
Applications of artificial intelligence in nuclear medicine image generation
Background
Artificial intelligence (AI) technology has been rapidly adopted in various fields (1-6). With the accumulation of medical data and AI technology development, especially deep learning (DL), data-driven based precision medicine has quickly progressed (7,8). Among the applications of AI in medicine, the most striking is for medical imaging. For example, radiomics is a method that extracts high-dimensional image features, either explicitly by traditional image analysis methods (such as textures) or implicitly by convolutional neural networks (CNNs). It is then used for different clinical applications, including diagnosis, treatment monitoring, and correlation analyses with histopathology or specific gene mutation status (9-13). In nuclear medicine imaging, AI has also focused on using the imaging data (14). Machine learning (ML) is an important branch of AI. Traditional ML methods have been widely used in medicine for a long time, including naive Bayes, support vector machines, and random forests. The applications of ML in nuclear medicine imaging include disease diagnosis [positron emission tomography (PET) (15), single-photon emission computed tomography (SPECT) (16,17)], prognosis [PET (18), SPECT (19)], lesion classification [PET (20), SPECT (21,22)], and imaging physics (23). In recent years, DL technologies such as CNNs, artificial neural networks (ANNs), and generative adversarial networks (GANs) have developed very fast and shown better performance than traditional ML in some cases. The applications of DL in nuclear medicine include disease diagnosis [PET (24), SPECT (25,26)], imaging physics [PET (27), SPECT (28)], image reconstruction [PET (29), SPECT (30)], image denoising [PET (31,32), SPECT (33)], image segmentation [PET (34), SPECT (35)], image classification [PET (36), SPECT (37)], and internal dose prediction (38,39).
More than 200 papers have been cited in this review. Due to the wide range of AI applications in nuclear medicine imaging, we did not try to cover all AI applications. We focused mainly on image generation development, including imaging physics, image reconstruction, image postprocessing, and internal dosimetry, over the last 5 years. We obtained the literatures by searching keywords in PubMed (“artificial intelligence”, “machine learning”, “deep learning”, “nuclear medicine imaging”, “SPECT”, “PET”, “correction”, “reconstruction”, “low-dose imaging”, “denoising”, “fusion”, or “dosimetry”, and so on). We also included conference recordings of SPIE (International Society for Optics and Phonics), NSS/MIC (IEEE Nuclear Science Symposium and Medical Imaging Conference), and MICCAI (Medical Image Computing and Computer-Assisted Intervention). The last search date was 18 September 2020. Although there were some existing reviews of nuclear medicine (especially for PET) in the literature (40-45), our review focused on AI applications in improving the quality of nuclear medicine imaging. The section “Imaging physics” provides AI applications in imaging physics, including the generation of attenuation maps and the estimation of scattered events. The section “Image reconstruction” reviews AI applications in image reconstruction, including the optimization of the reconstruction algorithm. The section “Image postprocessing” covers AI applications in image postprocessing, including the generation of high-quality reconstructed images (in full-dose or low-dose imaging) and image fusion. The section “Internal dosimetry” reviews AI applications in internal dose prediction. The section “Discussion and conclusions” provides a discussion and summary.
Imaging physics
As the two most widely used nuclear medicine imaging technologies, both PET and SPECT quantify radionuclides’ distribution in a recipient by measuring the gamma photons emitted from that recipient. In practice, gamma photons are attenuated due to tissue absorption in the recipient. The attenuation effect causes the number of photons to be less than expected and results in nonuniform deviations in the radioactive distribution due to the different attenuation paths from the tracer to the detector (46). Another factor affecting image quality is scattered photons. Scattering events will cause severe artifacts and quantitative errors. The emergence of AI technology is not a complete replacement of traditional methods but rather an auxiliary means to find function mapping relationships and largely depends on the model structure, data range, and training process. Recently, Wang et al. (23) and Lee et al. (47) separately summarized the wide application of ML and DL in PET attenuation correction (AC), and they explored the performance of AI in PET AC under different input conditions. Here, we extended the search scope to nuclear medicine imaging (PET/SPECT) and evaluated the AI technology from two angles, namely AC and scatter correction. In each part, we discussed the application of different types of AI structures under different imaging methods.
AC
Stand-alone nuclear medicine imaging
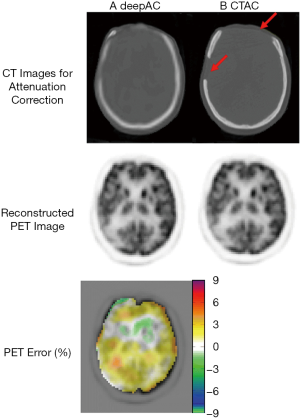
The content included in this part mainly focuses on PET. Usually, due to space limitations in the scanner, only PET is built for some applications such as ambulatory microdose PET (48) and helmet PET (49). The attenuation coefficients have usually been obtained via scanning external radiation sources (such as X-ray or barium sources), which is typically time-consuming and introduces additional radiation exposure (50). With AI’s help, pseudo-CT (pCT) images/corrected nuclear medical images can be obtained quickly for AC. Once the training is completed, the label image will no longer be needed, which will avoid additional costs and radiation risks. In recent years, some researchers have used convolutional autoencoder (CAE) and convolutional encoder-decoder (CED) structures to predict pCT images from attenuation uncorrected PET images, as shown in Figure 1. The CAE structure was originally proposed for unsupervised feature learning and later widely used in image denoising and other fields (51). The CED structure is similar to the CAE structure. The most well-known structure is the U-net. Unlike CAE, U-net augments the contracting path that enables high-resolution features to be combined in the output layers. Liu et al. (52) used a CED structure to predict pCT from uncorrected 18F-fluordeoxyglucose (18F-FDG) PET images. Their method’s quantitative PET results showed that the average error in most brain regions was less than 1%. However, for certain areas, such as the skull’s cortical area, significant errors were observed. An abnormal situation is shown in Figure 2, where the predicted pCT showed obvious differences in the skull (red arrow). Similarly, in the work from Hwang et al. (53), DL was employed to reconstruct activity and attenuation maps from PET raw data simultaneously. The results showed that the combination of CAE and CED could achieve better results than CAE or CED alone. In contrast to the above scheme, Shiri et al. (54) and Yang et al. (55) used a CED structure to produce AC PET from non-AC PET in the image space for brain imaging. The difference between the two studies was that the latter also took scatter correction into consideration. The input and output images can be similar and have a uniform structure and edge information; however, when the test data pattern is not represented in the training cohort, a significant error will be seen. For example, in the study by Yang et al. (55), the average skull density of the 34 subjects was 685.6±61.1 Hounsfield units (HU; min: 569.6 HU, max: 805.1 HU), whereas 1 subject had an uncommonly low skull density (475.1 HU). This was also translated into a major quantitative difference of 48.5%. With the information of time-of-flight, the estimation of AC factors could be further improved (56).

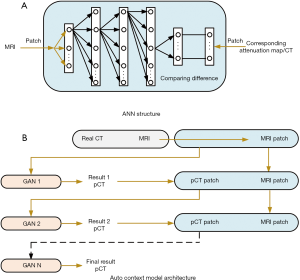
In comparison with other DL, GAN is more popular in attenuation map generation. Generally, the generator network is used to predict an attenuation map, and the discriminator network is used to distinguish the predicted pCT image and the attenuation map. These two networks are in a competitive relationship. If the discriminator network can distinguish the estimated and real images well, then the generator network needs to perform better; otherwise, the discriminator network should be strengthened. Shi et al. (57) designed a GAN to indirectly produce the attenuation map of SPECT from the emission data. Their inputs were the photopeak window and scatter window SPECT images. This approach can effectively learn the hidden information related to attenuation in the emission data.
Further, Armanious et al. (58) established and evaluated a conditional GAN (cGAN) method for the AC of 18F-FDG PET images of the brain without using anatomical information. Cycle-consistency network (cycle-GAN) is composed of two mirror-symmetric GANs and has been used for whole-body PET AC [Dong et al. (59,60)]. Dong et al. (60) combined a U-net structure and a residual block to form a new cycle-GAN generator in which the residual structure is important for learning. The method showed similar quantitative performance on heart, kidney, liver, and lesions with the gold-standard CT-based method, and the average whole body error was only 0.62%±1.26%.
In comparison with brain scanning, the AC for whole-body imaging is more challenging due to more unexpected factors such as truncation and body motion. Dong et al. (59,60) demonstrated a GAN network’s feasibility in predicting whole-body pCT/corrected PET images. The use of cycle-GAN makes the cycle introduce inverse transformation, which adds more constraints to the generator. This effectively prevents the model from crashing and can help the generator find the unique mapping. The proposed method may avoid the quantization bias caused by CT/magnetic resonance imaging (MRI) truncation or registration error. Besides, Shiri et al. (61) designed a couple of 2D and 3D deep residual networks to achieve joint attenuation and scatter correction for the whole uncorrected body. It is worth noting that they used more than 1,000 patients for training, which was useful for network training. On the test set of 150 patients, the voxel-wise relative errors (%) were –1.72%±4.22%, 3.75%±6.91%, and –3.08±5.64 for 2D slices input, 3D slices input, and 3D patches input, respectively. The diversity of cases in this large data training set (1,150 patients), including disease-free and pathological patients with various indications, such as age, body weight, and disease type, ensured the comprehensiveness of the data, which brings more reliability to the prediction results, and training directly in the image domain can ensure accurate lesion conspicuity and quantitative accuracy.
Compared with predicting the pCT image required for AC, it seems more convenient to directly predict the attenuation-corrected activity image. Besides, the network’s input and output have similar anatomical structures, which is beneficial for training. However, the method of directly predicting the corrected active image has some obvious limitations (54,55,58). It is a data-driven method that skips the attenuation/scattering correction related to the imaging’s physical properties, which brings some uncertainty to the reconstructed activity images. The quality of the prediction results will completely depend on the quality of the training data (e.g., the number of training sets, the choice of labels, whether the training data is suitable for different radiotracers). A more complete and larger data set offers more comprehensive variability, which is a prerequisite for determining prediction accuracy. Also, whether the training set contains enough pathological patterns has a significant impact on the accuracy and robustness of clinical predictions (the same applies to predicting pCT images). Although there are many AI solutions, further evaluation of the clinical benefit is needed. It is very important to develop a clinically useful model for combining domain knowledge and AI technology.
Hybrid nuclear medicine imaging
For PET/CT or SPECT/CT, CT-based AC (CTAC) has been the standard technique. However, the risk of CT radiation exposure is a concern of the public, which should be minimized for young subjects (62). Besides, when the CT structure might be truncated, CTAC may not provide satisfactory results due to the missing attenuation information for this part. Traditional approaches to solving this problem rely on prior information and non-attenuation corrected images (63,64). Recently, Thejaswi et al. (65) proposed an inpainting-based context encoder structure for SPECT/CT imaging, which inferred the truncated CT image’s missing information from the untruncated CT image to obtain the attenuation map. Thus, they provided a new way to solve the truncation problem.
In comparison with PET/CT, the AC in PET/MRI is more challenging as the voxel intensity of MRI cannot reflect the photon attenuation characteristics; therefore, the attenuation map of PET cannot be obtained directly (66,67). Traditional MR-based AC (MRAC) methods mainly include segmentation-based methods, atlases-based methods, template-based methods, emission, and transmission-based methods. Among them, the atlases-based method is easily affected by individual differences in the anatomical structure; the template-based method is more sensitive to individual differences in anatomy, case differences, and organ movement; emission and transmission-based methods are prone to slow imaging speed and long operation time due to the use of alternating iterative algorithm; and segmentation-based method has relatively good performance in speed and robustness, and anatomical structure (68,69). Nowadays, the segmentation-based method is the default AC method for the commercial PET/MR scanner. A leading challenge in generating attenuation maps from MR images is distinguishing between bone and air regions (e.g., mastoid of the temporal bone and the bone-fat of pelvic regions). With the development of ultrashort echo time (UTE) sequences such as zero echo time (ZTE) and UTE, the challenge could be alleviated for brain imaging but with high noise and image artifacts (68). Researchers have developed several ML-based methods to improve the segmentation-based method of MRAC, including random forest classifiers (70,71), support vector machines (72), Markov random fields (73), and clustering (74). The application of segmentation-based ML in MRAC for brain PET imaging has been well summarized by Mecheter et al. (75). Here we supplemented AI applications, especially DL, to their work and included the application for non-brain PET/MR imaging.
