
Multi-channel multi-task deep learning for predicting EGFR and KRAS mutations of non-small cell lung cancer on CT images
Introduction
Lung cancer is the most malignant tumor, with the highest morbidity and mortality rates worldwide. The 5-year survival rate of patients with stage IV lung cancer is less than 5%. Lung cancer is divided into non-small cell lung cancer (NSCLC) and small cell carcinoma, and lung adenocarcinoma and lung squamous cell carcinoma are common types of NSCLC (1,2). Although considerable progress has been made over the past 2 decades in the study of NSCLC, the overall cure rate and survival rate remain low (3). The emergence of targeted therapy has substantially increased the survival rate of NSCLC patients. Mutations of essential pathogenic genes should be identified before targeted therapy. The epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma (KRAS) genes are key genes in NSCLC (4,5).
Biopsy by endoscopy or puncture typically provides criteria for detecting EGFR and KRAS mutations. However, there are several limitations in the application of these methods: (I) patients with a lower Karnofsky Performance Score (KPS) are less likely to undergo the invasive procedure repeatedly; (II) not all tumors of all sizes or locations are suitable for biopsy; (III) biopsy increases the risk of cancer metastasis; (IV) repeated biopsies for the monitoring of genetic mutations throughout the treatment process are less practical. Hence, exploring non-invasive and easy-to-use methods for predicting EGFR and KRAS mutations is necessary.
Researchers have used CT images in recent years to predict gene mutations, mostly by traditional radiomics, machine learning, or statistical methods. Most researchers use radiomics to predict gene mutations (6-11). Digumarthy et al. (6) explored whether CT could be used to predict EGFR mutations in NSCLC. The radiomics features of CT images from 93 NSCLC patients were extracted, followed by quantitative and Pearson correlation analyses. The experimental results demonstrated that the combination of radiomics and clinical features could increase the performance of predicting EGFR mutations. Other studies (7,8) have also explored the associations of imaging genomics in NSCLC, demonstrating significant associations between imaging features and gene mutations, as well as evaluating the feasibility of predicting EGFR/KRAS mutations from CT images. Liu et al. (9) studied the correlations between radiomics features and EGFR mutations in lung adenocarcinoma. They concluded that the combination of radiomics features and clinical information could effectively improve EGFR mutation prediction. Rios Velazquez et al. (10) studied the associations between imaging phenotypes and key gene mutations in lung cancer. They suggested that the combination of imaging features with clinical models could increase the accuracy (ACC) of prediction. The highest-performing features could distinguish between EGFR-mutant tumors and KRAS-mutant tumors. Park et al. (11) studied the relationships between imaging characteristics and EGFR mutations, KRAS mutations, and ALK recombination in stage IIIB-IV lung cancer. Several other studies (12-19) have also demonstrated the feasibility of predicting genetic mutations from imaging characteristics.
Studies have also used machine learning or statistics to predict genetic mutations from imaging features (20,21). Gevaert et al. (20) used machine learning to predict key EGFR and KRAS mutations by extracting semantic information, which showed that semantic information and EGFR mutations were statistically significant, whereas KRAS mutations and semantic information were not statistically significant. Li et al. (21) extracted radiomics features and established a logistic regression model by using a cross-validation strategy to predict EGFR mutations in NSCLC.
Although the above radiomics, machine learning, and statistical methods have successfully predicted genetic mutations, they require complicated and strict procedures. They are time-consuming and need full guidance from an experienced imaging physician, from detection to segmentation to feature extraction and feature selection. Furthermore, radiomics features are sensitive to segmentation results obtained by manual segmentation and are not repeatable. The extraction of semantic information in a small number of studies requires experienced physicians, and the selection of semantic characteristics is difficult even in the medical field.
In recent years, deep learning has realized substantial success in the artificial intelligence field due to powerful feature extraction and classification capabilities, enabling users to avoid tedious manual feature extraction (22-25). Deep learning methods have also been gradually developed in the study of image prediction of gene mutations. Wang et al. (26) proposed CT images to train deep learning models and predict EGFR mutations in lung adenocarcinoma. First, they collected CT images of 844 patients with lung adenocarcinoma. Then, they constructed end-to-end deep learning models for the prediction of EGFR mutations from CT images. This approach is a non-invasive and easy-to-implement deep learning method. Other studies (27-29) also show that deep learning models can identify gene mutations in lung cancer.
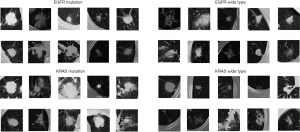
Although these deep learning methods have demonstrated higher classification performance than traditional radiomics or machine learning methods, they did not demonstrate similar performances for EGFR and KRAS mutation-based detection and for the ImageNet and Cifar10 datasets. The main reasons are as follows: (I) the deep learning models with superior performance rely on large-scale datasets; however, medical image datasets are often difficult to obtain on a large-scale (30,31); (II) extensive changes occur in the morphology, texture, and visual similarity of lung nodules among gene mutation types, as shown in Figure 1. From Figure 1, we can conclude that it is difficult for the typical convolutional neural network (CNN) to conduct feature extraction tasks for all types of nodules.
Many studies have been proposed to address the limitations of small medical datasets (32-34). Firstly, image representation ability that has been learned from larger-scale natural images can be transferred to medical small-sample images through transfer learning (32-34). Esteva et al. (32) pre-trained the GoogleNet Inception v3 CNN on Large-Scale ImageNet to classify skin cancer. Secondly, multiple slices can be extracted from 3D nodules, and 2D CNN can be extended to slice-by-slice medical images (35,36). Setio et al. (36) suggested decomposing the multi-view architecture from 3D nodules and training 2D CNN in each view. Thirdly, studies have used improved deep learning models to solve small-sample problems with medical images (37,38). Kang et al. (37) used the inception-resnet network for benign and malignant lung nodule classification. The experimental results demonstrated that the multiscale convolution kernel and the residual structure of inception-resnet were highly useful for various lung nodule feature extraction tasks. Guan et al. (38) proposed a residual attention learning network for the multi-classification of chest X-ray images. The method introduced an attention mechanism into the CNN, which enabled CNN to focus on the lesion area related to the prediction task, thereby substantially improving the CNN’s feature extraction performance. The experimental results demonstrate the advantages of using the attention mechanism in the multi-classification tasks of chest X-ray images. Fourthly, studies have used multi-task learning to overcome the small-sample problem of medical images (39). Additionally, patient medical record information has been added into deep learning models (40). In our previous work (41), domain knowledge was incorporated into our deep model, and the classification ACC on small samples was increased.
