
Texture analysis based on quantitative magnetic resonance imaging to assess kidney function: a preliminary study
Introduction
Chronic kidney disease (CKD) is a global public health problem with an unabated rise in prevalence and mortality (1). The early detection of renal function impairment and prediction of the likelihood of a progressive decline in the glomerular filtration rate (GFR) is important for timely therapeutic management (2). It has been established that decreased perfusion, chronic hypoxia, and renal fibrosis play a critical role in causing kidney damage (3). Thus, researchers have made efforts to develop novel biomarkers that can accurately assess these renal changes to detect and evaluate renal function impairment early and potentially predict disease progression (4).
Magnetic resonance imaging (MRI) has been applied as a useful tool to assess chronic kidney disease noninvasively, and different MRI techniques have shown great potential in the evaluation of renal function (5). Diffusion weighted imaging (DWI), one of the most studied techniques, has been demonstrated to characterize renal function, with decreased apparent diffusion coefficient (ADC) values in patients with renal dysfunction (6). Other studies have evaluated T1 and T2 mapping, which have already been applied in clinical practice to quantify myocardial edema or fibrosis in patients with myocardial infarction or cardiomyopathy (7,8). It has been shown that T1 mapping can detect the severity of acute kidney injury and predict further outcomes and that T2 values are sensitive markers of early cystogenesis in polycystic kidney disease (9,10). However, the study of T1 and T2 mapping to evaluate humans' renal function is limited (11). Also, DWI and T1 and T2 mapping measure the average value within a particular lesion and do not reflect the tissue’s signal heterogeneity.
Texture analysis (TA) is a novel technique that performs an ensemble of mathematical computations on conventional images to quantify tissue heterogeneity (12,13). It has been applied as new imaging biomarkers in oncology to classify tumors, predict prognosis, and monitor treatment responses (14-16). Previous studies have also demonstrated that TA based on T2-weighted images offers an approach to refine autosomal dominant polycystic kidney disease (ADPKD) and that TA based on DWI, blood oxygen level-dependent (BOLD) MRI, and susceptibility-weighted imaging (SWI) can assist in evaluating renal dysfunction (17,18). However, renal function evaluation in humans using TA based on ADC, T1, and T2 values has not yet been explored.
Therefore, the purpose of this study was to investigate the feasibility of using TA based on DWI and T1 and T2 mapping to detect GFR decline and discriminate different degrees of renal function impairment.
Methods
Patients
This prospective study was approved by the Medical Ethics Committee of Peking Union Medical College Hospital (Ethical No.: ZS-1271), and written informed consent was obtained from each participant. From February 2017 to May 2017, patients were randomly and consecutively recruited from the Department of Nephrology in our hospital. The inclusion criteria were as follows: (I) age ≥18 years old; and (II) biopsy-proven or clinically confirmed renal diseases, including Gitelman syndrome, immunoglobulin (Ig) A nephropathy, CKD, IgG4 nephropathy, malignant hypertension, acute kidney injury, and antineutrophil cytoplasmic antibodies (ANCA)-associated systemic vasculitis. The exclusion criteria included pregnancy, lactation, malignancies, hemodialysis, renal tumors with maximal diameter >1 cm or number of renal tumors >5 in each kidney, and patients unable to hold their breath for over 10 seconds, and general contradictions for MRI examination.
All participants followed 4-hour pre-examination fasting. Clinical information was collected from the medical database of our hospital. Estimated GFR (eGFR) was calculated using the Chronic Kidney Disease Epidemiology Collaboration equation as follows: GFR (mL/min/1.73 m2)=141×min (Scr/κ,1)α×max (Scr/κ,1)1.209×0.993age×1.018 (if female) ×1.157 (if black), where κ is 0.9 for males and 0.7 for females, α is −0.411 for males and −0.329 for females, min indicates the minimum value and max indicates the maximum value, age is in years, weight is in kilograms, and Scr is the serum creatinine level in micromoles per liter. The enrolled subjects were classified into four different groups according to their eGFR: (I) a normal renal function group (nRF), eGFR ≥90 mL/min/1.73 m2; (II) a mildly impaired renal function group (mi-IRF), 60≤ eGFR <90 mL/min/1.73 m2; (III) a moderately impaired renal function group (mo-IRF), 30≤ eGFR <60 mL/min/1.73 m2; and (IV) a severely impaired renal function group (se-IRF), eGFR <30 mL/min/1.73 m2.