AI (mainly DL) is now actively used to train mapping relationships to predict pCT data/attenuation map from MR data. Through extensive training of CT and MR images with good registration, the link between MR images and HU in the CT images can be established, thus eliminating the need for CT scan for AC. Variant DL methods such as ANN (76), CAE (77,78), CED (79), and GAN techniques (80) have been explored to perform MRAC in brain PET imaging. An example using ANNs to map the MRI image to the corresponding attenuation map is shown in Figure 3A, which is a feedforward neural network with five layers (76), They have demonstrated that the ANN model trained with one subject and BrainWeb phantom data can be applied well to other subjects. Most CAE and CED models were used to learn the mapping relationship between 2D MRI slices and 2D CT due to the heavy computation burden. Directly learning the 3D model is a challenge for computing; however, this approach is unnecessary because 2D slices contain a large amount of contextual information. Bradshaw et al. (81) designed a 3D deep CNN and demonstrated that DL of pelvic MRAC using only diagnostic MRI sequences is feasible. Beyond CNN, GAN applications are also gradually increasing, but they are limited to the brain and pelvic area (82). Nie et al. (83,84) implemented a fully convolutional network (FCN) training in pelvic imaging by using an antagonistic training strategy. It is worth noting that an auto context model is applied to iterate the network output continuously, and a GAN can sense the context, which improves the network modeling ability to some extent, as shown in Figure 3B. Jang et al. (85) inputted UTE images into a CNN network to achieve a robust estimation of pCT images.
It should be noted that the MR image in this study was obtained by dual-echo ramped hybrid encoding. Ladefoged et al. (86,87) used UTE images to obtain attenuation maps, and it is gratifying that they extended this method to the field of children (88). Similarly, Dixon MR images and ZTE MR images were inputted into the CNN framework separately or together to synthesize corresponding pCT images (89-92). Compared with the direct use of MRI images, The MR sequence has high signal intensity to the bones and can achieve better performance in MRAC, but it requires longer scanning time and has limited diagnostic value.
Although AI has shown great potential in CTAC/MRAC, most of the applications were limited to the brain (few to pelvic). Even if the same network structure as the brain is used, it is difficult to realize the whole body’s AC directly. This is mostly due to the high anatomical heterogeneity and inter-individual variability of the whole body. The only whole body AC based on AI has been presented by Dong et al. (59,60) and Shiri et al. (61); their work was focused on predicting the corrected PET image/pCT from the uncorrected PET image, as mentioned in the previous section. The major obstacles of AI in MRAC for whole-body scanning are insufficient representative training data sets and registration errors between training pairs (40). Wolterink et al. (93) explored the use of cycle-GAN to train unpaired MR and CT images, and the quantitative results on the test set of six images [compared to the reference CT, the peak-signal-to-noise ratio (PSNR) of the synthetic CT] was 32.3±0.7, which shows that this idea is feasible. With the continuous improvement of accumulating public datasets comes the gradual promotion of unpaired data training technology in the MRAC field. We believe this will reduce the impact of registration accuracy on the application of AI in MRAC, and we have reason to expect more universal AC methods (especially for whole-body) will appear.
Scatter correction
Traditional scatter correction methods have limitations in accuracy and noise characteristics (94,95). In general, scatter correction includes direct measurement or modeling to estimate scatter events. One of the traditional scatter correction methods is to use a lower energy window to measure the scatter image. Although double or triple energy window subtraction techniques have been continuously designed, the performance has not been significantly improved (96,97). The Monte Carlo simulation-based method is quite accurate but very time-consuming. The section “Stand-alone nuclear medicine imaging” outlined the use of AI technology to predict the activity image after attenuation directly and scatter correction, which is beneficial for independent nuclear medicine equipment. Once the AI prediction is proven effective, this type of method does not require obtaining the corresponding anatomical image, which effectively reduces cost (55,61). For PET scatter correction, Berker et al. (98) used the U-net to obtain single-scatter profiles. For brain imaging, this method has achieved high accuracy, but for beds where the high-absorption bladder extends beyond the axial field of view, the results showed poor performance. Qian et al. (99)proposed two CNNs to estimate scattering correction for PET. The first network had only 6 layers, of which the convolutional layer and the fully connected layer were used to predict multiple scatter profiles from a single scatter profile. The second network was used to obtain the total scattering distribution (both single and multiple scattering) directly from the emission and attenuation sinograms. The network structure, in this case, was unchanged. Monte Carlo simulation of scattering was used as a training label. Similar to the input of the second network used in Qian et al. (99), Xiang et al. (28) investigated a deep CNN (DCNN) structure (a 13-layer deep structure consisting of separate paths for emission and attenuation projections) for SPECT/CT scatter estimation for Y-90 nuclides. As shown in Figure 4, the DCNN and Monte Carlo dosimetry results for 90YPET showed a high degree of consistency.
Compared with whole-body imaging, the application of AI technology in specific regions (such as the brain and lungs) have better scatter correction (28,98). Although the time-consuming Monte Carlo simulation is required during the training process, it only needs to be performed once. Basically, with the increase of simulation data, scatter correction accuracy will be improved for all methods, either for non-AI or AI methods. The current method was only investigated for a single radioactive source, and the applicability for different radioactive sources has not yet been explored.
Image reconstruction
The image reconstruction from raw projection data is an inverse problem. The reconstruction algorithm in nuclear medicine includes an analytical filter back-projection (FBP) algorithm, algebraic reconstruction techniques (ARTs) algorithm, maximum likelihood algorithm [maximum likelihood expectation maximization (MLEM), ordered subset expectation maximization (OSEM)], and maximum a posterior (MAP) algorithm (100,101). The analytical methods are simple and fast, but there is a trade-off between high resolution and low noise, especially in adjacent parts where the radioactive distribution changes sharply. The maximum likelihood algorithm can simulate the physical characteristics in the process of data acquisition and better control the reconstruction quality, but this will take a longer time cost. Researchers have applied AI technology to nuclear medicine image reconstruction with AI technology development, mostly in PET reconstruction (102). The application of AI technology cannot solve the inverse problem. It essentially provides a mapping relationship to solve specific key problems in reconstruction with a data-driven solution, such as completing the transformation between the sinogram domain and the image domain or replacing traditional algorithms’ regularization. To a certain extent, the emergence of AI technology has made it possible to obtain better imaging quality without increasing hardware costs. Reader et al. (29) summarized the basic theory of PET reconstruction and the key paradigm shift used by DL in PET reconstruction. They strictly focused on raw PET data. Here, we focused the search scope on nuclear medicine image reconstruction (PET/SPECT) and introduced the application of AI to three different systems, namely static scan (shown in Figure 5), dynamic scan, and hybrid fusion.
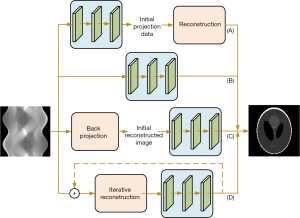
Image reconstruction in the static scan
AI applications in the projection domain
Detectors used in PET and SPECT are comprised of scintillators and photomultipliers [e.g., position-sensitive photomultiplier tubes (PSPMTs), silicon photomultipliers (SiPMs)] (103,104). Generally, large crystal arrays lead to low-resolution projection information, and thin crystal arrays can produce better visual quality; however, this approach also costs more because the detector cutting process is limited. In addition to the significant impact of low-resolution detectors on image quality, gaps or local failures of the detector due to the detector design [such as the octagonal configuration of the HRRT with eight gaps (105)] can also cause significant loss of projection data. The most common methods to complete sinogram data are interpolation-based methods (106) and penalized regression methods such as dictionary learning and discrete cosine changes. As shown in Figure 5A, compensation of the missing data in the projection space can improve the quality of recovery images (107).
Hong et al. (108) proposed a residual CNN method to predict higher resolution PET sinogram data from low-resolution sinogram data, making learning local feature information more efficient. The transfer learning scheme was incorporated into the method of dealing with poor labels and small training data sets. However, the network was gradually trained on the number of analytical simulations and Monte Carlo simulations and did not simulate attenuation and scattering events. The scheme only provided qualitative information on real data (because there was no ground truth), the result of which is shown in Figure 6. In contrast, Shiri et al. (109) used the CED model to achieve end-to-end mapping of high- and low-resolution PET images. The encoder part extracts the features of low-resolution images and effectively compresses them so that the decoder finally outputs higher-quality images. Also, Shiri et al. (110) used a similar structure to generate high-resolution PET images similar to point spread function (PSF) modeling, which can accelerate the reconstruction without complex spatial resolution modeling. The CED model was shown to be effective in image detail recovery. Besides, anatomical image-guided nuclear medicine image reconstruction technology [Schramm et al. (111)] can obtain more prior details to improve imaging resolution, which will be introduced in the section “Anatomical image-guided nuclear medicine image reconstruction”. Compared with a reconstructed image, the difference between different resolution data in the sinogram domain comes from sampling, which is only related to local information. In contrast, the difference between different resolution images in the reconstructed image comes from more complex sources. Therefore, it is desirable to obtain high-resolution projection data as much as possible in the sinogram domain.
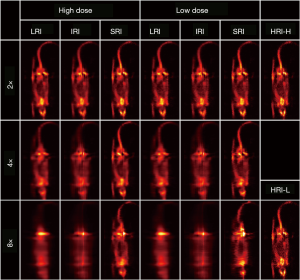
Shiri et al. (112) used a residual neural network (ResNet) to predict the full-time projection (the acquisition time is reduced from 20 to 10 s) and the full-angle projection (32 full projections reduced to 16 half projections) when using a dedicated dual-head cardiac SPECT camera with a fixed 90-degree angle for SPECT imaging. The results showed that reducing the acquisition angle can produce better predictive indicators [root mean square error (RMSE), structural similarity index measure (SSIM), and PSNR] than reducing the acquisition time. Ryden et al. (113) used the U-net structure to generate 177Lu-SPECT full projection data from sparse projections. The experiments of these works have certain guiding significance for clinical acquisition. However, reducing the acquisition time at each angle will inevitably introduce more errors. It has been shown that DL can effectively correct this error, but further shortening acquisition time is a key issue. Besides, Shiri et al. (105) used the CED structure to complement the sinogram gap generated by the HRRT scanner. The same structure was used by Whiteley et al. (114) to complement the sinogram generated by the local block failure of the detector. Liu et al. (115) combined the U-net structure with the residual structure to predict the full-loop data of the sinogram domain and the PET image domain from the partial loop data. These three tasks are similar, complementing the incomplete sinogram, and have important practical significance.
For the pre-reconstruction processing, the CNN’s task is to realize automatic learning of image features and end-to-end mapping between different images through fast reasoning. By introducing the residual learning into images, the degradation problem caused by an increase in network depth can be reduced, and the learning capability can be improved. The abundant application of AI in natural image super-resolution/generation proves that AI technology can obtain higher quality images. It is undeniable that medical images have higher requirements for intensity accuracy, especially in the sinogram domain. An obstacle that cannot be ignored is that it is often difficult for researchers to obtain enough sinogram data. Although transfer learning or data enhancement techniques can help fit the model better, the robustness and general clinical verification of the network need to be further verified. There is hope that CNN may assist in supplementing/generating high-quality sinogram images. It aims to obtain high-quality projection data with the help of AI for less sampling angle, ray beam, and acquisition time.
AI applications in direct reconstruction
As shown in Figure 5B, some studies have found that AI could be used to could obtain reconstructed images directly from projections, although this approach ignores some physics-related issues. The technology of AI can learn the mapping relationship between sinogram data and reconstructed images with a large amount of training data, which is composed of millions of parameters and garner an approximate solution to the inverse problem. However, once the training is completed, direct AI reconstruction is computationally efficient. Direct reconstruction based on AI can avoid the inaccurate assumption modeling present in traditional methods.
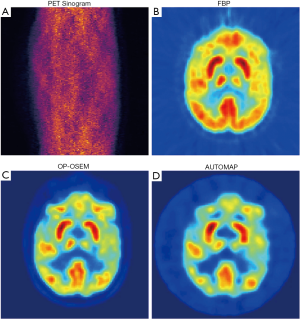
In 2018, Zhu et al. (116) reported that reconstruction was reencoded as a data-driven supervised learning task via manifold approximation automatic transformation (AUTOMAP). The network is composed of three fully connected layers and a CAE structure. AUTOMAP can learn a reconstruction function to improve artifact reduction and reconstruction accuracy for sinogram data from noisy and under-sampled acquisitions. Zhu et al. applied AUTOMAP to 18F-FDG PET data and obtained images comparable to the standard reconstruction methods, as shown in Figure 7. Later, Häggström et al. (117) applied the inverse-Radon transform to the PET data set. The authors designed a deep CED architecture called DeepPET. The encoder imitated the VGG16 network architecture by modification, shrinking the input data in a CNN-specific way; the decoder samples the shrinkage feature from the encoder to the PET image. DeepPET can achieve similar results to the traditional iterative method.