This paper proposes a multi-channel and multi-task deep learning (MMDL) model for predicting EGFR and KRAS mutation statuses simultaneously from CT images and patient personal information. First, we decomposed each 3D lung nodule cube into 9 fixed views. Then, we built an inception-attention-resnet model for extracting deep features from the 9 views. Finally, we combined 9 inception-attention-resnet models for multi-task learning to predict EGFR and KRAS mutations simultaneously. Furthermore, patient personal information was incorporated into the inception-attention-resnet model to include more prior knowledge of the mutations. An adaptive weighting scheme was adopted in the training process so that the proposed MMDL model could be trained in an end-to-end manner.
The proposed MMDL model’s main contributions are as follows: (I) to the best of our knowledge, this is one of the first studies to use deep learning models for simultaneous multi-gene mutation prediction; (II) the MMDL models can characterize the features of nodules more comprehensively and enable a single prediction task to benefit from multi-task learning, which increases the ACC of prediction. Moreover, the complete model can be trained in an end-to-end approach, thereby avoiding manual intervention; (III) the proposed model can incorporate multiple aspects of patient personal information related to the predictive task into the model’s learning process; (IV) the proposed inception-attention-resnet model can efficiently extract features from various types of lung nodules.
Methods
The proposed model consisted of 3 steps: (I) obtaining 9 2D nodule views from 3D cubes with nodules and extracting regions of interest (ROIs) from 2D nodule views; (II) Constructing pre-trained inception-attention-resnet models, which use the extracted 2D nodule ROIs and patient personal information as inputs for multi-task learning; (III) Combining 9 pre-trained inception-attention-resnet models for multi-task learning with adaptive weighted decision-level fusion. The overall flow of the algorithm is illustrated in Figure 2. Figure 2A shows the extraction of 9 view slices; Figure 2B is the multi-channel feature extraction; Figure 2C shows details of pre-trained inception-attention-resnet; Figure 2D is the multi-channel fusion process.
Obtaining 9 2D nodule slices and extracting ROIs
The basic strategy of multi-channel learning is to adopt the consistency and differentiation from various channels to achieve better performance. Since CT images are of variable spatial resolutions, we resampled them using spline interpolation to a uniform size of 1.0×1.0×1.0 mm3. First, 2D slices were stacked into 3D by the linear interpolation method. Then, 2 radiologists with more than 8 years of experience at the co-operative hospital labeled the nodule’s center. To ensure that for the 3D cube, all nodules (≤30 mm) were fully visible in the 2D view and sufficient contextual information was included, we considered the nodule center that was specified by the radiologists as the center and extracted a cube of size 64×64×64, which could contain the nodule completely (34). It is the 3D cube-containing nodules that we extracted. Although 3D deep learning models are already available for medical images (35,36), our model uses 2D deep learning multi-channel models for each 3D cube, providing more training samples.
We extracted 9 2D slices from 3 planes (transverse, sagittal, and axial) and 6 diagonal planes of the 3D cube containing lung nodules, which were used to characterize the nodule information fully. Each diagonal plane was obtained by cutting the 2 opposite faces of the cube diagonally, which contained 2 opposite edges and 4 vertices (as illustrated in Figure 2A). Therefore, we obtained 9 slices for each nodule, different slices entered different channels, and the channels could coordinate and promote each other.
Data augmentation techniques enhance the dataset, which can alleviate the overfitting of deep learning models. Augmented data were generated for each slice by conducting image translation, rotation, and vertical and horizontal flipping (40). The rotation angle was selected from 90°, 180°, 270°, and the translation was horizontal or vertical. All images were then uniformly adjusted to a size of 299×299 to accommodate the following model input. We standardized the data before passing it to the deep neural network, using Z-score standardization (42). After standardization, the data were adjusted to a normal distribution that conformed to the average of 0, and the variance was 1. Normalization is performed to return the increasingly biased distribution to the standardized distribution before activating the function.
Deep learning model
The inception-resnet-v2 model (Figure 3) (43) consists of a steam module, 3 reduction modules, and 3 inception-resnet modules. The steam module is used to preprocess the data before it enters the inception module and performs multiple convolutions and pooling operations. The reduction module plays a role in pooling and adopts a parallel structure to prevent the bottleneck problem. The inception-resnet module is the integration of the inception module and the resnet module, which can conduct multiple types of convolution operations (1×1, 3×3, or 7×7 convolution kernels) or pooling operations on the input image and can stitch all outputs into a deep feature map. Since convolution kernels that differ in terms of size also differ in terms of receptive field size, different types of information from the input image can be obtained. Furthermore, parallel convolution and stitching for various kinds of output feature maps will enable the model to achieve better image representation. The residual module solves gradient disappearance during training and avoids the performance degradation caused by the deep network.
Our inception-attention-resnet module (Figure 2C) adopts the above 3 types of inception-resnet modules as the basic units, where the design of the attention mechanism uses an hourglass attention structure similar to that in (44), as illustrated in Figure 4. Each inception-attention-resnet module is composed of 2 branches: a mask branch and a trunk branch. The trunk branch is composed of multiple stacked sequential inception-resnet modules, which are used for feature extraction. The mask attention branch adopts a full convolution structure similar to the bottom-up and top-down structures of the fully convolutional network (FCN) (45). First, for input, the bottom-up structure is used to conduct max pooling multiple times to quickly enlarge the receptive field after passing through multiple inception-resnet module units. The resulting output is fed to the top-down structure (which is symmetrical to the bottom-up structure). Multiple linear differences are calculated after the residual units to enlarge the input feature map, where the number of linear interpolations is the same as the number of max pooling, thereby ensuring that the inputs are the same size as the output. Then, 2 consecutive 1×1 convolutional layers are connected, and a sigmoid layer is connected for normalizing the output to [0,1]. Simultaneously, we added the skip connections between the bottom-up and top-down structures to capture various information proportions. Therefore, the mask attention branch will act as a feature selector to generate attention perception, thereby enhancing the useful features and suppressing the noise of the features that are generated by the trunk branch to produce a more discriminative feature representation.