MRI protocol
All MRI images were obtained on a MAGNETOM Skyra 3T MR scanner (Siemens Healthcare, Erlangen, Germany) using an 18-channel phased-array body coil combined with a 32-channel spine coil with imaging parameters in each sequence described in Table S1. To reduce the respiratory motion artifacts, patients held their breath during T1 mapping scanning. During T2 mappin scanning, motion artifacts were reduced by using respiratory triggerin through synchronizing the measurement with the breathing cycle of the patient. We also asked patients to use thoracic breathing and take shallow breaths during the scanning. Axial and coronal T2-weighted images were acquired for the kidney structure's anatomical identification, and then axial DWI and coronal T1 and T2 mapping of both kidneys were performed. T1 mapping based on the inversion recovery SNAPSHOT-FLASH sequence with respective reconstruction was described in detail in the work of Deichmann and Haase (19). For T1 mapping, free relaxation of the longitudinal magnetization after a 180° inversion is modeled by
Image analysis
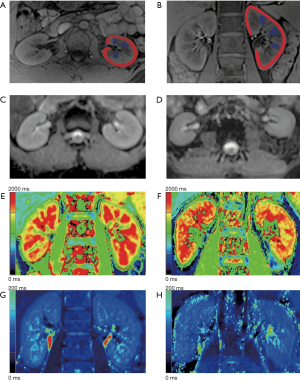
The images were anonymized before being reviewed by radiologists who were blinded to the participants’ clinical information, including renal function. One genitourinary radiologist (13 years of experience) selected the slice at the renal hilum level for each ADC and T1 and T2 map. Anatomical landmarks and visual coregistration were used to find corresponding slices among different maps. The selected images were transmitted to a workstation for TA afterward. TA was performed by a trained radiologist with 6 years of experience in renal imaging and 4 years of experience in TA using the commercially available research software TexRAD (TexRAD Ltd., www.texrad.com, part of Feedback Plc, Cambridge, UK). On each selected image, regions of interest (ROIs) for the cortex and medulla were placed in both kidneys (Figure 1A,B). For the cortex, the ROI was delineated along the cortex’s outline to cover the entire cortex. For the medulla, since many patients with impaired renal function had diminished corticomedullary differentiation (CMD), it was difficult to draw the medulla outline accurately. Thus, we placed at least three pyramidal ROIs in each kidney to represent the entire medulla, and the average of the values in these pyramidal ROIs was subsequently used for analysis.
Texture quantification by histogram analysis was performed after an image filtration process using a Laplacian Gaussian spatial bandpass filter (22). The spatial scaling factor (SSF) represented the size of the image features highlighted by the filter, and ranged between object radii of 0, 2, 3, 4, 5, and 6 mm. An SSF of 0 indicated no filtration, an SSF 2 represented fine, an SSF 3–5 represented medium, and an SSF 6 represented coarse texture scales. At each SSF, six texture features were extracted. The six texture features generated using the histogram were mean gray-level intensity (mean), standard deviation (sd), entropy, mean of positive pixels (mpp), skewness, and kurtosis. The value of each texture parameter across each SSF on each slice was recorded for both the cortex and medulla of the left, right, and both kidneys, respectively, on ADC and T1 and T2 maps. The values of the parameters for the cortex and medulla of both kidneys were automatically provided by the software. The process was repeated three times, and the average values of three measurements for each parameter on the ADC and T1 and T2 maps were used for statistical analysis.
Statistical analysis
The texture feature analysis and prediction model construction were implemented with scikit-learn (https://scikit-learn.org/) on Python 3.5.4 (https://www.python.org/). For each patient, a total of 6 (mean, sd, mpp, entropy, skewness, kurtosis) ×6 (SSF 0, 2, 3, 4, 5, 6) ×6 (cortex of right, left, both kidneys; medulla of right, left, both kidneys) ×3 (ADC, T1 mapping, T2 mapping) =648 features were extracted. Spearman's rank correlation coefficients were calculated between all features and prediction labels. We removed features that were strongly correlated with each other (those exhibiting a high correlation coefficient over 0.75). Features with a low correlation coefficient (<0.3) or a corresponding P value greater than 0.05 were removed accordingly. The least absolute shrinkage and selection operator (LASSO) algorithm was used to implement next-step feature dimensionality reduction. The remaining features were then used to train a prediction model in the training cohort.