Similarly, Chrysostomou et al. (118) applied CED structure to SPECT reconstruction. Shao et al. (119) established the mapping from the sinogram image to the compressed image domain-containing less output (e.g., 16×16 bits) (a structure consisting of seven convolutional layers and two fully connected layers). They then decompressed the result into a normal (128×128 bits) SPECT image (training the CAE unsupervised network so that the goal of its output was as close to its input as possible). This kind of network was carried out in a reverse sequence, and neural networks converged faster. The input of the structure was the sinogram data and the attenuation map for this system. Compared with ordinary Poisson ordered subsets expectation maximization (OP-OSEM), this system is less sensitive to noise, but this only entails the training of 2D data.
Additionally, Hu et al. (120) extended the improved Wasserstein GAN version of the CED network. The use of multiple loss functions can effectively avoid the loss of details in the reconstructed image. Besides, Shao et al. (121) also explored the feasibility of reconstruction with small viewing angles (reducing by 1/2 and 1/4, respectively), which has positive significance for exploring clinically reducing acquisition time.
Direct conversion from sinograms to images usually requires a large number of parameters. Most of the current research focused on 128×128 images. The amazing parameters benefit from using one or more fully connected layers. Although DeepPET relinquished the fully connected layer, it still needed to decode the sinogram to complete the feature extraction deeply. These operations’ benefit is the computational cost of converting between the sinogram domain and the image domain, which is completely different from image reconstruction. Recently, Whiteley et al. (122) designed an efficient Radon inversion layer to perform domain conversion. They only focused on the sinogram corresponding to each patch in the image domain and performed a small range of fully connected operations, which completely avoided excessive calculation of whole data. This strategy may be a new inspiration for reducing the number of parameters and increasing the image size. In their work, time-of-flight list-mode data is acquired and histogrammed into sinograms. The quantitative results better than OSEM + PSF are obtained under low count input, t average absolute deviation is 1.82%, the maximum value is 4.1%, and the negative deviation of OSEM + PSF reaches 50%. In the direct prediction, physics factors including attenuation and scatter are not explicitly modeled, which potentially leads to uncertain reconstruction [e.g., DeepPET may produce false results in low count imaging (117)].When encountering new radiopharmaceuticals and new equipment, it may need to restart training and require extensive clinical validations. Besides, most of the direct predictions were investigated with 2D data, how to extrapolate it to 3D in a computationally coefficient way will be the research focus in the future. How to obtain the mapping of two different domains (and break through the limitation of image size) at the minimum computational cost may become a future concern.
Applying AI to back-projection data
Unlike directly predicting the reconstructed image from the sinogram domain, as shown in Figure 5C, some researchers use back-projected data as network input to obtain reconstructed images. This method has advantages in reconstruction time and image quality. The back-projection data has the same structural information as the output, which will effectively avoid applying the fully connected layer in the Radon inversion layer and greatly reduce the number of training parameters. Jiao et al. (123) used a multiscale fully CNN (msfCNN) structure, which takes the back-projection image of sinogram data as the network input and makes full use of the large-scale context information to reconstruct PET images. In the network design, to have a large receiving field with better calculation efficiency, the reduced scale’s downscaling-upscaling structure and the extended convolution are designed.
Additionally, the application of subpixel convolution ensures resolution without loss. Similarly, Dietze et al. (124,125) proposed a deep CED structure to enhance the SPECT image reconstructed by fast-filtering back-projection, and the result showed that it was equivalent to the Monte Carlo reconstruction result. This was the first time this approach was used in the SPECT field, although it was only a qualitative validation. Furthermore, Xu et al. (126) extended the 2D CED structure to 3D and used it in dual tracer PET imaging, aiming to find the time and space information in training data. Their training data came from Monte Carlo simulation.
In this part, the AI structure was used as part of the fast back-projection reconstruction method or post-processing, which was proven to obtain reconstruction results equivalent to the Monte Carlo simulation. It must be mentioned that when combining reconstruction and AI structure, compared with the innovation of network architecture, a wider range of clinical training data verification algorithms may be required. In a specific task, the method of creating synthetic volumes can help augment the training data. For example, Dietze et al. (124) placed balls with random diameters at random locations in the liver and filled other locations with different patients’ active distribution blocks. In this way, they expanded the original 100 ground truth data to 1,000. Besides, adding more prior information and using multichannel input will collect effective information more efficiently in future work. Whiteley et al. (127) used the time-of-flight PET scanner’s timing resolution to combine the most likely annihilation position histogrammer with the U-net structure to achieve deblurring of time-of-flight-back-projected images. This method was used to reduce the position uncertainty of annihilation events effectively. In particular, AC was also considered herein.
Combination of iterative reconstruction and AI
Normalization (e.g., total variation) is often used in nuclear medicine image reconstruction to suppress noise artifacts while retaining edges, especially for sparse reconstruction, but it requires a large time overhead. Some researchers have applied the trained network to the iterative reconstruction framework, using penalty design or a variable re-parameterization method. As shown in Figure 5D, the combination of neural networks (especially U-net structure) and iterative reconstruction frameworks considered data consistency and can restore more image details. Gong et al. (128) combined a residual network with a U-net structure in the maximum likelihood framework to remove PET images’ noise by using the concept of iterative CNN reconstruction. The alternating direction method of the multiplier algorithm is used to optimize the reconstruction of the objective function. It is necessary to select an appropriate penalty parameter ρ. Unlike adding the network to the iterative loop, Kim et al. (129,130) designed an iterative reconstruction framework based on the denoising CNN (DNCNN) using the reconstructed image of low-dose data sampling six times as input and the label as the standard-dose image. To enhance the image quality and to avoid unnecessary deviation, they combined a local fitting function with the DNCNN. The experiment found that the noise interference was weakened or even eliminated, which greatly reduced the image reconstruction time.
Besides, some researchers have focused on implementing unrolled reconstruction using neural networks different from the above schemes. Gong et al. (131) combined the U-net structure with the updated EM algorithm steps (U-net is used to replace the penalty item’s gradient) to obtain better PET images. To solve the possible inconsistency between the CNN results and the penalty gradient, they further expanded the MAP-EM update step and combined it with CNN to obtain higher contrast in the matched noise situation (132). Lim et al. (133,134) proposed a recurrent framework to penalize the difference between the unknown image and the image obtained by the network. Multiple trained networks combined this framework, namely block coordinate descent network (including convolution, soft threshold, and deconvolution layer). An advantage of their method is a lower demand for computational memory. They focused their attention on low-count PET imaging and achieved a high contrast-to-noise ratio superior to non-trained regularizers reconstruction methods (total variation and non-local means). For low-count PET reconstruction, they demonstrated the reliable generalization ability of this method on small data sets. Mehranian et al. (135) proposed an optimization algorithm for Bayesian image reconstruction with a residual learning unit to constrain the step of regularization of the previous image estimate. In particular, they verified the effectiveness of PET input alone and PET/MR combined input in low-dose stimulation and short-term in vivo brain imaging.
Furthermore, to learn the relationship between the sinogram and each pixel in the reconstructed image, the ANN was introduced to SPECT to replace the iterative estimation framework (136). Inspired by this, Wang et al. (137) used an ANN to fuse images from the maximum likelihood and post-smoothed the maximum likelihood reconstruction to enhance PET images’ quality of myocardial perfusion. Similarly, Yang et al. (138)used an ANN to fuse image versions with different regularization weights reconstructed from the MAP algorithm for quantitative improvement. Their proposed method eliminated the need for parameter adjustment. Subsequently, the authors established a multilayer perceptron model based on back-propagation to improve Bayesian PET imaging (139) quantitatively. This structure can learn the structural information, size, texture, and edges of 3D images from the data, which is significant for brain image enhancement; however, for other parts of the body under different tracers, a more general study is needed. Attention must be given to the performance of ANN being affected by the number of hidden layers and the number of neurons.
Compared with inputting projection data into the network, combining the traditional iterative method combines reliable imaging physics knowledge and noise model, which will reduce the dependence on huge data sets and avoid the difficulty of training the network from 0. Compared with traditional reconstruction, using the AI structure to learn the regularization term in iterative reconstruction or directly replacing the unrolled formula’s potential function brings more constraints to the network training and can eliminate more noise. However, the former needs to avoid the uncertainty caused by selecting key parameters, and the latter still needs to consider the cost of memory and time. No matter what kind of scheme is tried, extensive clinical validity verification (and comparison between different AI schemes) is still missing, and higher quality matched ground tags are still lacking. Compared with the extensive exploration of CNN, the previously mentioned cycle-GAN (59,60), which has been shown to avoid the quantization deviation caused by registration, might be a new direction in the future.
Image reconstruction in dynamic imaging
In contrast to static imaging, dynamic imaging requires data detection in consecutive frames. The frame number and frame duration need to be determined for each application. However, there is a trade-off between frame number and duration. Therefore, it is challenging to obtain optimal results with traditional modeling methods to include MAP, or the maximum likelihood estimation and penalty weighted least squares (PWLS) model. The kernel-based iterative method can be equivalent to a 2-layer neural network structure, which requires the use of prior information based on anatomy. Wang et al. (140) first proposed a maximum likelihood estimation algorithm based on the kernel expectation-maximization method by taking the PET image intensity itself as prior information. Unlike the traditional maximum likelihood method, this approach has a better bias-variance tradeoff and higher contrast recovery in dynamic PET image reconstruction. Boudjelal et al. (141) further developed a kernel MLEM regularization (κ-MLEM) method, removing background noise while retaining the edge and suppressing image artifacts. Ellis et al. (142) proposed a method of using kernel expectation-maximization in research using PET with dual data sets, using AI technology to construct spatial basis functions for PET reconstruction for subsequent reconstruction. Spencer et al. (143) proposed a dynamic PET reconstruction method based on a highly constrained back-projection (HYPR) kernel, which can produce a better region of interest (ROI) accuracy. Spencer (144) employed the dual kernel method to fully explore the possibility under the kernel framework by combining a nonlocal kernel with a local convolution kernel. Nevertheless, the existing kernel methods only consider the spatial correlation. Wang (145) extended the spatial kernel method to the spatial-temporal domain, which can effectively reduce noise in the space and temporal domain for dynamic PET imaging.
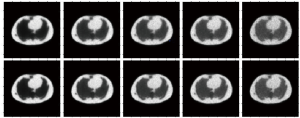
Besides, Cui et al. (146) described the dynamic reconstruction problem by combining MLEM with a stacked sparse autoencoder structure. This model was composed of multiple encoders and a decoder. The authors used the images of adjacent layers as prior knowledge and can recover more details in areas such as boundaries. A major issue with this approach is tissue specificity. Since the network parameters are pretrained, when the model extracts features from new test data, it may not be able to recognize the features, which will affect the reconstruction. As shown in Figure 8, compared to the MLEM algorithm, their method performs better with Zubal phantoms. However, since only the phantom body’s patches are used in the training phase, the results may not be as good as those obtained by the Monte Carlo simulation results. Yokota et al. (147) used random noise as input and realized dynamic PET reconstruction through the combination of non-negative matrix factorization and a deep image prior (DIP) framework. It is worth noting that U-net is used in parallel combination to extract the spatial factor after matrix decomposition, and the reconstruction result has a higher signal-to-noise ratio.
Furthermore, inspired by Gong et al. (128), the use of pre-trained noise reduction models can effectively perform a constrained maximum likelihood estimation. Xie et al. (148) upgraded and improved a GAN structure, which performed well in the trade-off between lesion contrast recovery and background noise. Compared with traditional kernel methods, integrating neural networks into the iterative reconstruction framework can maximize data consistency. In general, a clearer boundary and less noise can be obtained by inputting multiple frames of images. Adjacent frames have similar structural information, which can be used as prior information for each other, not for static imaging. In addition to minimizing the data distribution difference between training and test data, we also need to heed the high training time consumption caused by multi-frame data. A computationally efficient neural network is desirable for this task.
Anatomical image-guided nuclear medicine image reconstruction
With the emergence of hybrid imaging systems, combining anatomical information can improve image quality, although anatomical image-guided PET/SPECT reconstruction has not been routinely used in clinical applications. Addressing the problem that traditional smoothing before the reconstruction algorithm leads to excessive smoothing in the reconstructed image, the application of the sparse signal representation method based on dictionary learning can learn the dictionary from the corresponding anatomical images and be used to form a preceding signal in the reconstruction of images [MAP (149), maximum likelihood estimation (150)]. Dictionary learning is often combined with sparse models and is widely used in image denoising or super-resolution imaging. The sparsity of patches in dictionaries provides reconstruction regularization, and the dictionary can train CT or MR images to provide the inherent anatomical structures. However, some models, such as sparse coding, patch extraction, and dictionary learning, are slower than MLEM methods. In PET/MRI imaging, prior knowledge is often used to enhance PET and MRI dependence at a very small scale of image gradient, so the large scale of inter-image correlation between images and intra-image texture patterns cannot be captured. The advantages of AI technology can utilize MRI/CT anatomical information and boost PET image quality.
