
Building a patient-specific model using transfer learning for four-dimensional cone beam computed tomography augmentation
Introduction
Cone-beam computed tomography (CBCT) has been widely used for patient positioning guidance in radiation therapy (1). However, when three-dimensional (3D)-CBCT is used to image thoracic or upper abdominal regions, respiratory motions produce severe artifacts on images, thus compromising the efficacy of 3D-CBCT in image-guided radiation therapy (IGRT). To address this issue, four-dimensional (4D)-CBCT was developed to produce respiratory phase-resolved volumetric images. In 4D-CBCT, all projections are first sorted into different respiratory phase bins according to the breathing signal, and the CBCT images are then separately reconstructed for each phase bin (2-4). 4D-CBCT is able to reduce motion artifacts and visualize the tumor trajectory; however, the insufficient number of projections sampled in each bin leads to severe noises and streaks in images reconstructed using the Feldkamp-Davis-Kress (FDK) (5) method. These artifacts, in turn, affect the accuracy of target localization. Thus, improving the quality of 4D-CBCT images reconstructed from undersampled projections is essential to ensuring the precision of radiation therapy delivery.
In recent years, deep learning-based methods have been introduced in the medical imaging field to perform tasks ranging from segmentation (6) and classification (7) to image augmentation (8) and restoration (9). Several works have focused on using deep learning methods to augment 4D-CBCT images by improving the image quality itself (10) or by improving the reconstruction algorithm (11). Recently, we developed a deep learning method to augment the quality of 4D-CBCT reconstructed using an iterative method (12). However, these deep learning methods were all trained using data from a group of patients, and thus displayed suboptimal performance when applied to individual patients (13). To address this issue, the present study sought to build a patient-specific network model by optimizing the model based on an individual patient’s information through transfer learning to enhance the abilities of deep learning to augment the quality of 4D-CBCT.
Transfer learning is a machine learning technique whereby a model trained on one task is repurposed to complete a second related task (14). In a classic deep learning scenario, when training a model for a specific task, the network needs to be fed labeled data in the domain that the task requires (15). Conversely, when applying a model to some other tasks that use data from a different domain, the network needs to be retrained using newly labeled data from that domain (16,17). Conveniently, under the transfer learning method, if the new data domain is related to the original data domain, much less data from the new data domain need to be used to retrain the model. This is because the knowledge acquired from the old model can be transferred to the new model for a related task, thus reducing the amount of data needed to gain new knowledge from a new data domain (18-20).
To conduct transfer learning, the most commonly used method is to fine-tune the entire network of a previously trained model using data from a new domain. All the network layers are included in the fine-tuning process to improve the network performance for the second specific task (21). This whole-layer fine-tuning method uses the parameters from the previous network as starting points and refines all these parameters in the new training process. Notably, the learning rate of the network is reduced, as the model is able to take advantage of the stored knowledge of the previous network, and the new network is prevented from overfitting to the limited new training data. However, the training time is prolonged due to the lower learning rate. To improve the efficiency of the whole-layer fine-tuning method, the layer-freezing method was proposed. When a new task is closely related to a previous task, parameters in some of the layers barely change during the transfer-learning training process, as these layers have already been adequately trained to extract the corresponding features (22). Consequently, only the layers that are responsible for extracting patterns specific to the new data set need to be retrained by the new data. Thus, the layer-freezing method freezes the layers that barely change to significantly reduce the training time for transfer learning and the frequency of overfitting in the transfer learning process. However, it should be noted that the frozen layers cannot be updated via transfer learning and thus cannot be improved further.
Given the advantages of transfer learning, we sought to explore the feasibility of transferring a general deep learning network trained using group data to a patient-specific network retrained using an individual patient’s data. The deep learning network used in our study was a standard U-Net model, which has been shown to be capable of producing satisfactory results in the medical imaging field (23). The proposed method was evaluated in terms of its ability to augment CBCT reconstructed using both digitally reconstructed radiography (DRR) data and real projection data. The results were compared to the original deep-learning results both qualitatively and quantitatively to evaluate any improvements in the augmented under-sampled CBCT image quality due to transfer learning.