We compared six machine-learning classifiers, including Ridge Classifier, LogisticRegression, LinearSVC, Perceptron, SGDClassifier, and PassiveAggressiveClassifier. Five-fold cross-validation was used to compare the performance (prediction accuracy) of different machine-learning classifiers and help to select the optimal one. Receiver operating characteristic (ROC) analysis was performed, and the area under the ROC curve (AUC) was calculated for each comparison. The Delong test was used to calculate the 95% confidence index (CI) of the ROC curves. A two-sided P<0.05 indicated a statistically significant difference.
Results
Patient characteristics
In total, 116 participants were included in this study. Figure 1C,D,E,F,G,H shows examples of ADC and T1 and T2 maps of a healthy volunteer and a patient with impaired renal function, respectively. Figure 2 depicts the flowchart of patient selection in this study. Of the 116 subjects, 94 were patients with renal diseases (male/female 57/37, mean age 42.3±14.1 years), and 22 were healthy volunteers (male/female 12/10, mean age 29.4±6.2 years). Of the 94 patients with renal diseases, 20 had genetic testing confirmed Gitelman syndrome, 26 had biopsy-proven IgA nephropathy, 26 had clinically confirmed CKD, 12 had biopsy-proven IgG4 nephropathy, eight had malignant hypertension, one had acute kidney injury, and one had biopsy-proven ANCA-associated systemic vasculitis. The etiologies of CKD were as follows: chronic glomerulonephritis (n=9), diabetic mellitus nephropathy (n=5), chronic interstitial nephritis (n=4), and unknown etiology (n=8).
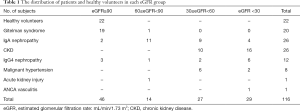
According to the eGFR, there were 46 patients with nRF (male/female 22/23, mean age 30.2±11.1 years, mean eGFR 117.3±14.0 mL/min/1.73 m2), 14 patients with mi-IRF (male/female 9/5, mean age 41.9±14.3 years, mean eGFR 72.0±6.5 mL/min/1.73 m2), 27 patients with mo-IRF (male/female 18/9, mean age 41.2±14.9 years, mean eGFR 46.4±8.9 mL/min/1.73 m2), and 29 patients with se-IRF (male/female 21/8, mean age 44.6±15.7 years, mean eGFR 19.1±7.5 mL/min/1.73 m2). Table 1 details the distribution of patients and healthy volunteers in each eGFR group. Patients in the nRF group were significantly younger than the patients in any of the IRF groups. The ages of the patients in the mi-IRF group were similar to those of the patients in the mo-IRF group, and both were slightly younger than the patients in the se-IRF group.
Full table
Detection and characterization of impaired renal function
None of the texture features quantified from the T2 map were selected to build classifiers to detect or evaluate renal function impairment. As for selecting the optimal classifier, RidgeClassifier was used to distinguish between nRF and IRF. Linear SVC and RidgeClassifier were further used to characterize the severity of impaired renal function: mi-IRF vs. mo-IRF and mo-IRF vs. se-IRF, respectively (Figure 3). Table 2 summarizes different classifiers’ performance built by texture features from ADC and T1 maps to differentiate between normal and abnormal, mildly impaired and moderately impaired, and moderately impaired and severely impaired renal function.


Full table
Normal vs. abnormal renal function
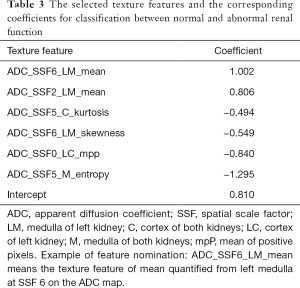
To distinguish between the normal and abnormal renal function groups, the patients were divided into two groups: nRF (n=46, eGFR ≥90 mL/min/1.73 m2) and IRF (n=70, eGFR <90 mL/min/1.73 m2). Data were randomly divided into training and testing datasets at a ratio of 2:1 (training n=77, testing n=39). The six texture features with the highest coefficients were selected to build the classifier. The selected features and corresponding coefficients are provided in Table 3. All of the selected texture features were quantified from ADC maps. The ADC-based texture features demonstrated favorable discrimination in both the training and testing datasets (AUC training: 0.877, testing: 0.835, P=0.63). The ROC curves for both datasets with five-fold cross validation are shown in Figure 4A,B. The performance of the classifier to distinguish between normal and abnormal renal function in both datasets was as follows: accuracy: training 0.821, testing 0.779; sensitivity: training 0.792, testing 0.761; specificity: training 0.867, testing 0.806; NPV: training 0.722, testing 0.694; and PPV: training 0.905, testing 0.854. The distribution plot showing the differences in the normal and abnormal function groups’ selected texture features is demonstrated in Figure 4C.