Sudarshan et al. (151) developed a patch-based joint dictionary method for PET/MRI to learn the regularity of a single patch and the correlation of corresponding spatial patches for Bayesian PET reconstruction with maximized expectations. Besides, Gong et al. (152) designed a reconstruction framework to train the reconstruction process based on the conditional DIP approach, named DIPRecon, which used a modified 3D U-net structure. No pretraining pairs were needed; in fact, only the patient’s prior information (T1-weighted MR) is needed, which is an unsupervised framework. Schramm et al. (111) used 3D OSEM PET and 3D structure MRI as input to train a residual network (a purely convolutional shift-invariant neural network). Interestingly, their network has achieved good performance on tracer data that has never been seen before, proving that the network has better learned the denoising operation of the input PET image. Compared with 2D image training, which can segment larger patches, 3D image training will become more cautious. In turn, smaller patches will no longer need to design more data collection. For most hybrid imaging, methods to improve the registration accuracy of the prior information will be a key factor affecting the quality of the network output. Pertinently, expertise in task solving can provide advantages over more complex AI technology structures. Anatomical information can assist low dose PET/SPECT imaging, which will be introduced in the section “Low-dose imaging”.
Image postprocessing
Low-dose nuclear medicine imaging is desirable in the clinic. One way to reduce image noise associated with low dose imaging is to apply a smoothing operation after iterative reconstruction. However, there is a trade-off between noise level and spatial resolution. With the development of GPU technology and AI’s outstanding performance in natural image denoising, AI is proven to achieve a better balance between noise level and image resolution. The AI technologies have been used to obtain high-quality nuclear medicine images, such as in image denoising and image fusion.
Low-dose imaging
Classical ML methods such as regression forest (153), sparse representation (154), canonical correlation analysis (155), and dictionary learning (156) have been investigated in reconstructing full-dose nuclear medicine images from low dose injection. Currently, the potential of DL-based low-dose nuclear medicine image denoising is still in its infancy. Here, we mainly summarized four structures: CNN, U-net structure, CAE, and GAN. The CAE/CNN structure here was essentially a low-pass filter, and the purpose of convolution was to extract useful features from the input, which also greatly improved the calculation efficiency. To ensure that the network output was the same size, a special design of padding and stride is essential. As shown in Figure 9A, the design of batch normalization layers is used to normalize the output operation. In natural images, DCNNs are widely used; Costa-Luis et al. (157) introduced them into the postprocessing step of PET imaging. The image generated by a DCNN can reduce the impact of ringing and obtain clearer edge information. Similar to Figure 9A (black line), Gong et al. (158,159) introduced skip connections in the CNN structure (improved VGG19) to denoise PET brain and lung images. This design can directly combine the early layer’s output with the deeper input of the same dimension, effectively avoiding the problem of gradient disappearance while obtaining more details. Nazari et al. (160) combined Alex-Net (161) and denoising autoencoders to achieve denoising/low-dose imaging of dopamine transporter SPECT images. By adding Gaussian white noise to achieve a 67% reduction in simulated scan time, the average absolute pixel difference of CNN denoising images and real images was 1.8%, which was much smaller than the 6.7% of noisy images. Unlike directly learning the end-to-end mapping between low-dose images and full-dose images, Xiang et al. (162) introduced structural T1 images into the network input layer and used an auto-context method to optimize the estimation, which undoubtedly made the model more robust. To obtain the spatial correlation of image voxels, the 3D convolutional layer was used by Song et al. in low-dose SPECT myocardial perfusion imaging (163,164). Such a structure can effectively suppress the noise level in the reconstructed myocardium, but the training of the 3D network usually requires many calculations.
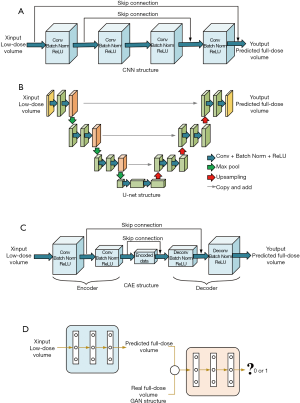
There is also a special U-net structure in the CED network. At each stage of the U-net, two overlapping convolutional layers are designed to provide a deeper network, the structure of which is shown in Figure 9B. However, it is often difficult to obtain satisfactory results by only inputting low-dose images. Such a network lacks sufficient knowledge to distinguish noise from useful information. Adjacent slices can be used as different input channels [PET-plus-MR or PET-only image input (165,166), three-layer PET image input (167,168), and five-layer SPECT image input (124,125)] can provide 2.5D structural information to the network, and this can be called a method of target feature enhancement. Compared with 3D convolution, calculation costs are high for this approach. Notably, 2.5D multichip input has its parameters and higher training effectiveness. The maximum value and residual learning have special advantages to improve training efficiency. Lu et al. (168) also predicted the deviation between images with different doses and obtained prediction results by adding them to the corresponding low-dose images. Liu et al. designed a 3U-net structure to combine the advantages of MRI and PET images fully. The PET and MRI images were entered into a 1U-net channel (to obtain initialization weights), and all outputs of multichip input were regarded as the third U-net network (169). The advantage of this network is that when PET and MRI images are incorporated into a 1U-net network, more features can be extracted under the premise of mutual interference. In particular, the data here need to be strictly registered. Subsequently, Hashimoto et al. extended the data to dynamic PET imaging (170,171). Importantly, the structure used was very similar to the U-net structure. Ramon et al. (172) used the CAE structure and demonstrated that regarding denoising, at a clinical dose input of 1/16, PET image quality was similar to that of the conventional 1/8 dose. Ramon et al. then went on to use a 3D CAE structure (33). The image results became less sensitive to the number of layers and filters by adding skip connections, which greatly alleviated some of the problems of overfitting; Figure 9C shows the CAE structure.
Compared with the supervised training mode, the unsupervised or semi-supervised learning mode is worth further exploration. The GAN structure is a general model with a more flexible framework. In Figure 9D, the generator network is used to create a full-dose image, and the discriminator network is used to distinguish predicted full-dose images and the actual full-dose images. Because the jump connection in the U-net structure can effectively combine deep and shallow features, it is widely used in generator networks [2D U-net (173) and 3D U-net (174,175)]. The pooling layer is usually not used in this case because the pooling layer is often used to reduce the dimensionality of the feature map (such as in classification tasks). The discriminator network is mostly a common CNN structure or an encoder structure, and the application of the residual structure in GANs has obvious advantages in improving the calculation efficiency (176-178). Xie et al. (179) expanded the input into five adjacent low-dose PET slices and introduced a self-attention gate to implicitly learn to suppress irrelevant regions in the input image while highlighting salient features. Unlike traditional GAN models, Kaplan et al. (178) inputted low-dose PET images to the generator network after low-pass filtering. This step can remove some noncritical noise to improve training efficiency.
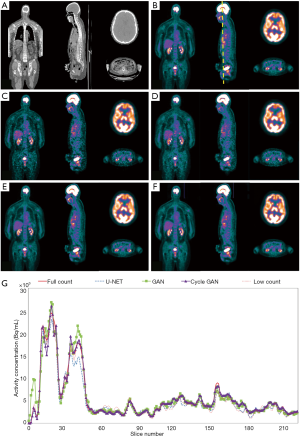
Furthermore, cGANs are widely used to learn the conditional model of data (173,174). To better avoid model failure, cycle-GANs introduce an inverse transform in a cyclic manner, which can better constrain the generator of training (177). Compared with U-net and GAN structures, the texture of the image generated by the cycle-GAN structure matches well with the full count image. The improvements in cycle-GAN are particularly evident in normal physiological uptake organs such as the brain, heart, liver, and kidneys are shown in Figure 10. For multimode image synthesis of full-dose PET, a local adaptive fusion network needs to be added prior to the generator network, and the fused image should be used as input to avoid adding more parameters to the generator but also to obtain richer structural information (175,176).
However, large amounts of training data are always hard to collect. The emergence of the DIP framework can use random noise as the input of the network to obtain denoised images without the need of previous training pairs, and some researchers have used a prior image of the patient (a previous image or a CT image) as the input of the network, which is similar to the 3D U-net framework; consequently, the PET image after denoising can be obtained through a certain iteration (152,180). Self-supervised learning has proven its wide practical value because the data does not need to be paired. Song et al. (181,182) proposed a dual-GAN to achieve super-resolution technology for PET images. The input was low-resolution PET images, high-resolution MR images and spatial information (axial and radial coordinates), and high-dimensional feature sets extracted from the auxiliary CNN, which used a pair of analog data sets for individual training in a supervised manner. Paired training data are not necessary here, enabling a new direction for clinical applications that lack paired data sets. In particular, a noise-to-noise approach was used for image denoising. The training pair’s input and target are both noise versions of the same unknown image, and the common signal component between the input and the target is effectively predicted. Chan et al. (183) combined the noise-to-noise approach with the residual network to achieve denoising of low-count PET images. Yie et al. (184) also used U-net to train a noise-to-noise net and the upgraded version, which showed that self-supervised denoising could effectively reduce the PET scan time or dose. Liu et al. (185) introduced the noise-to-noise training method into SPECT-MPI denoising. Furthermore, a 3D coupled U-net design can improve learning efficiency by reusing feature maps. This solution is better for perfusion defect detection than non-local means, CAE, and conventional 3D Gaussian filtering.
As the dose continues to decrease, the image quality will drop significantly. Compared to directly training the mapping relationship between low- and high-dose images, adding one or more MR images to the network’s input will help obtain high-resolution features, and such a model is also suitable for the denoising process. In most cases, real images under ideal conditions cannot be obtained in clinical practice, and due to the lack of paired data, self-supervised learning focusing on noise becomes a better choice. It should be noted that any single measurement standard is difficult for judging a certain algorithm’s effectiveness, and they often produce large deviations from human perception. How to design a more clinically meaningful task-based evaluation standard will be a very important task.
Image fusion
Another way to obtain higher quality nuclear medicine images is image fusion, which combines CT/MRI images with structural information to form a new clear image. Traditional methods mainly include component substitution and multi-resolution analysis, but it is often difficult to extract edge details (186). Subsequently, contourlet transform, non-subsampled contourlet transform, and wavelet transform were gradually proposed, but the cost of time needs to be considered (187,188). Due to the global coupling and pulse synchronization of neurons, pulse-coupled neural networks (PCNNs) are widely used in image fusion tasks. There is a fundamental difference between traditional neural networks and PCNNs. Coming from a biological background, PCNNs are based on the phenomenon of synchronous pulse firing in the cerebral cortex of cats and other animals. This structure can extract useful information from complex backgrounds without learning or training. Panigrahy et al. (189) proposed a new nonsubsampled shearlet transform domain medical fusion method based on weighted parameter adaptive dual-channel PCNN, which fuses MRI and SPECT images of AIDS and Alzheimer’s disease patients. This model is used to fuse high-pass sub-bands, and a weighting rule based on multiscale morphological gradients is used to fuse low-pass sub-bands. Similarly, nonsubsampled shearlet transform and nonsubsampled contourlet transform were combined with PCNNs to fuse PET/MRI and SPECT/CT tasks, respectively (190,191). Compared with other methods, it can retain more details of the source image.
Besides, CNNs have certain applications in multimodal medical image fusion. The CNN model focuses more on information near the tumor and ignores the impact of tumor location changes on feature learning. Kumar et al. (192) designed a supervised CNN to obtain the features of multimodal images and used the relativity of complementary information to fuse each multimodal feature of spatial position. DL often faces the problem of small sample sizes. Deep polynomial networks have good feature representations and perform well using small data sets. Shi et al. (193) designed a two-level multimodal stacked deep polynomial network algorithm to learn PET and MRI’s feature information and obtained remarkable results on datasets of different scales.