The attention module was designed using an hourglass attention structure, as shown in Figure 4. Assume that the input image is x and the feature map that is generated by the steam module is F, which will be fed into the trunk branch and the mask attention branch, respectively. Suppose the feature map generated by the trunk branch is Ti,c(x,φ), where φ is the trunk branch’s parameter. The mask attention branch obtains the mask Mi,c(x,θ) with the same size as the trunk output, and θ is a parameter of the mask attention branch. By normalizing Mi,c(x,θ) using the sigmoid function, the normalized result is obtained:
Since the mask attention branch can generate an attention perception function (38), Si,c(x) can be used to add soft weight to the feature map Ti,c(x) the trunk branch generates that. Therefore, the weighted output feature map is:
Where i represents the position of the i-th pixel on the c-th channel of the feature map, and
is the c-th channel of the feature map, in which C represents the total number of channels for the generated feature map. Mask attention branches act as feature selectors during forward-propagation and as filters for gradient updates during backpropagation to conduct parameter updates with the trunk branch. The mask’s gradient for the input feature is:Where θ is the parameter of the mask branch and φ is the parameter of the trunk branch.
To prevent performance degradation that is caused by applying soft weights to the trunk features Ti,c(x,φ), the output feature maps Oi,c(x,φ) and Ti,c(x,φ) are weighted for element-wise addition. Therefore, the final output of the attention module is:
Our inception-attention-resnet module (as shown in Figure 5) adopts 3 types of inception-resnet modules as the basic units, and various types of attention can be captured extensively. As part of the constructed model, 3 attention model can simulate the fast bottom-up feed-forward process and top-down attention feedback during a single feed-forward process, thereby enabling our model to be trained in an end-to-end approach.
Our proposed inception-resnet-attention model consists of a stem module, 3 inception-resnet-attention modules, 2 reduction modules, an average pooling layer, a dropout (keep =0.8), and a fully connected layer. As illustrated in Figure 2C, except for the inclusion of 3 inception-resnet-attention modules, the remaining module adopts the original inception-resnet-v2 structure.
To avoid overfitting in our deep model, we first pre-trained the inception-resnet-attention model on the large-scale public datasets and saved the model weights. Then, we transferred the pre-trained model to our medical image datasets. The CNN model is commonly pre-trained on the ImageNet datasets (46), including approximately 1.2 million images and 1,000 categories. However, it is not easy to obtain the pre-trained weights on ImageNet for a model that has been built from scratch because training on such a large dataset requires powerful equipment, including many GPUs. Since our task is a binary classification task, we chose a smaller dataset, cat vs. dog (https://www.kaggle.com/c/dogs-vs-cats/data), from the Kaggle competition to classify cats and dogs. The dataset contains a training set and a test set, where the training set contains 25,000 images, with cats and dogs each constituting half, and the test set contains 12,500 unlabelled pictures. The inception-resnet-attention model was pre-trained on the cat vs. dog dataset. The network uses end-to-end training to minimize the cross-entropy loss function. A mini-batch random gradient descent algorithm with a batch size of 16 is used as an optimizer. The maximum number of epochs is set to 100, and the learning rate is set to 0.001.
Furthermore, to prevent overfitting of the network, we adopted an early stopping strategy during the training process. First, we randomly selected 10% of the images from the training set as the validation set. If the network error on the training set continued to drop during training, but the error on the validation set stopped dropping more than 5 times, we terminated the training process early, even before the maximum number of epochs was reached. We then saved the weights of the convolution and pooling layers of the pre-trained network (except the fully connected layer) and used them to initialize the network’s convolution and pooling layers for subsequent construction.
To utilize the inception-resnet-attention model to predict gene mutations, we removed its last fully connected layer and added 2 fully connected (FC) layers to the network (as illustrated in Figure 2C). Each branch includes 3 FC layers: FC256 (256 neurons), FC128 (128 neurons), and FC2 (2 neurons). We added patient information factors (age, gender, and smoking status) to each branch’s FC256 layer. Finally, the last FC layer (FC2) of the 2 branches is used to predict EGFR and KRAS mutations. The weights of these 3 fully connected layers are initialized randomly, and the activation function of the last layer uses the sigmoid function. First, only the newly added fully connected layers are adjusted (FC256, FC128, and FC) in the network, and then all layers are adjusted. There are 2 reasons: (I) avoid overfitting due to small amounts of data; (II) features of the deep network’s first layers contain more general characteristics (e.g., edge information). Our prediction involved multi-tasks: the last 2 FC2 layers predict the mutation probabilities of EGFR and KRAS. Although multi-tasks increase the computational burden of the network, it can realize the objective of simultaneously performing multiple tasks.
Since our CT image was grey and the pre-trained inception-resnet-attention network takes 3 channels as input for colour images, we inserted a 3×3 kernel convolution layer after the input image to convert the input monochrome image into a 3-channel image that is most suitable for the network.