Full table
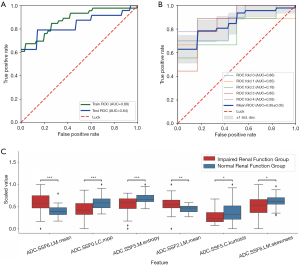
Mildly vs. moderately impaired renal function
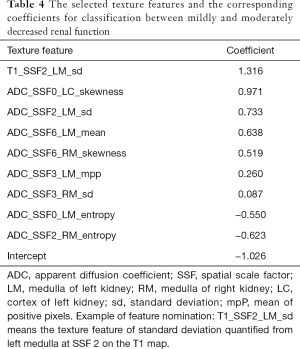
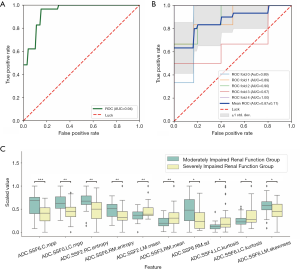
To distinguish between mi-IRF (n=14, 60≤ eGFR <90 mL/min/1.73 m2) and mo-IRF (n=27, 30≤ eGFR <60 mL/min/1.73 m2), nine texture features were selected, including seven features quantified from the ADC map and one feature quantified from the T1 map to build the classifier (Table 4). The classifier produced an AUC of 0.937 [95% confidential interval (CI): 0.864–1], an accuracy of 0.878, a sensitivity of 0.889, a specificity of 0.857, an NPV of 0.800, and a PPV of 0.923 to differentiate between mi-IRF and mo-IRF (Table 2). The ROC curves with cross validation are shown in Figure 5A,B, and the distribution plot of the selected texture features in the mildly and moderately impaired renal function groups is demonstrated in Figure 5C.
Full table
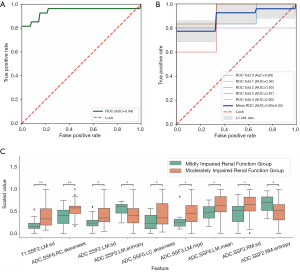
Moderately vs. severely impaired renal function
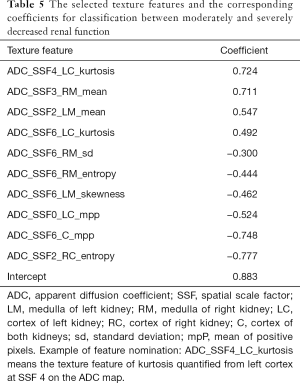
For separating mo-IRF (n=27, 30≤ eGFR <60 mL/min/1.73 m2) from se-IRF (n=29, eGFR <30 mL/min/1.73 m2), we found that the classifier with 10 selected features quantified from ADC maps could yield an AUC of 0.940 (95% CI: 0.883–1), an accuracy of 0.821, a sensitivity of 0.759, a specificity of 0.889, an NPV of 0.774, and a PPV of 0.880 (Table 2). The selected features and corresponding coefficients are provided in Table 5. The ROC curves with cross validation are shown in Figure 6A,B, and the distribution plot of the selected texture features of the moderately and severely impaired renal function groups is shown in Figure 6C.
Full table
Discussion
This study demonstrated that texture features quantified from ADC and T1 maps were related to eGFR, and a higher number of texture features on ADC maps showed a correlation with eGFR than those based on T1 maps. Texture features quantified from the renal medulla seemed to be more related to renal function than features quantified from the renal cortex. Combinations of different texture features could enable the detection of eGFR decline, and the discrimination between different degrees of eGFR decreases with relatively satisfactory performance.
Consistent efforts have been made to reveal DWI's potential for evaluating renal diseases, and promising results have been achieved for estimating fibrosis in CKD and guiding biopsy in acute graft dysfunction (23,24). Of the DWI models, the monoexponential model with ADC calculation is the most robust and widely used. Numerous studies have confirmed that ADC can differentiate between normal and impaired kidneys and correlates with renal function estimated by creatinine values (25-29). A few studies have also revealed that cortical ADC is relatively well correlated with cortical fibrosis and chronic lesions (30-32). In the present study, certain texture features quantified from the ADC map showed good correlation with eGFR changes and were selected to build models for the classification of different degrees of renal impairment, which achieved satisfactory performance. All of the selected texture features to classify normal, abnormal, moderately, and severely impaired renal function were quantified from the ADC map, indicating the potentially significant role of the ADC map in evaluating renal function. However, as ADC is a single parameter in the monoexponential model, and since renal impairment is a rather complicated process, it remains unclear whether alterations in ADC values reflect the decline of renal function alone, the degree of tissue fibrosis, or both.