The PCNN is a neural network structure with a single cortical feedback signal. It does not require a training process to obtain useful information, which means that increasingly complex parameters are required. Also, the relationship between real-time processing speed and fusion effectiveness is not positive. Therefore, how to improve operational efficiency while obtaining better fusion effects will become the biggest problem. Challenges still exist regarding the CNN image in nuclear medicine image fusion. The sample demand is large (need expert annotation), training time is long, network framework is simple, and there are many convergence problems. We believe that combining CNN with traditional methods can enable the retention of more information. For example, Hermessi et al. (194) proposed that combining CNN with the wavelet fusion combination realizes the fusion of CT and MRI, and we believe that the same idea applies to nuclear medicine imaging. In general, although medical image fusion methods have developed from the spatial domain, transformation domain to DL applications, most of the methods can only be regarded as improvements on the original methods, and they have not completely resolved the fundamental problems in fusion. Such problems include the effective extraction of feature information, and some rely on the accuracy of registration. Most of the current fusions focus on the fusion of two modal images, and for wider clinical applications, the fusion of more modal images is still challenging.
Internal dosimetry
In addition to focusing on obtaining higher-quality nuclear medicine images, obtaining more accurate dose maps is also important because internal dosimetry is the key to personalized treatment in nuclear medicine. Personalized dose estimation can minimize the risk of radiation-induced toxicity (195). In personalized therapy, Monte Carlo simulation is used as the gold standard for dosimetry, although it has not been applied in clinical practice (196). The factors that limit its conventional application include its huge amount of calculation and computing time. The most widely used dosimetry method in clinical practice comes from the Medical Internal Radiation Dose (MIRD) committee (197) model and is an organ-based metrology method. The premise of this method is to assume that the activity in each organ is evenly distributed. The voxel-based method [dose point kernel (198), voxel S-value (197)] considers this deficiency but ignores the heterogeneity of different media. In the previous part, we demonstrated that the application of AI in nuclear medicine imaging is ubiquitous. In this part, we will focus on its application in dose distribution prediction.
Lee et al. (38) input PET and CT images into a 3D U-net to perform the internal dose prediction. Their reference image comes from the truth dose rate map simulated by Monte Carlo. The tissue density information of the CT image is organically combined with the activity distribution of the PET image. Compared with the traditional voxel S-value method, the dose rate map obtained by this method performs well in areas such as lungs, bones, and organ borders and is stable in whole-body dose determination. It is noteworthy that they only used 10 patient data, and they needed to retrain for different tracers or applications in PET/SPECT. Similarly, Götz et al. (39) combined U-net and empirical mode decomposition to achieve a dose map of patients who had undergone 177Lu-PSMA therapy where the input was SPECT and CT. The small number of patients, especially the acquisition of the ground truth data required an astonishing time, and the results mentioned above can be used as proof of concept. Unlike the above method, Akhavanallaf et al. (199) proposed a new whole-body element dosimetry method, which does not require a whole-body dose map of the training step. They used a 20-layer ResNet algorithm to implement a prediction medium-specific S value kernel. The network’s input is the density map generated by the CT image, and the reference is the corresponding dose distribution kernel of the point source at different locations. The simulation time in this process only needs 1/8,000 of the Monte Carlo simulation time. Compared with the MC-based kernel, this method has an average relative absolute error of 4.5%±1.8% and good consistency. The single-input network ratio is obtained by the network prediction multi-input network parameter volume, the training cost is smaller, and the voxel-level MIRD dosimetry idea is effectively expanded. Götz et al. (200) proposed that a DL strategy based on the U-net architecture can learn accurate dose voxel kernels (DVKs) (discretizing continuous dose point kernels), which is faster than the gold standard Monte Carlo simulation. However, the accuracy of prediction is limited by time-integrated activity distribution estimation.
The application of DL in internal dosimetry has just begun. Current researchers have adopted the U-net structure and U-net due to its unique down-sampling and up-sampling steps, and jumper connections at each level of spatial resolution greatly retain the spatial resolution information. This effectively avoids the problem of vanishing gradient. Compared with the network structure, how to quickly generate accurate ground truth dose maps is particularly important. Besides, the accuracy of image registration is also one of the factors affecting the final prediction results. Akhavanallaf et al. (199) have shown that it is possible to generate a whole body element dose map within a few minutes, making it possible to quickly generate ground truth data. Also, most of the direct end-to-end predicted dose maps do not involve physical factors. It would be more meaningful if physical factors (such as Compton scattering) can be considered. The current research is only for the specific tracer/imaging modality, and further training is needed for more extensive clinical verification.
Discussion and conclusions
In recent years, due to the explosive development of AI in the field of computer vision and image processing, AI, especially DL, is being increasingly used in nuclear medicine imaging. As described in this article, the application of AI in nuclear medicine imaging has demonstrated potential in promoting the nuclear medicine imaging system and paving the way for precision medicine. With the rapid development of hardware, especially GPU technology, AI can quickly process, mine, and analyze large amounts of data. Once training is completed, AI can usually provide faster and better resolution of specific tasks than traditional methods. In particular, the data-driven end-to-end mapping method provides new opportunities for many traditional tasks, such as the prediction of attenuation maps, improvement of reconstruction quality, prediction of internal dose map, and AI has demonstrated improved image quality and quantification for multiple tasks. Unlike traditional methods that require more human participation, evaluating the effectiveness of AI performance often depends on training data, network structure, and hyperparameter settings.
For the clinical application of AI, we need to pay attention to the following aspects. First, for different topics, what kind of network structure is the best? Zeng et al. (201) believed that the structure of a neural network is unnecessary. In AI, training data set pairs are used as black-box input and output. Almost all algorithms contain parameters that need to be adjusted according to tasks. By constantly updating the parameter set’s values to find the optimal parameters in learning, this process is repeated many times until the result is satisfactory. In existing research, the practicability of performance depends on the design of the structure. What they have in common is that they need enough data sets as dependencies. Therefore, how to break the restriction of network structure and provide an interpretable network structure will still require future development. Because of the limitations of memory, time, and the network’s immense weight, larger images are difficult to manage. However, if training data are scarce, one should consider whether such an approach is meaningful, especially when making some pure predictions (e.g., reconstruct the image directly from the projection image). How to avoid the unpredictability of training is problematic.
Secondly, we cannot guarantee that the data pairs involved in training contain almost all possible situations. The promotion of data integration and sharing should be the focus of further research. Also, using only a limited number of cases is not always convincing, so we need to pay attention to the existence of abnormal data. Although transfer learning and data expansion can be used to improve this situation, a large amount of training data will double the amount of calculation required. Here, the acquisition of training data is more critical than the training of network structure, and the results of using more professional training data can better resemble the actual effect. Compared with the limited paired training set, how to train unpaired training ensemble as a current hot spot, cycle-GAN emergence that does not require paired data provides more directions for this topic. This problem is evident not only in a single field but in almost all AI-related research.
Thirdly, we should thoughtfully consider whether we should risk using the good results obtained by the AI method in clinical practice; accordingly, ways to verify the proposed method in general practice may be the next step to be investigated. The indicators RMSE, PSRN, and SSIM, are often used to evaluate the quality of composite images, but studies have shown that the interpretation of similar indicators may not match clinical task evaluation (202).In addition to the commonly used evaluation indicators, professional evaluations are particularly important. At present, most of the existing AI applications have been developed for specific tasks. Although the application of contextual information makes AI more intelligent, it is not realistic to let AI completely replace physicians and complete tasks automatically. Deficiencies of AI include a lack of real baseline data, lack of label data, and provision of an insufficient interpretation of models and methods. Compared with traditional methods, the research community still seems to be exploring how to better utilize AI technology, which should involve a wider range of situations. In actual applications, more evaluation indicators are needed to determine the effectiveness of these methods.
Fourthly, compared with independent system imaging, more training knowledge can be obtained by hybrid imaging during network training. We found that multimode imaging, as well as prediction, may be a new research direction. However, the most important problem is the registration of multi-mode images. The training method performed in combination with unpaired data may be the reasonable direction. Taking the brain as an example, there is still the possibility of head drift between the acquisition time windows of different modal systems. Also, the use of multi-mode images as multiple inputs to the network will inevitably bring more parameters, increasing the difficulty of network convergence and training time, which requires additional attention in the network design.
Finally, we would like to sketch the landscape of the AI technology advancements that offer improvements in nuclear medicine imaging quality. We mainly focused on four aspects: imaging physics (AC and scatter correction), image reconstruction (a static system, dynamic system, and hybrid system), image postprocessing (low-dose imaging and image fusion), and internal dosimetry. Once the learning is complete, AI prediction will take less time than the traditional methods. Researchers are still actively investigating the possibility of AI technology in improving the quality of nuclear medicine imaging and its application in the clinic.
Acknowledgments
Funding: This work was supported in part by NSAF (U1830126); the National Key R&D Program of China (2016YFC0105104); the National Natural Science Foundation of China (81671775, 81830052); Construction Project of Shanghai Key Laboratory of Molecular Imaging (18DZ2260400); Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018-2020).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1078). The authors have no conflicts of interest to declare.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Mont MA, Krebs VE, Backstein DJ, Browne JA, Mason JB, Taunton MJ, Callaghan JJ. Artificial intelligence: influencing our lives in joint arthroplasty. J Arthroplasty 2019;34:2199-200. [Crossref] [PubMed]
- Ting DSW, Pasquale LR, Peng L, Campbell JP, Lee AY, Raman R, Tan GSW, Schmetterer L, Keane PA, Wong TY. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol 2019;103:167-75. [Crossref] [PubMed]
- Hessler G, Baringhaus KH. Artificial intelligence in drug design. Molecules 2018;23:2520. [Crossref] [PubMed]
- Vaishya R, Javaid M, Khan IH, Haleem A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab Syndr 2020;14:337-9. [Crossref] [PubMed]
- Li W, Li Y, Qin W, Liang X, Xu J, Xiong J, Xie Y. Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy. Quant Imaging Med Surg 2020;10:1223-36. [Crossref] [PubMed]
- Subramanian M, Wojtusciszyn A, Favre L, Boughorbel S, Shan J, Letaief KB, Pitteloud N, Chouchane L. Precision medicine in the era of artificial intelligence: implications in chronic disease management. J Transl Med 2020;18:472. [Crossref] [PubMed]
