on Biomedical Imaging; 2020 Apr 3-7; Iowa City, IA, USA. New York: IEEE, 2020:1-4.
- Korolev S, Safiullin A, Belyaev M, Dodonava Y. Residual and plain convolutional neural networks for 3D brain MRI classification. ISBI 2017: 2017 IEEE 14th International Symposium on Biomedical Imaging; 2017 Apr 18-21; Melbourne, Australia. New York: IEEE, 2017:835-8.
- Liu M, Zhang J, Nie D, Yap P, Shen D. Anatomical landmark based deep feature representation for MR images in brain disease diagnosis. IEEE J Biomed Health Inform 2018;22:1476-85. [Crossref] [PubMed]
- Liu M, Zhang J, Adeli E, Shen D. Landmark-based deep multi-instance learning for brain disease diagnosis. Med Image Anal 2018;43:157-68. [Crossref] [PubMed]
- Liu M, Zhang J, Adeli E, Shen D. Deep multi-task multi-channel learning for joint classification and regression of brain status. MICCAI 2017: Proceedings of the 20th international conference on medical image computing and computer-assisted intervention; 2017 Sep 11-13; Quebec City, QC, Canada. Berlin: Springer, 2017:3-11.
- Liu M, Zhang J, Adeli E, Shen D. Joint classification and regression via deep multi-task multi-channel learning for Alzheimer's disease diagnosis. IEEE Trans Biomed Eng 2019;66:1195-206. [Crossref] [PubMed]
- Lian C, Liu M, Zhang J, Shen D. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer's disease diagnosis using structural MRI. IEEE Trans Pattern Anal Mach Intell 2020;42:880-93. [Crossref] [PubMed]
- Qiu S, Joshi PS, Miller MI, Xue C, Zhou X, Karjadi C, Chang GH, Joshi AS, Dwyer B, Zhu S, Kaku M, Zhou Y, Alderazi YJ, Swaminathan A, Kedar S, Saint-Hilaire M, Auerbach SH, Yuan J, Sartor EA, Au R, Kolachalama VB. Development and validation of an interpretable deep learning framework for Alzheimer’s disease classification. Brain 2020;143:1920-33. [Crossref] [PubMed]
- Long B, Yu CP, Konkle T. Mid-level visual features underlie the high-level categorical organization of the ventral stream. Proc Natl Acad Sci U S A 2018;115:E9015-24. [Crossref] [PubMed]
- Jin D, Xu J, Zhao K, Hu F, Yang Z, Liu B, Jiang T, Liu Y. Attention-based 3D convolutional network for Alzheimer’s disease diagnosis and biomarkers exploration. ISBI 2019: 2019 IEEE 16th International Symposium on Biomedical Imaging; Apr 8-11; Venice, Italy. New York: IEEE, 2019:1047-51.
- Li Q, Xing X, Sun Y, Xiao B, Wei H, Huo Q, Zhang M, Zhou X S, Zhan Y, Xue Z, Shi F. Novel iterative attention focusing strategy for joint pathology localization and prediction of MCI progression. MICCAI 2019: Proceedings of the 22nd international conference on medical image computing and computer-assisted intervention; 2019 Oct 13-17; Shenzhen, China. Berlin: Springer, 2019:307-15.
- Lian C, Liu M, Wang L, Shen D. End-to-end dementia status prediction from brain MRI using multi-task weakly-supervised attention network. MICCAI 2019: Proceedings of the 22nd international conference on medical image computing and computer-assisted intervention; 2019 Oct 13-17; Shenzhen, China. Berlin: Springer, 2019:158-67.
- Zhang L, Wang L, Zhu D. Jointly Analyzing Alzheimer's Disease Related Structure-Function Using Deep Cross-Model Attention Network. ISBI 2020: 2020 IEEE 17th International Symposium on Biomedical Imaging; 2020 Apr 3-7; Iowa City, IA, USA. New York: IEEE, 2020:563-7.
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. CVPR 2016: Proceedings of the IEEE conference on computer vision and pattern recognition; June 27-30; Las Vegas, NV, USA. New York: IEEE, 2019:770-8.
- Yang Z, Luo T, Wang D, Hu Z, Gao J, Wang L. Learning to navigate for fine-grained classification. ECCV 2018: Proceedings of the European Conference on Computer Vision; Sep 8-14; Munich, Germany. Berlin: Springer, 2018:420-35.
- Wen J, Thibeau-Sutre E, Diaz-Melo M, Samper-González J, Routier A, Bottani S, Dormont D, Durrleman S, Burgos N, Colliot O. Convolutional neural networks for classification of Alzheimer's disease: Overview and reproducible evaluation. Med Image Anal 2020;63:101694 [Crossref] [PubMed]
- Fonov VS, Evans AC, Mckinstry RC, Almli CR, Collins DL. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage 2009;47:S102. [Crossref]
- Fonov VS, Evans AC, Botteron KN, Almli CR, Mckinstry RC, Collins DL. Unbiased average age-appropriate atlases for pediatric studies. NeuroImage 2011;54:313-27. [Crossref] [PubMed]
- Jayachandran A, Dhanasekaran R. Multi class brain tumor classification of MRI images using hybrid structure descriptor and fuzzy logic based RBF kernel SVM. Iranian Journal of Fuzzy Systems 2017;14:41-54.
- Grandini M, Bagli E, Visani G. Metrics for multi-class classification: an Overview. arXiv:2008.05756 [preprint]. Available online: https://arxiv.org/abs/2008.05756
- Qian N. On the momentum term in gradient descent learning algorithms. Neural Netw 1999;12:145-51. [Crossref] [PubMed]
- Goyal P, Dollár P, Girshick R, Noordhuis P, Wesolowski L, Kyrola A, Tulloch A, Jia Y, He K. Accurate, large minibatch sgd: Training imagenet in 1 hour. arXiv:1706.02677v2 [preprint]. 2018 [cited 2021 Jan 24]: [12 p.]. Available online: https://arxiv.org/abs/1706.02677
- Hu J, Shen L, Sun G. Squeeze-and-excitation networks. CVPR 2018: Proceedings of the IEEE conference on computer vision and pattern recognition; June 18-22; Salt Lake City, UT, USA. New York: IEEE, 2019:7132-41.
- Woo S, Park J, Lee JY, Kweon IS. Cbam: Convolutional block attention module. ECCV 2018: Proceedings of the European conference on computer vision; 2018 Sep 8-14; Munich, Germany. Berlin: Springer, 2018:3-19.
- Saied I, Arslan T, Chandran S, Smith C, Spires-Jones T, Pal S, Non-Invasive RF. Technique for Detecting Different Stages of Alzheimer’s Disease and Imaging Beta-Amyloid Plaques and Tau Tangles in the Brain. IEEE Trans Med Imaging 2020;39:4060-70. [Crossref] [PubMed]
- Zhang X, Han L, Zhu W, Sun L, Zhang D. An Explainable 3D Residual Self-Attention Deep Neural Network for Joint Atrophy Localization and Alzheimer's Disease Diagnosis using Structural MRI. IEEE J Biomed Health Inform 2021; Epub ahead of print. [Crossref] [PubMed]
- Lian C, Liu M, Wang L, Shen D. Multi-Task Weakly-Supervised Attention Network for Dementia Status Estimation With Structural MRI. IEEE Trans Neural Netw Learn Syst 2021; Epub ahead of print. [Crossref] [PubMed]
- Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579-605.