Several studies have investigated renal T1 mapping for evaluating renal transplants and renal function. T1 CMD could be used to evaluate renal interstitial fibrosis in allografts, and T1 values are sensitive to possible acute kidney injury changes in patients with lung transplantation (33-35). Cortical T1 and T1 CMD are moderate to strongly correlated with the severity of renal impairment (36,37). Our study revealed that standard deviation quantified from the left kidney's medulla at SSF2 on the T1 map showed a good correlation with eGFR decline and was selected to build the classifier to differentiate between mildly and moderately impaired renal function. However, no other texture features quantified from the T1 map were selected to build classifiers to detect or evaluate renal function impairment. One possible explanation for this is that ADC-based texture features outperformed T1 map-based texture features in the assessment of renal function; thus, T1 map-based texture features were not selected when building the classifiers. The only selected T1 map-based texture feature was quantified from the renal medulla. However, as previous studies have demonstrated that cortical T1 is sensitive to oxygenation level changes, the implication of medullary T1 modulations has yet to be determined. Thus, this result should be interpreted with caution, and whether specific texture features quantified from medullary T1 values truly correlate with eGFR decline still needs to be further clarified and remains a subject for future exploration.
The role of T2 mapping in evaluating renal function in humans is also under investigation, but studies involving in vivo measurements of renal T2 values are relatively scarce. Elevated T2 values were observed in patients with early-stage ADPKD, and T2 mapping may potentially improve the assessment of early-disease progression compared with total kidney volume (17). It has also been shown that renal T2 values measurements have the potential to assess ischemia-reperfusion injury (38). In the present study, we attempted to identify the relation of in vivo renal T2 values to eGFR and clarify the role of T2 mapping in evaluating renal function. Our results, however, showed that none of the texture features quantified from the T2 map were selected to detect eGFR decline or discriminate between different degrees of renal function impairment. It seems that renal T2 values are not correlated with eGFR, and it is possible that T2 mapping may not be of great value in evaluating renal dysfunction. However, further research is needed to confirm our results and explore the potential of T2 mapping in assessing renal diseases.
TA has become a novel research focus on rapid development in recent years, especially in oncological imaging. Quantitative texture features are promising biomarkers for pathological changes or the response to treatment (14). Several studies have explored the potential of TA to evaluate kidney diseases. Kline et al. demonstrated that TA of T2-weighted MRI images could be a significant prognostic biomarker for the subsequent eGFR decline and disease progression in ADPKD (17). Researchers used the stability of texture features as the basis for feature selection, and entropy was selected as a result that showed a strong correlation with the subsequent percentage change in eGFR. Ding et al. performed TA on DWI, BOLD, and SWI to evaluate renal dysfunction and found that BOLD and SWI (but not DWI) may be suitable for assessing renal dysfunction in the early stages (18). They showed that entropy was correlated with eGFR, which was similar to our results; however, skewness and kurtosis were not significantly correlated with eGFR, which differed from our findings. This discrepancy may be attributable to the fact that they quantified texture features from the renal parenchyma instead of our study, which quantified texture features from the renal cortex or medulla, respectively. Like the study by Kline et al., entropy quantified from BOLD also showed the capability to differentiate non-severe renal function impairment from normal renal function (17). In line with these two studies, our results support the conclusion that TA based on MRI could evaluate renal dysfunction. In addition to the ADC map, we explored the potential of texture features quantified from T1 and T2 maps to evaluate renal function, and our results implied that T1 mapping might be of value for the evaluation of renal function. However, our findings need to be confirmed in future studies.
The present study has several limitations that should be noted. First, since the subjects were divided into four groups according to eGFR, each group's sample size was small, especially the mi-IRF group (n=14). The dataset was imbalanced when differentiating mi-IRF from mo-IRF (n=27). A greater number of subjects with more balanced grouping should be enrolled to validate the results further. Second, since we focused on evaluating renal function based on eGFR rather than underlying renal diseases, patients with different renal pathologies were included. Our results showed that TA based on quantitative MRI could detect and characterize impaired renal function regardless of the cause of renal impairment. However, we did not assess TA’s potential to distinguish among these renal pathologies, which is worth investigating in the future.