Methods
Theoretical basis
In our experiment, we want to build up a model that uses limited-projection CBCT images as input and output the augmented CBCT images. In this model, r is used to denote a group of patients’ CBCT images composed of pixels that are reconstructed from limited projections, and r* is used to denote the corresponding fully sampled CBCT or planning CT images that can be used as the ground-truth images. The main goal of the deep learning method is to find an image restoration deep-learning network, g, that satisfies the condition:
[1]
The network, g, is trained using the paired data. However, the neural network trained on the group data is not optimized to augment individual patient’s data. To transfer the group-trained network, g, to a patient-specific network, the individual patient’s CBCT data are used for the transfer learning. Specifically, dataset, s, is used to denote a specific patient’s undersampled CBCT images with the corresponding ground truth, s*.
The main goal of transfer learning is to retrain the original network, g, into a patient-specific network that satisfies the condition:
[2]
This means that the network parameters’ optimization process will have the starting point, g and reach an end point,
Network architecture
U-Net architecture (see Figure 1) was previously implemented for biomedical imaging segmentation in Ronneberger’s work (23). It starts with a contracting path and is then followed by an expansive path. The contracting path repeatedly calls the application of two convolutions, each followed by a rectified linear unit (ReLU), and a max-pooling operation for down-sampling. Each down-sampling operation also doubles the feature channels. Similarly, every step in the expansive path consists of two convolutions and a ReLU. The difference is that the down-sampling operations in the contracting path are replaced by up-sampling operations that reduce the feature channels by half. The number on the top of each layer denotes the number of feature channels in each layer. Additionally, concatenation layers are added after each up-sampling operation to compensate for the loss of border pixels within every convolution. Finally, a convolution is added to the network to reduce the feature channels to the desired number.
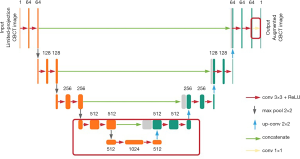
Transfer learning based on U-Net architecture
Figure 2 shows the overall workflow of the transfer learning scheme. The U-Net model is first trained to augment undersampled CBCT to match with fully sampled CBCT or planning CT images using group data. The model is then retrained as a patient-specific model using an individual patient’s CT or prior CBCT data and transfer learning to optimize its performance for that individual. The network can be updated adaptively by adding the most recent day’s CBCT images to the training data.
The present study sought to investigate two transfer-learning methods: the whole-network fine-tuning method and the layer-freezing method (see the description in the introduction section above). The whole-network fine-tuning method uses new patient data to retrain all the layers in the network, and a lower learning rate to ensure that the parameter changes slowly from the starting point. This method changes the original knowledge stored in the network model by adjusting the parameters in the network based on the new patient data.
Similarly, the layer-freezing method starts with the trained model; however, it retrains only the bottom and final layers of the network (see Figure 1). As the contracting path goes deeper toward the middle of the network, the feature channels’ number increases, which means the extracted features’ size decreases. In lung imaging, the main clinical interest is in the bronchus and nodules, which all appear in images as small textures. Thus, we chose to retrain the six convolution layers in the bottom of the network, the convolution layer at the end of the network, together with one down-sampling, one up-sampling, and the one concatenating layer that is attached to the six convolution layers (see the layers inside the red rectangle in Figure 1). The parameters in the upper layers are frozen. Due to the lower number of parameters being retrained, the training time of the layer-freezing method is shorter than that of the whole-network fine-tuning method.