Multi-model decision-level fusion
The basic strategy of multi-channel learning is to utilize the consistency and differentiation from multiple channels to achieve higher performance (see Figure 2D). For the j-inception-resnet-attention model,
represents the training set and Xi represents the i-th subject. The EGFR mutation class labels are denoted as and the KRAS mutation class labels as . E and K are the numbers of EGFR and KRAS mutation categories, respectively. The 2 branches of the fully connected layer of each inception-resnet-attention model conduct the EGFR and KRAS mutation prediction tasks (see Figure 2D).We defined the outputs of the first and second branches as A and B, respectively, which can be expressed as follows:
Where, respectively,
and represent the weight and bias of the j-th inception-resnet-attention model in the first task, and and represent the weight and bias of the j-th inception-resnet-attention model in the second task. Note that the 2 tasks share only the convolution layers’ weights of the inception-resnet-attention, while the weights of the fully connected layer are non-shared; thus, and are not equal. represents the sigmoid activation function. We combined 9 inception-resnet-attention models for multi-task learning to make the final prediction, as illustrated in Figure 2B. Each inception-resnet-attention model has 2 neuron output layers in 1 branch of the FC2 layer that is connected to the same neuron classification layer. This layer is the final classification layer, namely, FC_last, which contains 2 sigmoid neurons. The 2 neurons’ outputs, and , of FC_last are predictions that are made by the MMDL model and can be expressed by the following formula:Where
and is a set of weights between the j-th inception-resnet-attention model’s output layer and the first neuron in the FC_last layer. The parameter m represents the m-th neuron in the output layers of one of the branches of each inception-resnet-attention model. The sum of m is the weighted sum of each branch output layer. The cumulative sum of j represents the weighted sum of the outputs of one branch of the 9 inception-resnet-attention models. is defined similarly as . represents the sigmoid activation function. The MMDL model objective function is as follows:The first item is the cross-entropy loss of multi-class classification, which is used to evaluate the difference between the predicted value and the actual value of the EGFR mutation of the input sample. The second term is used to evaluate whether the input sample belongs to the difference between the predicted value and the actual value of the KRAS mutation, where
is the indicator function such that if is true, then , and is the probability that the network will correctly predict the subject as belonging to category . is defined similarly.The label of the KRAS mutation class is
, and K is the number of KRAS mutation classes. In our study, both the EGFR mutant class label and the KRAS mutant class label were used in the backpropagation process to update the network weights in the convolution layer and learn the most relevant features in the FC layer. The proposed network model was used to learn a non-linear mapping. from the input image to the EGFR spatial mapping of mutant labels and KRAS mutant labels. The EGFR and KRAS mutations were handled using the same approach.Results
Our participating hospital collected the training dataset at Shanxi Province, which included 363 patients from 2017 to 2018 with mutations in EGFR and KRAS. The institutional review board approved this retrospective study of the participating center (Shanxi Province Cancer Hospital), and the need for informed patient consent was waived. There were 156 males and 207 females. Their ages ranged from 43 to 80, with a median age of 62. There were 236 smokers and 127 non-smokers, 164 cases of EGFR mutations and 199 wild type cases, and 84 cases of KRAS mutations, and 279 wild type cases. The validation data were from the public dataset, The Cancer Imaging Archive (TCIA; http://www.cancerimagingarchive.net/). The NSCLC radiogenomics data in TCIA consisted of 211 patients, including 162 patients who satisfied the experimental requirements: CT and clinical information integrity. There were 119 males and 43 females, among which 137 were smokers, and 25 were non-smokers. The age range was 43 to 87 years old, with a median age of 69 years. There were 25 cases of EGFR mutations, 102 wild type cases, and 32 unknown or not collected cases. There were 30 cases of KRAS mutations, 94 wild type cases, and 38 unknown or not collected cases. The clinical features of the training dataset and the validation dataset are presented in Table 1.
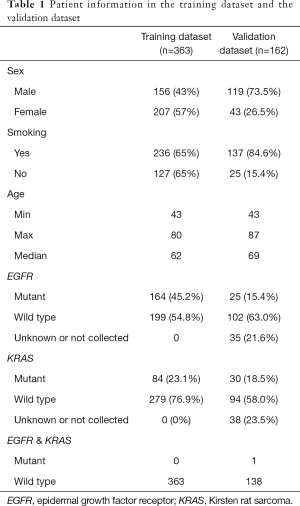
Full table
Comparison with traditional methods
First, we compared our model with the traditional clinical and radiomics models to predict EGFR and KRAS mutations. Most previous studies (7-9) used clinical features and radiomics features to predict gene mutations. The clinical features mainly included age, gender, smoking status, and tumor stage. The radiomics feature extraction process was as follows: first, the nodule region was manually segmented by a radiologist with more than 8 years of experience. The radiomics features were extracted using PyRadiomics (http://PyRadiomics.readthedocs.io/en/latest/) (39), and feature selection was conducted. We selected 1,108 radiomics features and used recursive feature elimination for feature selection. Finally, a random forest of 100 trees was constructed in the radiomics and clinical models to predict EGFR and KRAS mutations.
We compared our proposed model with the radiomics model and the clinical model in the traditional method, and we used the area under the curve (AUC), accuracy (ACC), sensitivity (SEN), and specificity (SPE) as the primary measurements. The results of the training and validation datasets are presented in Tables 2,3, respectively. Tables 2,3 suggest that our model can outperform the traditional models in prediction. The AUC value reached 81.29%, and the ACC value was 75.06% in the validation dataset when EGFR mutations were predicted. The AUC value reached 74.23%, and the ACC value was 69.64% in the validation dataset when KRAS mutations were predicted.
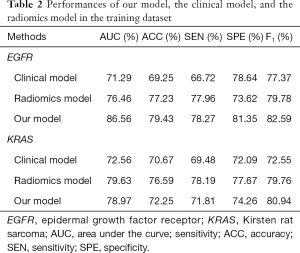
Full table
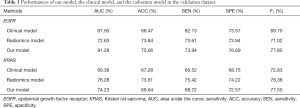
Full table
Then, we compared the prediction results of the radiomics model with those of the clinical model. The radiomics model realized relatively satisfactory performance, with 73.84% ACC of the validation data when predicting EGFR mutations and 73.61% ACC of the validation data when predicting KRAS mutations. The reason is that more features were extracted from the original CT images in the radiomics model. However, the radiomics models require the radiologist to sketch tumor regions, which takes more time.
The EGFR mutation prediction performance was better than that of KRAS in the 3 models (our deep model, the radiomics model, and the clinical model). This may be because the EGFR mutation information was better reflected in the features of the image, and the KRAS mutation information was less well reflected in the image. This is why most studies (6-11) on CT image prediction of gene mutations in lung cancer focus on predicting EGFR mutations. Moreover, according to the literature (20), it is impossible to predict KRAS mutations using semantic information. Therefore, we did not compare semantic information to predict EGFR/KRAS gene mutations with our method.