Third, the data were not divided into training and validation datasets when differentiating between mildly and moderately impaired renal function and between moderately and severely impaired renal function because of the limited number of patients in each group. Further investigation with an independent validation cohort should be conducted to validate our results. Fourth, we only selected the slice at the renal hilum level for analysis, given the time-saving nature of this method and the fact that not all the slices of the included patients exhibited sufficiently good image quality for TA. We plan to perform whole kidney analysis in the future to validate our study. Fifth, since many patients with impaired renal function in this study had diminished CMD, it was hard to draw the medulla outline accurately. Thus, we adopted a compromise approach by placing at least three pyramidal ROIs on the medulla to represent the whole medulla, which may have introduced bias. A greater number of patients with visible CMD should be enrolled to allow the entire medulla’s delineation for further analysis. Sixth, undoubtedly, TA based on MRI is not time- or cost-effective for evaluating renal function; however, our study demonstrated the possibility of using this novel technique to evaluate renal dysfunction or disorders.
In conclusion, TA based on quantitative MRI offers an opportunity to monitor renal dysfunction. Compared with those from T2 maps, texture features quantified from ADC and T1 maps may be more suitable for detecting and characterizing renal dysfunction with relatively satisfactory performance. It is promising that texture features based on quantitative MRI may serve as imaging biomarkers to reveal renal impairment and potentially act as a tool to evaluate renal pathologies with further exploration in the future noninvasively.
Acknowledgments
Funding: This study has received funding from the Beijing Municipal Natural Science Foundation under Grant No.7192176; the National Natural Science Foundation of China under Grant No. 81901742; and the Clinical and Translational Research Project of the Chinese Academy of Medical Sciences under Grant No. 2019XK320028.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-842). JS reports that he is an employee of Philips Healthcare in China. JA reports that she is an employee of Siemens Healthcare in China. The other authors have no conflicts of interest to declare. None of the authors serves as a current Editorial Board Member or Section Editor for this journal.
Ethical Statement: This prospective study was approved by the Medical Ethics Committee of Peking Union Medical College Hospital (Ethical No.: ZS-1271), and written informed consent was obtained from each participant.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Webster AC, Nagler EV, Morton RL, Masson P. Chronic Kidney Disease. Lancet 2017;389:1238-52. [Crossref] [PubMed]
- Andrassy KM. Comments on 'KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease'. Kidney Int 2013;84:622-3. [Crossref] [PubMed]
- Fine LG, Norman JT. Chronic hypoxia as a mechanism of progression of chronic kidney diseases: from hypothesis to novel therapeutics. Kidney Int 2008;74:867-72. [Crossref] [PubMed]
- Zhang WR, Parikh CR. Biomarkers of Acute and Chronic Kidney Disease. Annu Rev Physiol 2019;81:309-33. [Crossref] [PubMed]
- Jiang K, Lerman LO. Prediction of Chronic Kidney Disease Progression by Magnetic Resonance Imaging: Where Are We? Am J Nephrol 2019;49:111-3. [Crossref] [PubMed]