- Adir O, Poley M, Chen G, Froim S, Krinsky N, Shklover J, Shainsky-Roitman J, Lammers T, Schroeder A. Integrating artificial intelligence and nanotechnology for precision cancer medicine. Adv Mater 2020;32:e1901989 [Crossref] [PubMed]
- Hatt M, Parmar C, Qi J, Naqa IE. Machine (deep) learning methods for image processing and radiomics. IEEE Trans Radiat Plasma Med Sci 2019;3:104-8. [Crossref]
- Ou X, Zhang J, Wang J, Pang F, Wang Y, Wei X, Ma X. Radiomics based on 18F-FDG PET/CT could differentiate breast carcinoma from breast lymphoma using machine-learning approach: a preliminary study. Cancer Med 2020;9:496-506. [Crossref] [PubMed]
- Shiri I, Maleki H, Hajianfar G, Abdollahi H, Ashrafinia S, Hatt M, Zaidi H, Oveisi M, Rahmim A. Next-generation radiogenomics sequencing for prediction of EGFR and KRAS mutation status in NSCLC patients using multimodal imaging and machine learning algorithms. Mol Imaging Biol 2020;22:1132-48. [Crossref] [PubMed]
- Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, Mattonen SA, El Naqa I. Machine and deep learning methods for radiomics. Med Phys 2020;47:e185-202. [Crossref] [PubMed]
- Attanasio S, Forte SM, Restante G, Gabelloni M, Guglielmi G, Neri E. Artificial intelligence, radiomics and other horizons in body composition assessment. Quant Imaging Med Surg 2020;10:1650-60. [Crossref] [PubMed]
- Nensa F, Demircioglu A, Rischpler C. Artificial intelligence in nuclear medicine. J Nucl Med 2019;60:29S-37S. [Crossref] [PubMed]
- Duffy IR, Boyle AJ, Vasdev N. Improving PET imaging acquisition and analysis with machine learning: a narrative review with focus on Alzheimer's disease and oncology. Mol Imaging 2019;18:1536012119869070 [Crossref] [PubMed]
- Magesh PR, Myloth RD, Tom RJ. An explainable machine learning model for early detection of Parkinson's disease using LIME on DaTSCAN imagery. Comput Biol Med 2020;126:104041 [Crossref] [PubMed]
- Martin-Isla C, Campello VM, Izquierdo C, Raisi-Estabragh Z, Baeßler B, Petersen SE, Lekadir K. Image-based cardiac diagnosis with machine learning: a review. Front Cardiovasc Med 2020;7:1. [Crossref] [PubMed]
- Toyama Y, Hotta M, Motoi F, Takanami K, Minamimoto R, Takase K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Sci Rep 2020;10:17024. [Crossref] [PubMed]
- Tang J, Yang B, Adams MP, Shenkov NN, Klyuzhin IS, Fotouhi S, Davoodi-Bojd E, Lu L, Soltanian-Zadeh H, Sossi V, Rahmim A. Artificial neural network-based prediction of outcome in Parkinson's disease patients using DaTscan SPECT imaging features. Mol Imaging Biol 2019;21:1165-73. [Crossref] [PubMed]
- Moazemi S, Khurshid Z, Erle A, Lütje S, Essler M, Schultz T, Bundschuh RA. Machine learning facilitates hotspot classification in PSMA-PET/CT with nuclear medicine specialist accuracy. Diagnostics (Basel) 2020;10:622. [Crossref] [PubMed]
- Huang GH, Lin CH, Cai YR, Chen TB, Hsu SY, Lu NH, Chen HY, Wu YC. Multiclass machine learning classification of functional brain images for Parkinson's disease stage prediction. Statistical Analysis and Data Mining 2020;13:508-23. [Crossref]
- Kaplan Berkaya S, Ak Sivrikoz I, Gunal S. Classification models for SPECT myocardial perfusion imaging. Comput Biol Med 2020;123:103893 [Crossref] [PubMed]
- Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Nye JA, Yang X. Machine learning in quantitative PET: a review of attenuation correction and low-count image reconstruction methods. Phys Med 2020;76:294-306. [Crossref] [PubMed]
- Zhang T, Shi M. Multi-modal neuroimaging feature fusion for diagnosis of Alzheimer's disease. J Neurosci Methods 2020;341:108795 [Crossref] [PubMed]
- Papandrianos N, Papageorgiou E, Anagnostis A, Feleki A. A deep-learning approach for diagnosis of metastatic breast cancer in bones from whole-body scans. Applied Sciences-Basel 2020;10:27. [Crossref]
- Liu Y, Xu Y, Meng X, Wang X, Bai T. A study on the auxiliary diagnosis of thyroid disease images based on multiple dimensional deep learning algorithms. Curr Med Imaging 2020;16:199-205. [Crossref] [PubMed]
- Hu Z, Li Y, Zou S, Xue H, Sang Z, Liu X, Yang Y, Zhu X, Liang D, Zheng H. Obtaining PET/CT images from non-attenuation corrected PET images in a single PET system using Wasserstein generative adversarial networks. Phys Med Biol 2020;65:215010 [Crossref] [PubMed]
- Xiang H, Lim H, Fessler JA, Dewaraja YK. A deep neural network for fast and accurate scatter estimation in quantitative SPECT/CT under challenging scatter conditions. Eur J Nucl Med Mol Imaging 2020;47:2956-67. [Crossref] [PubMed]
- Reader AJ, Corda G, Mehranian A. Costa-Luis Cd, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci 2021;5:1-25. [Crossref]
- Hoeschen C. Use of artificial intelligence for image reconstruction. Radiologe 2020;60:15-23. [Crossref] [PubMed]
- Bal A, Banerjee M, Chaki R, Sharma P. An efficient method for PET image denoising by combining multi-scale transform and non-local means. Multimed Tools Appl 2020;79:29087-120. [Crossref]
- Klyuzhin IS, Cheng JC, Bevington C, Sossi V. Use of a Tracer-specific deep artificial neural net to denoise dynamic PET images. IEEE Trans Med Imaging 2020;39:366-76. [Crossref] [PubMed]
- Ramon AJ, Yang Y, Pretorius PH, Johnson KL, King MA, Wernick MN. Improving diagnostic accuracy in low-dose SPECT myocardial perfusion imaging with convolutional denoising networks. IEEE Trans Med Imaging 2020;39:2893-903. [Crossref] [PubMed]
- Blanc-Durand P, Jégou S, Kanoun S, Berriolo-Riedinger A, Bodet-Milin C, Kraeber-Bodéré F, Carlier T, Le Gouill S, Casasnovas RO, Meignan M, Itti E. Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur J Nucl Med Mol Imaging 2020; Epub ahead of print. [Crossref] [PubMed]
- Wolterink JM. Left ventricle segmentation in the era of deep learning. J Nucl Cardiol 2020;27:988-91. [Crossref] [PubMed]
- Qiu S, Joshi PS, Miller MI, Xue C, Zhou X, Karjadi C, Chang GH, Joshi AS, Dwyer B, Zhu S, Kaku M, Zhou Y, Alderazi YJ, Swaminathan A, Kedar S, Saint-Hilaire MH, Auerbach SH, Yuan J, Sartor EA, Au R, Kolachalama VB. Development and validation of an interpretable deep learning framework for Alzheimer's disease classification. Brain 2020;143:1920-33. [Crossref] [PubMed]
- Hsu SY, Yeh LR, Chen TB, Du WC, Huang YH, Twan WH, Lin MC, Hsu YH, Wu YC, Chen HY. Classification of the multiple stages of Parkinson's disease by a deep convolution neural network based on Tc-99m-TRODAT-1 SPECT images. Molecules 2020;25:17. [Crossref]
- Lee MS, Hwang D, Kim JH, Lee JS. Deep-dose: a voxel dose estimation method using deep convolutional neural network for personalized internal dosimetry. Sci Rep 2019;9:10308. [Crossref] [PubMed]
- Götz TI, Schmidkonz C, Chen S, Al-Baddai S, Kuwert T, Lang EW. A deep learning approach to radiation dose estimation. Phys Med Biol 2020;65:035007 [Crossref] [PubMed]
- Zaharchuk G. Next generation research applications for hybrid PET/MR and PET/CT imaging using deep learning. Eur J Nucl Med Mol Imaging 2019;46:2700-7. [Crossref] [PubMed]
- Kaji S, Kida S. Overview of image-to-image translation by use of deep neural networks: denoising, super-resolution, modality conversion, and reconstruction in medical imaging. Radiol Phys Technol 2019;12:235-48. [Crossref] [PubMed]
- Visvikis D, Cheze Le Rest C, Jaouen V, Hatt M. Artificial intelligence, machine (deep) learning and radio(geno)mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging 2019;46:2630-7. [Crossref] [PubMed]
- Porenta G. Is there value for artificial intelligence applications in molecular imaging and nuclear medicine? J Nucl Med 2019;60:1347-9. [Crossref] [PubMed]
- Aktolun C. Artificial intelligence and radiomics in nuclear medicine: potentials and challenges. Eur J Nucl Med Mol Imaging 2019;46:2731-6. [Crossref] [PubMed]
- Gong K, Berg E, Cherry SR, Qi J. Machine learning in PET: from photon detection to quantitative image reconstruction. Proceedings of the IEEE 2020;108:51-68. [Crossref]
- Thompson RC. CT attenuation correction for thallium SPECT MPI and other benefits of multimodality imaging. J Nucl Cardiol 2019;26:1596-8. [Crossref] [PubMed]
- Lee JS. A review of deep learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Radiat Plasma Med Sci 2021;5:160-84. [Crossref]
- Melroy S, Bauer C, McHugh M, Carden G, Stolin A, Majewski S, Brefczynski-Lewis J, Wuest T. Development and design of next-generation head-mounted ambulatory microdose positron-emission tomography (AM-PET) system. Sensors (Basel) 2017;17:1164. [Crossref] [PubMed]
- Tashima H, Yamaya T. Proposed helmet PET geometries with add-on detectors for high sensitivity brain imaging. Phys Med Biol 2016;61:7205-20. [Crossref] [PubMed]
- Mannheim JG, Schmid AM, Schwenck J, Katiyar P, Herfert K, Pichler BJ, Disselhorst JA. PET/MRI hybrid systems. Semin Nucl Med 2018;48:332-47. [Crossref] [PubMed]
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006;313:504-7. [Crossref] [PubMed]
- Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for (18)F-FDG PET attenuation correction. EJNMMI Phys 2018;5:24. [Crossref] [PubMed]
- Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, Lee JS. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med 2018;59:1624-9. [Crossref] [PubMed]
- Shiri I, Ghafarian P, Geramifar P, Leung KH, Ghelichoghli M, Oveisi M, Rahmim A, Ay MR. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC). Eur Radiol 2019;29:6867-79. [Crossref] [PubMed]
- Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Phys Med Biol 2019;64:075019 [Crossref] [PubMed]
- Arabi H, Zaidi H. Deep learning-guided estimation of attenuation correction factors from time-of-flight PET emission data. Med Image Anal 2020;64:101718 [Crossref] [PubMed]
- Shi L, Onofrey JA, Liu H, Liu YH, Liu C. Deep learning-based attenuation map generation for myocardial perfusion SPECT. Eur J Nucl Med Mol Imaging 2020;47:2383-95. [Crossref] [PubMed]
- Armanious K, Kustner T, Reimold M, Nikolaou K, La Fougere C, Yang B, Gatidis S. Independent brain (18)F-FDG PET attenuation correction using a deep learning approach with Generative Adversarial Networks. Hell J Nucl Med 2019;22:179-86. [PubMed]
- Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, Mao H, Nye JA, Yang X. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol 2019;64:215016 [Crossref] [PubMed]
- Dong X, Lei Y, Wang T, Higgins K, Liu T, Curran WJ, Mao H, Nye JA, Yang X. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys Med Biol 2020;65:055011 [Crossref] [PubMed]
- Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, Ay MR, Zaidi H. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging 2020;47:2533-48. [Crossref] [PubMed]
- Fahey FH, Goodkind A, MacDougall RD, Oberg L, Ziniel SI, Cappock R, Callahan MJ, Kwatra N, Treves ST, Voss SD. Operational and dosimetric aspects of pediatric PET/CT. J Nucl Med 2017;58:1360-6. [Crossref] [PubMed]
- Sourbelle K, Kachelriess M, Kalender WA. Reconstruction from truncated projections in CT using adaptive detruncation. Eur Radiol 2005;15:1008-14. [Crossref] [PubMed]
- Heußer T, Brehm M, Ritschl L, Sawall S, Kachelrieß M. Prior-based artifact correction (PBAC) in computed tomography. Med Phys 2014;41:021906 [Crossref] [PubMed]
- Thejaswi A, Nivarthi A, Beckwith DJ, Johnson KL, Pretorius PH, Agu EO, King MA, Lindsay C. A deep-learning method for detruncation of attenuation maps. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-3.
- Wagenknecht G, Kaiser HJ, Mottaghy FM, Herzog H. MRI for attenuation correction in PET: methods and challenges. Magma 2013;26:99-113. [Crossref] [PubMed]
- Su Y, Rubin BB, McConathy J, Laforest R, Qi J, Sharma A, Priatna A, Benzinger TL. Impact of MR-based attenuation correction on neurologic PET studies. J Nucl Med 2016;57:913-7. [Crossref] [PubMed]
- Mehranian A, Arabi H, Zaidi H. Vision 20/20: Magnetic resonance imaging-guided attenuation correction in PET/MRI: challenges, solutions, and opportunities. Med Phys 2016;43:1130-55. [Crossref] [PubMed]
- Ladefoged CN, Law I, Anazodo U, St Lawrence K, Izquierdo-Garcia D, Catana C, Burgos N, Cardoso MJ, Ourselin S, Hutton B, Merida I, Costes N, Hammers A, Benoit D, Holm S, Juttukonda M, An H, Cabello J, Lukas M, Nekolla S, Ziegler S, Fenchel M, Jakoby B, Casey ME, Benzinger T, Hojgaard L, Hansen AE, Andersen FL. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage 2017;147:346-59. [Crossref] [PubMed]
- Stone JR, Wilde EA, Taylor BA, Tate DF, Levin H, Bigler ED, Scheibel RS, Newsome MR, Mayer AR, Abildskov T, Black GM, Lennon MJ, York GE, Agarwal R, DeVillasante J, Ritter JL, Walker PB, Ahlers ST, Tustison NJ. Supervised learning technique for the automated identification of white matter hyperintensities in traumatic brain injury. Brain Inj 2016;30:1458-68. [Crossref] [PubMed]
- Bonte S, Goethals I, Van Holen R. Machine learning based brain tumour segmentation on limited data using local texture and abnormality. Comput Biol Med 2018;98:39-47. [Crossref] [PubMed]
- Rincón M, Díaz-López E, Selnes P, Vegge K, Altmann M, Fladby T, Bjørnerud A. Improved automatic segmentation of white matter hyperintensities in MRI based on multilevel lesion features. Neuroinformatics 2017;15:231-45. [Crossref] [PubMed]
- Ahmadvand A, Daliri MR, Zahiri SM. Segmentation of brain MR images using a proper combination of DCS based method with MRF. Multimed Tools Appl 2018;77:8001-18. [Crossref]
- Polly FP, Shil SK, Hossain MA, Ayman A, Jang YM. Detection and classification of HGG and LGG brain tumor using machine learning. In: 2018 International Conference on Information Networking (ICOIN). Chiang Mai: IEEE, 2018:813-7.