Comparison with deep methods
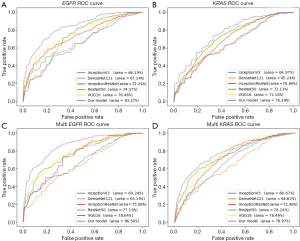
Although our model adopts inception-resnet-attention as the main component, it can be replaced with other popular deep learning models. We embedded popular deep learning networks (VGG16, ResNet50, DenseNet121, InceptionV3, and Inception-ResNet-v2) into our model for comparison and conducted single-task training (prediction of KRAS and EGFR separately) and multi-task training (simultaneous prediction of KRAS and EGFR). We obtained the receiver operating characteristic (ROC) curves (as shown in Figure 6), AUC, SEN, and SPE (as presented in Table 4).

Full table
The results in Figure 6 suggest that the inception-resnet-attention model that we selected achieved the best performance. When predicting EGFR mutation in single-task, our model’s AUC value was 8.78% higher than VGG16, 11.53% higher than ResNet50, 23.88% higher than DenseNet121, 25.65% higher than InceptionV3, and 13.56% higher than Inception-ResNet-v2. When predicting EGFR mutation in multi-task, our model’s AUC value was 10.07% higher than VGG16, 12.14% higher than ResNet50, 32.78% higher than DenseNet121, 25.01% higher than InceptionV3, and 15.29% higher than Inception-ResNet-v2. When predicting single-task and multi-task EGFR mutations, the AUC values that were obtained by our model were 13.56% and 15.29% higher, respectively than those of Inception-ResNet-v2. Hence, the addition of the attention mechanism significantly improved the performance of the model.
Table 4 presents the models’ ACC, SEN, and SPE when predicting EGFR and KRAS in a single-task and multi-task. Table 4 suggests that in almost all models, the multi-task results were better than the single-task results. Taking our MMDL model as an example, for single-task prediction of EGFR, the ACC was 73.09%, and for multi-task prediction, the prediction ACC was 75.06%. The reason is that in deep learning in a multi-task prediction, the tasks will promote and influence each other, and the results will be better than the prediction result of a single task. According to the overall results, the mutation prediction results for EGFR were better than those for KRAS, which is probably due to the sensitive of EGFR to images. Taking our MMDL model as an example, the ACC in single-task prediction of EGFR was 7.06% higher than that of KRAS, and the ACC in multi-task prediction of EGFR was 7.78% higher than that of KRAS.
Visualization of the models
Our model aims at examining the relationship between tumor image features and EGFR/KRAS mutations. We used deep learning visualization to explain the prediction process and to identify the tumor regions that are most relevant to the detection of EGFR mutations and KRAS mutations. Since our model is an end-to-end model, the visualization of tumor areas can facilitate clinicians’ focus on the tumor areas that require attention.
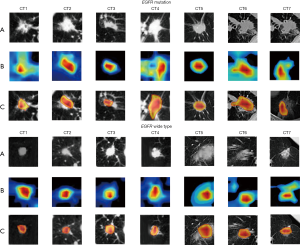
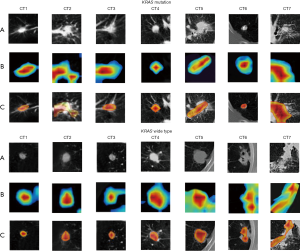
To evaluate our proposed MMDL model’s feature extraction performance and understand why excellent feature extraction performance can be realized on CT images, we visualized the middle layer of our deep learning model, as illustrated in Figures 7,8. We randomly selected 7 nodules between the EGFR mutant/wild type and the KRAS mutant/wild type (the first row). The class activation map (CAM; the second row) of the last convolutional layer was visualized, where CAM is a heat map that highlights the decision maker’s attention when making the decision, which reveals areas that are related to the prediction (47). Furthermore, we overlaid the CAM on the original nodule image to more intuitively display the nodule area associated with the prediction (the third row). Figures 7,8 present the activation results of EGFR and KRAS mutations and the wild types, respectively. Rows (A), (B), and (C) in Figures 7,8 represent the tumor area, CAM and suspicious area, respectively. Figures 7,8 suggest that the proposed MMDL model can correctly activate the corresponding nodule area for EGFR or KRAS.
Comparison of results among multiple views
To evaluate the advantages of multiple views, we compared different single views and various combinations of views. The comparison results are presented in Figure 9 and Table 5. Following our hypothesis, multi-views outperformed every single view, as multi-views contained sufficient nodule information. In predicting the single view, views 4, 6, 7, and 9 achieved superior performance as they obtained more information than the remaining views.
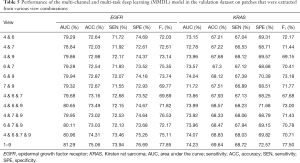
Full table
We also compared the prediction results under various combinations of views 4, 6, 7, and 9 to explore the prediction performances under various combinations, as presented in Table 5. The results demonstrated that even the combination of the most accurate prediction, that is, the combination of views 4, 6, 7, and 9, could not reach the prediction performance of all views.
Comparison with the state of the art methods
Most studies have proven that CT image features can predict EGFR mutations. The single-task prediction of EGFR mutations in our study was compared with the state of the art methods. The comparison results are presented in Table 6. Studies A, B, C, and D predicted gene mutations based on deep learning methods, and the remaining studies utilized radiomics or traditional statistical methods.

Full table
Among the deep learning methods, A used pathological images to predict mutations in multiple genes (STK11, EGFR, FAT1, SETBP1, KRAS, and TP53), with a predictive average AUC of 0.754. A had a larger dataset and was an earlier study in the literature on using deep learning to predict gene mutations. In B, CT images were used to predict gene mutations using deep learning methods. An end-to-end deep learning model was constructed based on 844 datasets, and a predicted AUC of 0.81 was obtained. C used 3D CNN to predict EGFR mutations in lung adenocarcinoma. Predicted AUC values of 0.776 and 0.838 were achieved without clinical data and with clinical data, respectively, which supported the necessity of adding clinical information and demonstrated that adding a small amount of clinical information to the training model could effectively improve the predictive performance of the model. D had a larger dataset, extracted richer features (deep features, clinical features, and radiomics features), had a higher AUC value of 0.834 and obtained higher SEN and SPE than methods A, B, and C.