- Caroli A, Schneider M, Friedli I, Ljimani A, De Seigneux S, Boor P, Gullapudi L, Kazmi I, Mendichovszky IA, Notohamiprodjo M, Selby NM, Thoeny HC, Grenier N, Vallée JP. Diffusion-weighted magnetic resonance imaging to assess diffuse renal pathology: a systematic review and statement paper. Nephrol Dial Transplant 2018;33:ii29-ii40. [Crossref] [PubMed]
- Duca F, Kammerlander AA, Panzenböck A, Binder C, Aschauer S, Loewe C, Agis H, Kain R, Hengstenberg C, Bonderman D, Mascherbauer J. Cardiac Magnetic Resonance T1 Mapping in Cardiac Amyloidosis. JACC Cardiovasc Imaging 2018;11:1924-6. [Crossref] [PubMed]
- Krittayaphong R, Zhang S, Saiviroonporn P, Viprakasit V, Tanapibunpon P, Rerkudom B, Yindeengam A, Wood JC. Assessment of Cardiac Iron Overload in Thalassemia With MRI on 3.0-T: High-Field T1, T2, and T2* Quantitative Parametric Mapping in Comparison to T2* on 1.5-T. JACC Cardiovasc Imaging 2019;12:752-4. [Crossref] [PubMed]
- Franke M, Baeßler B, Vechtel J, Dafinger C, Höhne M, Borgal L, Göbel H, Koerber F, Maintz D, Benzing T, Schermer B, Persigehl T. Magnetic resonance T2 mapping and diffusion-weighted imaging for early detection of cystogenesis and response to therapy in a mouse model of polycystic kidney disease. Kidney Int 2017;92:1544-54. [Crossref] [PubMed]
- Adams LC, Bressem KK, Scheibl S, Nunninger M, Gentsch A, Fahlenkamp UL, Eckardt KU, Hamm B, Makowski MR. Multiparametric Assessment of Changes in Renal Tissue after Kidney Transplantation with Quantitative MR Relaxometry and Diffusion-Tensor Imaging at 3 T. J Clin Med 2020;9:1551. [Crossref] [PubMed]
- Wolf M, de Boer A, Sharma K, Boor P, Leiner T, Sunder-Plassmann G, Moser E, Caroli A, Jerome NP. Magnetic resonance imaging T1- and T2-mapping to assess renal structure and function: a systematic review and statement paper. Nephrol Dial Transplant 2018;33:ii41-ii50. [Crossref] [PubMed]
- Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59:1061-9. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Miller TA, Robinson KR, Li H, Seiwert TY, Haraf DJ, Lan L, Giger ML, Ginat DT. Prognostic value of pre-treatment CT texture analysis in combination with change in size of the primary tumor in response to induction chemotherapy for HPV-positive oropharyngeal squamous cell carcinoma. Quant Imaging Med Surg 2019;9:399-408. [Crossref] [PubMed]
- Shi L, Zhou XL, Sun JJ, Huang JH, Wang X, Li K, Pang PP, Xu YJ, Chen M, Zhang MM. Whole-tumor perfusion CT using texture analysis in unresectable stage IIIA/B non-small cell lung cancer treated with recombinant human endostatin. Quant Imaging Med Surg 2019;9:968-75. [Crossref] [PubMed]
- Chamming’s F. Ueno Y, Ferré R, Kao E, Jannot AS, Chong J, Omeroglu A, Mesurolle B, Reinhold C, Gallix B. Features from Computerized Texture Analysis of Breast Cancers at Pretreatment MR Imaging Are Associated with Response to Neoadjuvant Chemotherapy. Radiology 2018;286:412-20. [Crossref] [PubMed]
- Kline TL, Korfiatis P, Edwards ME, Bae KT, Yu A, Chapman AB, Mrug M, Grantham JJ, Landsittel D, Bennett WM, King BF, Harris PC, Torres VE, Erickson BJ. CRISP Investigators. Image texture features predict renal function decline in patients with autosomal dominant polycystic kidney disease. Kidney Int 2017;92:1206-16. [Crossref] [PubMed]
- Ding J, Xing Z, Jiang Z, Zhou H, Di J, Chen J, Qiu J, Yu S, Zou L, Xing W. Evaluation of renal dysfunction using texture analysis based on DWI, BOLD, and susceptibility-weighted imaging. Eur Radiol 2019;29:2293-301. [Crossref] [PubMed]
- Deichmann R, Haase A. Quantification of T1 Values by SNAPSHOT-FLASH NMR Imaging. Journal of Magnetic Resonance 1992;96:608-12.