- Mecheter I, Alic L, Abbod M, Amira A, Ji J. MR image-based attenuation correction of brain PET imaging: review of literature on machine learning approaches for segmentation. J Digit Imaging 2020;33:1224-41. [Crossref] [PubMed]
- Yang B, Tang J. Learning-based attenuation correction for brain PET/MRI using artificial neural networks. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-4.
- Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology 2018;286:676-84. [Crossref] [PubMed]
- Lee KS, Tao L, Best-Devereux J, Levin CS. Study of a convolutional autoencoder for automatic generation of MR-based attenuation map in PET/MR. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-3.
- Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. [Crossref] [PubMed]
- Arabi H, Zeng G, Zheng G, Zaidi H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur J Nucl Med Mol Imaging 2019;46:2746-59. [Crossref] [PubMed]
- Bradshaw TJ, Zhao G, Jang H, Liu F, McMillan AB. Feasibility of deep learning-based PET/MR attenuation correction in the pelvis using only diagnostic MR images. Tomography 2018;4:138-47. [Crossref] [PubMed]
- Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, Kerkmeijer LGW, van den Berg CAT. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol 2018;63:185001 [Crossref] [PubMed]
- Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng 2018;65:2720-30. [Crossref] [PubMed]
- Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with context-aware generative adversarial networks. In: MICCAI 2017: Medical Image Computing and Computer Assisted Intervention. Quebec City: Springer, 2017:417-25.
- Jang H, Liu F, Zhao G, Bradshaw T, McMillan AB. Technical note: deep learning based MRAC using rapid ultrashort echo time imaging. Med Phys 2018;45:3697-704. [Crossref] [PubMed]
- Ladefoged CN, Benoit D, Law I, Holm S, Kjær A, Højgaard L, Hansen AE, Andersen FL. Region specific optimization of continuous linear attenuation coefficients based on UTE (RESOLUTE): application to PET/MR brain imaging. Phys Med Biol 2015;60:8047-65. [Crossref] [PubMed]
- Ladefoged CN, Andersen FL, Kjær A, Højgaard L, Law I. RESOLUTE PET/MRI Attenuation Correction for O-(2-(18)F-fluoroethyl)-L-tyrosine (FET) in Brain Tumor Patients with Metal Implants. Front Neurosci 2017;11:453. [Crossref] [PubMed]
- Ladefoged CN, Marner L, Hindsholm A, Law I, Højgaard L, Andersen FL. Deep learning based attenuation correction of pet/mri in pediatric brain tumor patients: evaluation in a clinical setting. Front Neurosci 2019;12:1005. [Crossref] [PubMed]
- Shi K, Fürst S, Sun L, Lukas M, Navab N, Förster S, Ziegler SI. Individual refinement of attenuation correction maps for hybrid PET/MR based on multi-resolution regional learning. Comput Med Imaging Graph 2017;60:50-7. [Crossref] [PubMed]
- Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, Hope TA, Larson PEZ. Zero-echo-time and Dixon deep pseudo-CT (ZeDD CT): direct generation of pseudo-CT images for pelvic PET/MRI attenuation correction using deep convolutional neural networks with multiparametric MRI. J Nucl Med 2018;59:852-8. [Crossref] [PubMed]
- Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol 2018;63:125011 [Crossref] [PubMed]
- Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, Soricelli A, Salvatore M, Malpica N, Catana C. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med 2019;60:429-35. [Crossref] [PubMed]
- Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT synthesis using unpaired data. In: SASHIMI 2017: Simulation and Synthesis in Medical Imaging. Québec City: Springer, 2017:14-23.
- Ibaraki M, Matsubara K, Sato K, Mizuta T, Kinoshita T. Validation of a simplified scatter correction method for 3D brain PET with (15)O. Ann Nucl Med 2016;30:690-8. [Crossref] [PubMed]
- Hutton BF, Buvat I, Beekman FJ. Review and current status of SPECT scatter correction. Phys Med Biol 2011;56:R85-112. [Crossref] [PubMed]
- Shao L, Freifelder R, Karp JS. Triple energy window scatter correction technique in PET. IEEE Trans Med Imaging 1994;13:641-8. [Crossref] [PubMed]
- Hirano Y, Koshino K, Iida H. Influences of 3D PET scanner components on increased scatter evaluated by a Monte Carlo simulation. Phys Med Biol 2017;62:4017-30. [Crossref] [PubMed]
- Berker Y, Maier J, Kachelrieß M. Deep scatter estimation in PET: fast scatter correction using a convolutional neural network. In: 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC). Sydney: IEEE, 2018:1-5.
- Qian H, Rui X, Ahn S. Deep learning models for PET scatter estimations. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-5.
- Gedik GK, Sari O. Influence of single photon emission computed tomography (SPECT) reconstruction algorithm on diagnostic accuracy of parathyroid scintigraphy: comparison of iterative reconstruction with filtered backprojection. Indian J Med Res 2017;145:479-87. [PubMed]
- Qi J, Leahy RM. Iterative reconstruction techniques in emission computed tomography. Phys Med Biol 2006;51:R541-78. [Crossref] [PubMed]
- Ravishankar S, Ye JC, Fessler JA. Image reconstruction: from sparsity to data-adaptive methods and machine learning. Proc IEEE Inst Electr Electron Eng 2020;108:86-109.
- Ma T, Xu T, Liu H, Wei Q, Peng F, Deng Z, Gong G, Gong H, Wang S, Liu Y. Development of a SiPM based preclinical PET SPECT imaging system imaging system. J Nucl Med 2017;58:abstr 397.
- Massari R, D’Elia A, Soluri A. Preliminary results on a small animal SPECT system based on H13700 PSMPT coupled with CRY018 array. Nucl Instrum Methods Phys Res A 2019;940:296-301. [Crossref]
- Shiri I, Sheikhzadeh P, Ay MR. Deep-fill: deep learning based sinogram domain gap filling in positron emission tomography. arXiv 2019:1906.07168v1.
- Jong HWAMd. Boellaard R, Knoess C, Lenox M, Michel C, Casey M, Lammertsma AA. Correction methods for missing data in sinograms of the HRRT PET scanner. IEEE Trans Nucl Sci 2003;50:1452-6. [Crossref]
- Shojaeilangari S, Schmidtlein CR, Rahmim A, Ay MR. Recovery of missing data in partial geometry PET scanners: Compensation in projection space vs image space. Med Phys 2018;45:5437-49. [Crossref] [PubMed]
- Hong X, Zan Y, Weng F, Tao W, Peng Q, Huang Q. Enhancing the image quality via transferred deep residual learning of coarse PET sinograms. IEEE Trans Med Imaging 2018;37:2322-32. [Crossref] [PubMed]
- Shiri I, Leung K, Ghafarian P, Geramifar P, Oveisi M, Ay MR, Rahmim A. HiResPET: high resolution PET image generation using deep convolution encoder decoder network. J Nucl Med 2019;60:abstr 1368.
- Shiri I, Leung K, Geramifar P, Ghafarian P, Oveisi M, Ay MR, Rahmim A. PSFNET: ultrafast generation of PSF-modelled-like PET images using deep convolutional neural network. J Nucl Med 2019;60:abstr 1369.
- Schramm G, Rigie D, Vahle T, Rezaei A, Van Laere K, Shepherd T, Nuyts J, Boada F. Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network. NeuroImage 2021;224:117399 [Crossref] [PubMed]
- Shiri I. AmirMozafari Sabet K, Arabi H, Pourkeshavarz M, Teimourian B, Ay MR, Zaidi H. Standard SPECT myocardial perfusion estimation from half-time acquisitions using deep convolutional residual neural networks. J Nucl Cardiol 2020; Epub ahead of print. [Crossref] [PubMed]
- Ryden T, Marin I, van Essen M, Svensson J, Bernhardt P. Deep learning generation of intermediate projections and Monte Carlo based reconstruction improves 177Lu SPECT images reconstructed with sparse acquired projections. J Nucl Med 2019;60:abstr 44.
- Whiteley W, Gregor J. CNN-based PET sinogram repair to mitigate defective block detectors. Phys Med Biol 2019;64:235017 [Crossref] [PubMed]
- Liu CC, Huang HM. Partial-ring PET image restoration using a deep learning based method. Phys Med Biol 2019;64:225014 [Crossref] [PubMed]
- Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487-92. [Crossref] [PubMed]
- Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder-decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal 2019;54:253-62. [Crossref] [PubMed]
- Chrysostomou C, Koutsantonis L, Lemesios C, Papanicolas CN. SPECT Imaging reconstruction method based on deep convolutional neural network. In: 2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Manchester: IEEE, 2019:1-4.
- Shao WY, Du Y. SPECT Image reconstruction by deep learning using a two-step training method. J Nucl Med 2019;60:abstr 1353.
- Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, Teng Y, Liu X, Yang Y, Liang D, Zhu X, Zheng H. DPIR-Net: direct PET image reconstruction based on the wasserstein generative adversarial network. IEEE Trans Radiat Plasma Med Sci 2021;5:35-43. [Crossref]
- Shao W, Leung K, Pomper M, Du Y. SPECT image reconstruction by a learnt neural network. J Nucl Med 2020;61:abstr 1478.
- Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham) 2020;7:032503 [Crossref] [PubMed]
- Jiao J, Ourselin S. Fast PET reconstruction using multi-scale fully convolutional neural networks. arXiv. 2017:1704.07244v1.
- Dietze M, Branderhorst W, Viergever M, De Jong H. Accelerated SPECT image reconstruction with a convolutional neural network. J Nucl Med 2019;60:abstr 1351.
- Dietze MMA, Branderhorst W, Kunnen B, Viergever MA, de Jong H. Accelerated SPECT image reconstruction with FBP and an image enhancement convolutional neural network. EJNMMI Phys 2019;6:14. [Crossref] [PubMed]
- Xu J, Liu H. Three-dimensional convolutional neural networks for simultaneous dual-tracer PET imaging. Phys Med Biol 2019;64:185016 [Crossref] [PubMed]
- Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, Gregor J. FastPET: near real-time reconstruction of PET histo-image data using a neural network. IEEE Trans Radiat Plasma Med Sci 2021;5:65-77. [Crossref]
- Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, El Fakhri G, Qi J, Li Q. Iterative PET Image Reconstruction Using Convolutional Neural Network Representation. IEEE Trans Med Imaging 2019;38:675-85. [Crossref] [PubMed]
- Kim K, Wu D, Gong K, Kim JH, Son YD, Kim HK, Fakhria GE, Li Q. Penalized PET reconstruction using CNN prior. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-4.
- Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, Kim HK, El Fakhri G, Li Q. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Trans Med Imaging 2018;37:1478-87. [Crossref] [PubMed]
- Gong K, Wu D, Kim K, Yang J, Georges El F, Seo Y, Li Q. EMnet: an unrolled deep neural network for PET image reconstruction. In: Proc.SPIE. San Diego: SPIE, 2019.
- Gong K, Wu D, Kim K, Yang J, Sun T, Georges El F, Seo Y, Li Q. MAPEM-Net: an unrolled neural network for Fully 3D PET image reconstruction. In: Proc.SPIE. Philadelphia: SPIE, 2019.
- Lim H, Huang Z, Fessler JA, Dewaraja YK, Chun IY. Application of trained Deep BCD-Net to iterative low-count PET image reconstruction. In: 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC). Sydney: IEEE, 2018:1-4.
- Lim H, Chun IY, Dewaraja YK, Fessler JA. Improved low-count quantitative PET reconstruction with an iterative neural network. IEEE Trans Med Imaging 2020;39:3512-22. [Crossref] [PubMed]
- Mehranian A, Reader AJ. Model-based deep learning PET image reconstruction using forward-backward splitting expectation maximisation. IEEE Trans Radiat Plasma Med Sci 2021;5:54-64. [Crossref]
- Floyd CR. An artificial neural network for SPECT image reconstruction. IEEE Trans Med Imaging 1991;10:485-7. [Crossref] [PubMed]
- Wang X, Yang B, Moody JB, Tang J. Improved myocardial perfusion PET imaging using artificial neural networks. Phys Med Biol 2020;65:145010 [Crossref] [PubMed]
- Yang B, Ying L, Tang J. Enhancing Bayesian PET image reconstruction using neural networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). Melbourne: IEEE, 2017:1181-4.