Methods E, F, G, and H used radiomics features to predict gene mutations. G had the largest dataset, extracted clinical characteristics, and achieved a higher AUC value of 0.775.
Method I extracted semantic features for prediction and obtained excellent classification performance (AUC=0.89), but the extraction of semantic features required professional radiomics physicians. Compared with method I, our method does not require specialized radiomics physicians to participate in the extraction of semantic features, while also achieving excellent classification performance. Method I also showed that semantic features could accurately predict EGFR mutations but not KRAS mutations. However, our model can simultaneously predict multiple gene mutations and do not require physicians to extract semantic features, which further demonstrates our deep learning model’s superior performance.
Method J used PET/CT to extract SUVmax and clinical features and constructed a multivariate analysis model with an AUC value of 0.77 and an ACC rate of 77.6%.
Our model learns nodules’ features from multiple views, inputs the multi-tasking deep learning model, and simultaneously predicts EGFR and KRAS mutations. The multi-task prediction tasks influence and promote each other. The results demonstrate that our model achieves higher ACC, SEN, and SPE than other available deep and radiomics models.
Discussion
In our study, an MMDL framework for predicting EGFR and KRAS mutations using non-invasive CT images in NSCLC was proposed. The results suggested that our model is a feasible model for predicting both EGFR and KRAS mutations in NSCLC. Our method’s main advantage is that it automatically learns CT image features, thereby eliminating the need to extract features manually. Furthermore, a small amount of patient personal information (age, gender, and smoking status) was included in our model’s learning process, meaning that more prior knowledge can be acquired, and the robustness of the learning model can be increased. The experimental results demonstrated that our method could simultaneously predict EGFR and KRAS mutations and outperform single predictions.
Our method is a non-invasive auxiliary detection method suitable for avoiding invasive damage when surgery and biopsy are not convenient. Also, CT images are easily available throughout the treatment period to monitor EGFR and KRAS genetic mutations. The acquisition of CT images is relatively inexpensive in terms of cost and time. Moreover, our method does not require physicians’ domain knowledge, facilitating the economical and convenient prediction of EGFR and KRAS mutations.
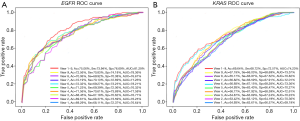
To demonstrate that transfer learning could improve the gene mutation prediction performance in terms of prediction ACC, we evaluated the performances of pre-trained inception-attention-resnet and scratch inception-attention-resnet with the same architecture. We used cat vs. dog to train inception-attention-resnet in 50 epochs. The results in Figure 10 suggest that pre-trained inception-attention-resnet achieved higher prediction performance than scratch inception-attention-resnet.

Furthermore, we examined the effect of the addition of clinical information on the prediction results in the multi-task prediction. The results (Tables 7,8) suggest that the addition of clinical information to the model significantly improved the performance. For predicting EGFR mutations, the ACC value increased by 8.50% in the training dataset and 5.82% in the validation dataset. The AUC value increased by 6.03% in the training dataset and 4.15% in the validation dataset. The ACC value increased by 7.77% in the training dataset and by 8.59% in the validation dataset for predicting KRAS mutations. The AUC value increased by 6.30% in the training dataset and by 3.20% in the validation dataset.
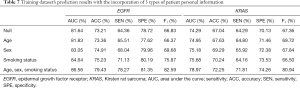
Full table
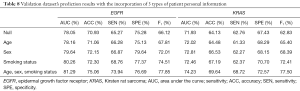
Full table
We examined the effects of adding various types of clinical information on the prediction results. The experimental results demonstrated that the types of information, in increasing order of influence on the prediction results, were age, gender, and smoking status, which we added to the clinical information. Age had the smallest effect among the 3 factors. The predicted AUC for EGFR mutations after adding age increased by 0.11, and ACC increased by 0.13. The predicted AUC value for KRAS mutations after the addition of age increased by 0.09, and the ACC increased by 0.35. The effect of gender was weaker than the effect of smoking status.
The incidence of EGFR mutations was significantly higher in women and non-smokers. Although EGFR mutations were more often detected in early patients than in advanced patients, the difference was not significant.
To illustrate our proposed model’s effectiveness after the addition of attention strategy, we randomly selected different types of nodules more intuitively for visualizing EGFR mutation or wild type, and KRAS mutation or wild type. We visualized their feature activation maps before and after adding the attention mechanism, as shown in Figures 11,12. Row A is the original nodule image, row B is the feature activation map without the attention mechanism, and row C is the feature activation map with the attention mechanism. As can be seen from Figures 11,12, adding the attention mechanism to the model significantly reduced the meaningless low-level background blue features and highlighted the yellow features of the high-level nodule region with the most discriminative value, thereby significantly improving the prediction performance of the model.

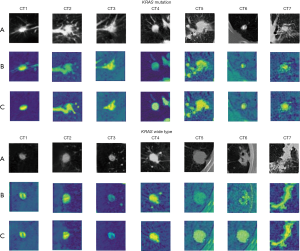
Our study used deep learning models to detect EGFR and KRAS mutations and to explore their relationship. In multi-task prediction in the validation dataset, the AUC for detecting EGFR mutations was 81.29%, the ACC was 75.06%, the SEN was 73.94%, and the SPE remained 76.69%. The overall radiomics model validation results were an AUC of 72.63%, an ACC of 73.84%, the SEN of 73.61%, the SPE of 73.94%. Therefore, our MMDL model outperformed the radiomics model in predicting EGFR and KRAS mutations. Furthermore, the radiomics model requires the radiologist to delineate the lesion area in advance. These results suggest that the proposed deep learning model can predict EGFR and KRAS mutations simultaneously. However, the use of non-invasive image analysis to predict gene mutations is never a substitute for biopsies.
Compared with pathological biopsy, the main advantage of image analysis is that it provides an alternative solution if the patient’s physical condition is not suitable for biopsy. In the process of exploring tumor treatment, the proposed prediction model can be repeatedly tracked.