- Sumpf TJ, Uecker M, Boretius S, Frahm J. Model-based nonlinear inverse reconstruction for T2 mapping using highly undersampled spin-echo MRI. J Magn Reson Imaging 2011;34:420-8. [Crossref] [PubMed]
- Block KT, Uecker M, Frahm J. Model-based iterative reconstruction for radial fast spin-echo MRI. IEEE Transactions on Medical Imaging 2009;28:1759-69. [Crossref] [PubMed]
- Goh V, Ganeshan B, Nathan P, Juttla JK, Vinayan A, Miles KA. Assessment of response to tyrosine kinase inhibitors in metastatic renal cell cancer: CT texture as a predictive biomarker. Radiology 2011;261:165-71. [Crossref] [PubMed]
- Steiger P, Barbieri S, Kruse A, Ith M, Thoeny HC. Selection for biopsy of kidney transplant patients by diffusion-weighted MRI. Eur Radiol 2017;27:4336-44. [Crossref] [PubMed]
- Inoue T, Kozawa E, Okada H, Inukai K, Watanabe S, Kikuta T, Watanabe Y, Takenaka T, Katayama S, Tanaka J, Suzuki H. Noninvasive evaluation of kidney hypoxia and fibrosis using magnetic resonance imaging. J Am Soc Nephrol 2011;22:1429-34. [Crossref] [PubMed]
- Emre T, Kilickesmez O, Buker A, Inal BB, Dogan H, Ecder T. Renal function and diffusion-weighted imaging: a new method to diagnose kidney failure before losing half function. Radiol Med 2016;121:163-72. [Crossref] [PubMed]
- Cakmak P, Yagci AB, Dursun B, Herek D, Fenkci SM. Renal diffusion-weighted imaging in diabetic nephropathy: correlation with clinical stages of disease. Diagn Interv Radiol 2014;20:374-8. [Crossref] [PubMed]
- Namimoto T, Yamashita Y, Mitsuzaki K, Nakayama Y, Tang Y, Takahashi M. Measurement of the apparent diffusion coefficient in diffuse renal disease by diffusion-weighted echo-planar MR imaging. J Magn Reson Imaging 1999;9:832-7. [Crossref] [PubMed]
- Xu X, Fang W, Ling H, Chai W, Chen K. Diffusion-weighted MR imaging of kidneys in patients with chronic kidney disease: initial study. Eur Radiol 2010;20:978-83. [Crossref] [PubMed]
- Thoeny HC, De Keyzer F, Oyen RH, Peeters RR. Diffusion-weighted MR imaging of kidneys in healthy volunteers and patients with parenchymal diseases: initial experience. Radiology 2005;235:911-7. [Crossref] [PubMed]
- Li Q, Li J, Zhang L, Chen Y, Zhang M, Yan F. Diffusion-weighted imaging in assessing renal pathology of chronic kidney disease: A preliminary clinical study. Eur J Radiol 2014;83:756-62. [Crossref] [PubMed]
- Razek AA, Al-Adlany M, Alhadidy AM, Atwa MA, Abdou NEA. Diffusion tensor imaging of the renal cortex in diabetic patients: correlation with urinary and serum biomarkers. Abdom Radiol (NY) 2017;42:1493-500. [Crossref] [PubMed]
- Zhao J, Wang ZJ, Liu M, Zhu J, Zhang X, Zhang T, Li S, Li Y. Assessment of renal fibrosis in chronic kidney disease using diffusion-weighted MRI. Clin Radiol 2014;69:1117-22. [Crossref] [PubMed]
- Friedli I, Crowe LA, Berchtold L, Moll S, Hadaya K, de Perrot T, Vesin C, Martin PY, de Seigneux S, Vallée JP. New Magnetic Resonance Imaging Index for Renal Fibrosis Assessment: A Comparison between Diffusion-Weighted Imaging and T1 Mapping with Histological Validation. Sci Rep 2016;6:30088. [Crossref] [PubMed]
- Huang Y, Sadowski EA, Artz NS, Seo S, Djamali A, Grist TM, Fain SB. Measurement and comparison of T1 relaxation times in native and transplanted kidney cortex and medulla. J Magn Reson Imaging 2011;33:1241-7. [Crossref] [PubMed]
- Peperhove M, Vo Chieu VD, Jang MS, Gutberlet M, Hartung D, Tewes S, Warnecke G, Fegbeutel C, Haverich A, Gwinner W, Lehner F, Bräsen JH, Haller H, Wacker F, Gueler F, Hueper K. Assessment of acute kidney injury with T1 mapping MRI following solid organ transplantation. Eur Radiol 2018;28:44-50. [Crossref] [PubMed]
- Gillis KA, McComb C, Patel RK, Stevens KK, Schneider MP, Radjenovic A, Morris ST, Roditi GH, Delles C, Mark PB. Non-Contrast Renal Magnetic Resonance Imaging to Assess Perfusion and Corticomedullary Differentiation in Health and Chronic Kidney Disease. Nephron 2016;133:183-92. [Crossref] [PubMed]
- Cox EF, Buchanan CE, Bradley CR, Prestwich B, Mahmoud H, Taal M, Selby NM, Francis ST. Multiparametric Renal Magnetic Resonance Imaging: Validation, Interventions, and Alterations in Chronic Kidney Disease. Front Physiol 2017;8:696. [Crossref] [PubMed]
- Ko SF, Yip HK, Zhen YY, Lee CC, Lee CC, Huang SJ, Huang CC, Ng SH, Lin JW. Severe bilateral ischemic-reperfusion renal injury: hyperacute and acute changes in apparent diffusion coefficient, T1, and T2 mapping with immunohistochemical correlations. Sci Rep 2017;7:1725. [Crossref] [PubMed]