- Yang B, Ying L, Tang J. Artificial neural network enhanced bayesian PET image reconstruction. IEEE Trans Med Imaging 2018;37:1297-309. [Crossref] [PubMed]
- Wang G, Qi J. PET image reconstruction using kernel method. IEEE Trans Med Imaging 2015;34:61-71. [Crossref] [PubMed]
- Boudjelal A, Messali Z, Elmoataz A. A novel kernel-based regularization technique for PET image reconstruction. Technologies 2017;5:37. [Crossref]
- Ellis S, Reader AJ. Kernelised EM image reconstruction for dual-dataset PET studies. In: 2016 IEEE Nuclear Science Symposium, Medical Imaging Conference and Room-Temperature Semiconductor Detector Workshop (NSS/MIC/RTSD). Strasbourg: IEEE, 2016:1-3.
- Spencer B, Qi J, Ramsey DB, Wang G. Dynamic PET image reconstruction for parametric imaging using the HYPR kernel method. In: Proc.SPIE. Orlando: SPIE, 2017.
- Spencer B, Wang G. Statistical image reconstruction for shortened dynamic PET using a dual kernel method. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017.
- Wang G. High temporal-resolution dynamic pet image reconstruction using a new spatiotemporal kernel method. IEEE Trans Med Imaging 2019;38:664-74. [Crossref] [PubMed]
- Cui J, Liu X, Wang Y, Liu H. Deep reconstruction model for dynamic PET images. PLoS One 2017;12:e0184667 [Crossref] [PubMed]
- Yokota T, Kawai K, Sakata M, Kimura Y, Hontani H. Dynamic PET image reconstruction using nonnegative matrix factorization incorporated with deep image prior. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV). Seoul: IEEE, 2019:3126-35.
- Xie Z, Reheman B, Gong K, Zhang X, Qi J. Generative adversarial networks based regularized image reconstruction for PET. In: Proc.SPIE. Philadelphia: SPIE, 2019.
- Tang J, Yang B, Wang Y, Ying L. Sparsity-constrained PET image reconstruction with learned dictionaries. Phys Med Biol 2016;61:6347-68. [Crossref] [PubMed]
- Chen S, Liu H, Shi P, Chen Y. Sparse representation and dictionary learning penalized image reconstruction for positron emission tomography. Phys Med Biol 2015;60:807-23. [Crossref] [PubMed]
- Sudarshan VP, Egan GF, Chen Z, Awate SP. Joint PET-MRI image reconstruction using a patch-based joint-dictionary prior. Med Image Anal 2020;62:101669 [Crossref] [PubMed]
- Gong K, Catana C, Qi J, Li Q. PET image reconstruction using deep image prior. IEEE Trans Med Imaging 2019;38:1655-65. [Crossref] [PubMed]
- Kang J, Gao Y, Shi F, Lalush DS, Lin W, Shen D. Prediction of standard-dose brain PET image by using MRI and low-dose brain [18F]FDG PET images. Med Phys 2015;42:5301-9. [Crossref] [PubMed]
- Wang Y, Zhang P, An L, Ma G, Kang J, Shi F, Wu X, Zhou J, Lalush DS, Lin W, Shen D. Predicting standard-dose PET image from low-dose PET and multimodal MR images using mapping-based sparse representation. Phys Med Biol 2016;61:791-812. [Crossref] [PubMed]
- An L, Zhang P, Adeli E, Wang Y, Ma G, Shi F, Lalush DS, Lin W, Shen D. Multi-level canonical correlation analysis for standard-dose PET image estimation. IEEE Trans Image Process 2016;25:3303-15. [Crossref] [PubMed]
- Wang Y, Ma G, An L, Shi F, Zhang P, Lalush DS, Wu X, Pu Y, Zhou J, Shen D. Semisupervised tripled dictionary learning for standard-dose PET image prediction using low-dose PET and multimodal MRI. IEEE Trans Biomed Eng 2017;64:569-79. [Crossref] [PubMed]
- Costa-Luis COd, Reader AJ. Deep learning for suppression of resolution-recovery artefacts in MLEM PET image reconstruction. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-3.
- Gong K, Guan J, Liu C, Qi J. PET image denoising using deep neural network. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-2.
- Gong K, Guan J, Liu C, Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci 2019;3:153-61. [Crossref] [PubMed]
- Nazari M, Kimiaei S, Ehrenburg M, Kluge A, Bucher R. Convolutional neural network-based denoising allows 67% reduction of scan time or tracer dose in dopamine transporter SPECT. Eur J Nucl Med Mol Imaging 2019;46:S218-9.
- Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Neural Information Processing Systems 2012;25:1097-105.
- Xiang L, Qiao Y, Nie D, An L, Lin W, Wang Q, Shen D. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing 2017;267:406-16. [Crossref] [PubMed]
- Song C, Yang Y, Wernick MN, Pretorius PH, King MA. Low-dose cardiac-gated spect studies using a residual convolutional neural network. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Venice: IEEE, 2019:653-6.
- Song C, Yang Y, Wernick MN, Pretorius PH, King MA. Approximate 4D reconstruction of cardiac-gated spect images using a residual convolutional neural network. In: 2019 IEEE International Conference on Image Processing (ICIP). Taipei: IEEE, 2019:1262-6.
- Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, Poston KL, Sha SJ, Greicius MD, Mormino E, Pauly JM, Srinivas S, Zaharchuk G. Ultra-low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI Inputs. Radiology 2019;290:649-56. Erratum in: Radiology 2020;296:E195. [Crossref] [PubMed]
- Spuhler K, Serrano-Sosa M, Cattell R, DeLorenzo C, Huang C. Full-count PET recovery from low-count image using a dilated convolutional neural network. Med Phys 2020;47:4928-38. [Crossref] [PubMed]
- Xu J, Gong E, Pauly J, Zaharchuk G. 200x low-dose PET reconstruction using deep learning. arXiv 2017:1712.04119v1.
- Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, Liu C. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol 2019;64:165019 [Crossref] [PubMed]
- Liu CC, Qi J. Higher SNR PET image prediction using a deep learning model and MRI image. Phys Med Biol 2019;64:115004 [Crossref] [PubMed]
- Hashimoto F, Ohba H, Ote K, Teramoto A, Tsukada H. Dynamic PET image denoising using deep convolutional neural networks without prior training datasets. IEEE Access 2019;7:96594-603.
- Hashimoto F, Ote K, Tsukada H. Dynamic PET image denoising using deep convolutional neural network without training datasets. J Nucl Med 2019;60:abstr 242.
- Ramon AJ, Yang Y, Pretorius PH, Johnson KL, King MA, Wernick MN. Initial investigation of low-dose SPECT-MPI via deep learning. In: 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC). Sydney: IEEE, 2018:1-3.
- Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med Phys 2019;46:3555-64. [Crossref] [PubMed]
- Wang Y, Yu BT, Wang L, Zu C, Lalush DS, Lin WL, Wu X, Zhou JL, Shen DG, Zhou LP. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018;174:550-62. [Crossref] [PubMed]
- Wang Y, Zhou L, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D. 3D auto-context-based locality adaptive multi-modality GANs for PET synthesis. IEEE Trans Med Imaging 2019;38:1328-39. [Crossref] [PubMed]
- Wang Y, Zhou L, Wang L, Yu B, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D. Locality adaptive multi-modality GANs for high-quality PET image synthesis. Med Image Comput Comput Assist Interv 2018;11070:329-37.
- Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, Mao H, Nye JA, Yang X. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Phys Med Biol 2019;64:215017 [Crossref] [PubMed]
- Kaplan S, Zhu YM. Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging 2019;32:773-8. [Crossref] [PubMed]
- Xie Z, Baikejiang R, Li T, Zhang X, Gong K, Zhang M, Qi W, Asma E, Qi J. Generative adversarial network based regularized image reconstruction for PET. Phys Med Biol 2020;65:125016 [Crossref] [PubMed]
- Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, Zheng K, Wu Z, Fu L, Xu B, Zhu Z, Tian J, Liu H, Li Q. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging 2019;46:2780-9. [Crossref] [PubMed]
- Song TA, Chowdhury SR, El Fakhri G, Li QZ, Dutta J. Super-resolution PET imaging using a generative adversarial network. J Nucl Med 2019;60:abstr 576.
- Song TA, Chowdhury SR, Yang F, Dutta J. PET image super-resolution using generative adversarial networks. Neural Netw 2020;125:83-91. [Crossref] [PubMed]
- Chan C, Zhou J, Yang L, Qi W, Asma E. Noise to noise ensemble learning for PET image denoising. In: 2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Manchester: IEEE, 2019:1-3.
- Yie SY, Kang SK, Hwang D, Lee JS. Self-supervised PET denoising. Nucl Med Mol Imaging 2020;54:299-304. [Crossref] [PubMed]
- Liu J, Yang Y, Wernick MN, Pretorius PH, King MA. Deep learning with noise-to-noise training for denoising in SPECT myocardial perfusion imaging. Med Phys 2021;48:156-68. [Crossref] [PubMed]
- Rahmani S, Strait M, Merkurjev D, Moeller M, Wittman T. An adaptive IHS pan-sharpening method. IEEE Geosci Remote Sens Lett 2010;7:746-50. [Crossref]
- Shabanzade F, Ghassemian H. Multimodal image fusion via sparse representation and clustering-based dictionary learning algorithm in NonSubsampled Contourlet domain. In: 2016 8th International Symposium on Telecommunications (IST). Tehran: IEEE, 2016:472-7.
- Seal A, Bhattacharjee D, Nasipuri M, Rodríguez-Esparragón D, Menasalvas E, Gonzalo-Martin C. PET-CT image fusion using random forest and à-trous wavelet transform. Int J Numer Method Biomed Eng 2018; [Crossref] [PubMed]
- Panigrahy C, Seal A, Mahato NK. MRI and SPECT image fusion using a weighted parameter adaptive dual channel PCNN. IEEE Signal Process Lett 2020;27:690-4. [Crossref]
- Ouerghi H, Mourali O, Zagrouba E. Non-subsampled shearlet transform based MRI and PET brain image fusion using simplified pulse coupled neural network and weight local features in YIQ colour space. IET Image Processing 2018;12:1873-80. [Crossref]
- Huang C, Tian G, Lan Y, Peng Y, Ng EYK, Hao Y, Cheng Y, Che W. a new pulse coupled neural network (PCNN) for brain medical image fusion empowered by shuffled frog leaping algorithm. Front Neurosci 2019;13:210. [Crossref] [PubMed]
- Kumar A, Fulham M, Feng D, Kim J. Co-learning feature fusion maps from PET-CT images of lung cancer. IEEE Trans Med Imaging 2019; Epub ahead of print. [Crossref] [PubMed]
- Shi J, Zheng X, Li Y, Zhang Q, Ying S. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer's disease. IEEE J Biomed Health Inform 2018;22:173-83. [Crossref] [PubMed]
- Hermessi H, Mourali O, Zagrouba E. Convolutional neural network-based multimodal image fusion via similarity learning in the shearlet domain. Neural Comput Appl 2018;30:2029-45. [Crossref]
- Stabin MG, Madsen MT, Zaidi H. Personalized dosimetry is a must for appropriate molecular radiotherapy. Med Phys 2019;46:4713-6. [Crossref] [PubMed]
- Zaidi H. Relevance of accurate Monte Carlo modeling in nuclear medical imaging. Med Phys 1999;26:574-608. [Crossref] [PubMed]
- Bolch WE, Bouchet LG, Robertson JS, Wessels BW, Siegel JA, Howell RW, Erdi AK, Aydogan B, Costes S, Watson EE, Brill AB, Charkes ND, Fisher DR, Hays MT, Thomas SR. MIRD pamphlet No. 17: the dosimetry of nonuniform activity distributions--radionuclide S values at the voxel level. Medical Internal Radiation Dose Committee. J Nucl Med 1999;40:11S-36S. [PubMed]
- Berger MJ. Distribution of absorbed dose around point sources of electrons and beta particles in water and other media. J Nucl Med 1971;5-23. [PubMed]
- Akhavanallaf A, Shiri I, Arabi H, Zaidi H. Whole-body voxel-based internal dosimetry using deep learning. Eur J Nucl Med Mol Imaging 2020; Epub ahead of print. [Crossref] [PubMed]
- Götz TI, Lang EW, Schmidkonz C, Kuwert T, Ludwig B. Dose voxel kernel prediction with neural networks for radiation dose estimation. Z Med Phys 2021;31:23-36. [Crossref] [PubMed]
- Zeng GL. Machine learning: any image reconstruction algorithm can learn by itself. In: 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Atlanta: IEEE, 2017:1-3.
- Yu Z, Rahman MA, Schindler T, Gropler R, Laforest R, Wahl R, Jha A. AI-based methods for nuclear-medicine imaging: Need for objective task-specific evaluation. J Nucl Med 2020;61:abstr 575.