Conclusions
In our study, a deep learning model that uses non-invasive CT images was proposed to predict EGFR and KRAS mutations in NSCLC. We used a 363-case dataset collected by our partner hospital to build our model, and the accuracies for predicting EGFR and KRAS mutations were 79.43% and 72.25%, respectively. Using 163 cases available in the public TCIA dataset to evaluate our model, the accuracies for detecting EGFR and KRAS mutations were 75.06% and 69.64%, respectively. The results suggest that our model is a feasible model for predicting EGFR and KRAS mutations in NSCLC simultaneously. Our method’s main advantage is that it is a non-invasive auxiliary detection method suitable for avoiding invasive damage if surgery and biopsy are not convenient. The acquisition of CT images is relatively inexpensive and fast. Moreover, our method does not require a physician’s domain knowledge, facilitating the economic and convenient prediction of gene mutations in EGFR and KRAS.
Although satisfactory prediction results have been obtained for predicting EGFR and KRAS mutations simultaneously, our model has several limitations. First, the dataset was small, and some data were incomplete. Furthermore, the proportion of gene mutation data was small in our collected dataset. Therefore, in future research, we will collect additional data and use them to build our model. Second, only 1 patient in our dataset had mutations in both EGFR and KRAS. Thus, we did not study this patient and whether it is possible to predict both genes simultaneously through further image study.
Additionally, pathological images were not used in our study, but other studies have shown that pathological images can be used to predict gene mutation information. Therefore, in our subsequent studies, we will also attempt to utilize pathological images. Finally, the effect of staging on the results was not reflected in our comparative experiments because it is not possible to conduct comparative training in deep models using so few cases of mutations in various stages. In subsequent studies, we will collect more data to compare among different stages and explore gene mutations’ prediction using images of different stages.
Acknowledgments
Funding: This work is partly supported by the National Natural Science Foundation of China (Grant number 61872261, 61972274), the Open Funding Project of State Key Laboratory of Virtual Reality Technology and Systems, Beihang University (Grant No. 2018-VRLAB2018B07), and the Research Project Supported by Shanxi Scholarship Council of China (201803D421036, 201801D121139). This work is also partly supported by the Key Research and Development (R&D) Projects of Shanxi Province (No.201803D31168) and the Applied Basic Research Programs of Shanxi Province (No. 201801D121307).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-600). The authors have no conflicts of interest to declare.
Ethical Statement: This retrospective study was approved by the institutional review board of the participating center (Shanxi Province Cancer Hospital), and the need for informed patient consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Motono N, Funasaki A, Sekimura A, Usuda K, Uramoto H. Prognostic value of epidermal growth factor receptor mutations and histologic subtypes with lung adenocarcinoma. Med Oncol 2018;35:22. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, Kurata T, Chiappori A, Lee KH, De Wit M. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N Engl J Med 2018;379:2342-50. [Crossref] [PubMed]
- Ettinger DS, Wood D, Akerley W, Bazhenova L, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, Damico TA, Dilling TJ. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J Natl Compr Canc Netw 2016;14:255-64. [Crossref] [PubMed]
- Nakatani K, Yamaoka T, Ohba M, Fujita K, Arata S, Kusumoto S, Takitakemoto I, Kamei D, Iwai S, Tsurutani J. KRAS and EGFR Amplifications Mediate Resistance to Rociletinib and Osimertinib in Acquired Afatinib-Resistant NSCLC Harboring Exon 19 Deletion/T790M in EGFR. Mol Cancer Ther 2019;18:112-26. [Crossref] [PubMed]
- Digumarthy SR, Padole AM, Gullo RL, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore) 2019;98:e13963 [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. Erratum in: Nat Commun 2014;5:4644. [Crossref] [PubMed]
- Rizzo S, Raimondi S, de Jong EEC, van Elmpt W, De Piano F, Petrella F, Bagnardi V, Jochems A, Bellomi M, Dingemans AM, Lambin P. Genomics of non-small cell lung cancer (NSCLC): Association between CT-based imaging features and EGFR and K-RAS mutations in 122 patients-An external validation. Eur J Radiol 2019;110:148-55. [Crossref] [PubMed]
- Liu Y, Balagurunathan Y, Garcia A, Stringfield O, Ye ZX, Gillies R. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-448.e6. [Crossref] [PubMed]
- Rios Velazquez E, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH, Gillies R, Quackenbush J, Aerts H. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. [Crossref] [PubMed]
- Park J, Kobayashi Y, Urayama KY, Yamaura H, Yatabe Y, Hida T. Imaging Characteristics of Driver Mutations in EGFR, KRAS, and ALK among Treatment-Naive Patients with Advanced Lung Adenocarcinoma. PLoS One 2016;11:e0161081 [Crossref] [PubMed]
- Zhang L, Chen B, Liu X, Song J, Fang M, Hu C, Dong D, Li W, Tian J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl Oncol 2018;11:94-101. [Crossref] [PubMed]
- Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, Fumagalli C, Spitaleri G, Rampinelli C, De Marinis F, Spaggiari L, Bellomi M. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol 2016;26:32-42. [Crossref] [PubMed]
- Guan J, Chen M, Xiao N, Li L, Zhang Y, Li Q, Yang M, Liu L, Chen L. EGFR mutations are associated with higher incidence of distant metastases and smaller tumor size in patients with non-small-cell lung cancer based on PET/CT scan. Med Oncol 2016;33:1. [Crossref] [PubMed]
- Yang Y, Yang Y, Zhou X, Song X, Liu M, He W, Wang H, Wu C, Fei K, Jiang G. EGFR L858R mutation is associated with lung adenocarcinoma patients with dominant ground-glass opacity. Lung Cancer 2015;87:272-7. [Crossref] [PubMed]
- Siegele BJ, Shilo K, Chao BH, Carbone DP, Zhao W, Ioffe O, Franklin WA, Edelman MJ, Aisner DL. Epidermal growth factor receptor (EGFR) mutations in small cell lung cancers: Two cases and a review of the literature. Lung Cancer 2016;95:65-72. [Crossref] [PubMed]
- Lee HJ, Kim YT, Kang CH, Zhao B, Tan Y, Schwartz LH, Persigehl T, Jeon YK, Chung DH. Epidermal growth factor receptor mutation in lung adenocarcinomas: relationship with CT characteristics and histologic subtypes. Radiology 2013;268:254-64. [Crossref] [PubMed]
- Yip SS, Kim J, Coroller TP, Parmar C, Velazquez ER, Huynh E, Mak RH, Aerts HJ. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non-Small Cell Lung Cancer. J Nucl Med 2017;58:569-76. [Crossref] [PubMed]
- Usuda K, Sagawa M, Motono N, Ueno M, Tanaka M, Machida Y, Matoba M, Taniguchi M, Tonami H, Ueda Y, Sakuma T. Relationships between EGFR mutation status of lung cancer and preoperative factors - are they predictive? Asian Pac J Cancer Prev 2014;15:657-62. [Crossref] [PubMed]
- Gevaert O, Echegaray S, Khuong A, Hoang CD, Shrager JB, Jensen KC, Berry GJ, Guo HH, Lau C, Plevritis SK, Rubin DL, Napel S, Leung AN. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep 2017;7:41674. [Crossref] [PubMed]
- Li S, Ding C, Zhang H, Song J, Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys 2019;46:4545-52. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton GE. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Liu C, Hu SC, Wang C, Lafata K, Yin FF. Automatic detection of pulmonary nodules on CT images with YOLOv3: development and evaluation using simulated and patient data. Quant Imaging Med Surg 2020;10:1917-29. [Crossref] [PubMed]
- Wang Y, Zhou L, Wang M, Shao C, Shi L, Yang S, Zhang Z, Feng M, Shan F, Liu L. Combination of generative adversarial network and convolutional neural network for automatic subcentimeter pulmonary adenocarcinoma classification. Quant Imaging Med Surg 2020;10:1249-1264. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, Ko JM, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Wang S, Shi JY, Ye ZX, Dong D, Yu DD, Zhou M, Liu Y, Gevaert O, Wang K, Zhu YB, Zhou HY, Liu ZY, Tian J. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019;53:11. [Crossref] [PubMed]
- Xiong JF, Jia TY, Li XY, Yu W, Xu ZY, Cai XW, Fu L, Zhang J, Qin BJ, Fu XL, Zhao J. Identifying epidermal growth factor receptor mutation status in patients with lung adenocarcinoma by three-dimensional convolutional neural networks. Br J Radiol 2018;91:20180334 [Crossref] [PubMed]
- Li XY, Xiong JF, Jia TY, Shen TL, Hou RP, Zhao J, Fu XL. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J Thorac Dis 2018;10:6624-35. [Crossref] [PubMed]
- Xiong J, Li X, Lu L, Schwartz LH, Fu X, Zhao J, Zhao B. Implementation Strategy of a CNN Model Affects the Performance of CT Assessment of EGFR Mutation Status in Lung Cancer Patients. IEEE Access 2019;7:64583-91.
- Oquab M, Bottou L, Laptev I, Sivic J, Ieee. Learning and Transferring Mid-Level Image Representations using Convolutional Neural Networks. IEEE Conference on Computer Vision and Pattern Recognition. New York: IEEE; 2014: 1717-24.
- Gondara L. Medical image denoising using convolutional denoising autoencoders. 2016 IEEE 16th International Conference on Data Mining Workshops. International Conference on Data Mining Workshops. New York: IEEE; 2016: 241-6.
- Esteva A, Kuprel B, Novoa RA. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Oncologie 2017;19:407-8.
- Shen W, Zhou M, Yang F, Dong D, Yang C, Zang Y, Tian J, editors. Learning from Experts: Developing Transferable Deep Features for Patient-Level Lung Cancer Prediction. 19th International Conference on Medical Image Computing and Computer-Assisted Intervention. Med Image Comput Comput Assist Interv 2016:124-131.
- Shin HC, Roth HR, Gao MC, Lu L, Xu ZY, Nogues I, Yao JH, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. [Crossref] [PubMed]
- Shen W, Zhou M, Yang F, Yu DD, Dong D, Yang CY, Zang YL, Tian J. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognit 2017;61:663-73. [Crossref]
- Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MMW, Naqibullah M, Sanchez CI, van Ginneken B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. [Crossref] [PubMed]
- Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 2017;12:e0188290 [Crossref] [PubMed]
- Guan QJ, Huang YP. Multi-label chest X-ray image classification via category-wise residual attention learning. Pattern Recognit Lett 2020;130:259-66. [Crossref]
- Liu M, Zhang J, Adeli E, Shen D. Deep Multi-Task Multi-Channel Learning for Joint Classification and Regression of Brain Status. Med Image Comput Comput Assist Interv 2017;10435:3-11.
- Xie YT, Zhang JP, Xia Y, Fulham M, Zhang YN. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inf Fusion 2018;42:102-10. [Crossref]
- Yang WK, Zhao JJ, Qiang Y, Yang XT, Dong YY, Du QQ, Shi GH, Zia MB. DScGANS: Integrate Domain Knowledge in Training Dual-Path Semi-supervised Conditional Generative Adversarial Networks and S3VM for Ultrasonography Thyroid Nodules Classification. Med Image Comput Comput Assist Interv 2019;558-66.
- Patro S, Sahu KK. Normalization: A preprocessing stage. arXiv preprint arXiv:1503.06462, 2015.
- Szegedy C, Ioffe S, Vanhoucke V, Alemi AA, Aaai. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. AAAI Conf. Artif. Intell., AAAI. 2017; 4278-84.
- Wang F, Jiang M, Qian C, Yang S, Li C, Zhang H, Wang X, Tang X. Residual Attention Network for Image Classification. Computer vision and pattern recognition; 2017; 6450-8.
- Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Computer vision and pattern recognition; 2015;3431-40.
- Deng J, Dong W, Socher R, Li L, Li K, Feifei L, editors. ImageNet: A large-scale hierarchical image database. Computer vision and pattern recognition; 2009.
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]