Experiment design
A comparison of the two transfer learning methods
The dataset used in this study included 18 non-small cell lung cancer (NSCLC) patients’ planning 4D-CT data from the Cancer Imaging Archive. Each patient in the dataset had 5 to 7 4D-CT sets that covered their periods of treatment, which ranged from 25 to 35 days. To reconstruct the CBCT image, we used the sixth phase of the CT image set for each patient and simulated 360-projection data sets from the 3D volume. From the 360-projection data sets, we then used 72 (20%) projections to reconstruct the limited-projection CBCT images using the FDK algorithm. The training sets for the basic U-Net model only used the first day’s CT image sets of 17 patients. In training the network, we shuffled the paired 4D-CT images and CBCT images and extracted 5% of the data for network validation. This corresponded to 2,040 slices of images for the training and validation data. The remaining patient’s data were used for transfer-learning and network-testing purposes. Written informed consent was obtained from the patient to publish the results and any accompanying images in this study.
In this study, a U-Net model was first trained with the group data of 17 patients. We then conducted experiments on the two different transfer learning methods mentioned above using the testing data. The first days’ 4D-CT images from the testing data and the corresponding limited-projection CBCT images were used for transfer learning. A basic U-Net model, a layer-freezing model, and a whole-network fine-tuning model were tested with the second day’s 4D-CBCT images reconstructed from 72 projections.
A comparison of sequential transfer learning and all-data transfer learning
In the sequential transfer learning experiment, the two trained transfer learning models from experiment were used as starting points for the whole-network fine-tuning method and layer-freezing method, respectively. The second day’s data were fed to the models for retraining, and the models were tested with the third day’s data. This process was then repeated with the 3rd- and 4th-day’s data and the 4th- and 5th-day’s data.
We also compared sequential transfer learning to all-data transfer learning to evaluate the effects of the sequential transfer learning method. In the experiment, all the data from previous days were packed together to retrain the basic U-Net model. For example, to test the model with the 3rd-day’s data, the first-two-day’s data were fed to the network together for retraining. The augmented CBCT images from the two methods were then compared to each other.
Effects of projection numbers
In this study, we further lowered the projection number used for undersampled CBCT reconstruction. Thirty-six (10%) projections were extracted from the 360-projection dataset to reconstruct the undersampled CBCT. Both the undersampled DRR data reconstructed from the 36- and 72-projection data were used to compare the effects of projection numbers on transfer learning. We chose the better method of the two transfer learning methods to evaluate this effect. The two models were trained using the basic U-Net model with 36- and 72-projection data, respectively. Starting with the two models, the 1st- and 2nd-day’s patient data were used as training data for transfer learning. The results for both the basic U-Net model and transfer learning model were evaluated using the planning CT image as a reference to determine the difference between the improvements from the transfer learning methods for CBCT images reconstructed with different numbers of projections.
Real-patient data evaluation
The data set used in this study included eight breath-hold lung patients’ data from the Duke Clinic acquired under an Institutional Review Board (IRB)-approved protocol. We also had access to 5 to 7 days of 3D-CBCT data for each patient. Each patient’s 3D-CBCT data were constructed from data comprised of 894 full-angle projections and had 100 slices of image. To simulate the 4D-CBCT image, which has limited projections, and to compare the augmentation results with the DRR study, we used 89 (10%) projections and 178 (20%) projections respectively, from the 894-projection dataset to reconstruct the limited-projection CBCT images. Among the data from the eight patients, data from seven patients were used for training and validation, and the data of the remaining patient was used for transfer learning and testing purposes. Thus, 1,400 slices of images were used for training, and 500 slices of images were used for transfer learning and testing. The fully sampled 3D-CBCT images were used as a reference to evaluate the results. Written informed consent was obtained from the patients to publish the results and any accompanying images in this study.
Evaluation methods
Visual inspection
As the main goal of transfer learning is to recover additional small details that are lost in traditional deep learning methods, checking small textures in the augmented images is of great significance. In this study, we used the same window level for all the augmented images and ground truth images, which was normalized from 0 to 1. Difference maps were also generated between augmented images and ground truth images to determine which part displayed the most difference. Additionally, to evaluate the performance of transfer learning, we extracted the lung area from the body area using a novel method from MathWorks (24) to view any improvements in different parts of the body.
Quantitative evaluation
In addition to the visual inspection, we also used the peak signal-to-noise ratio (PSNR), which is defined as follows:
[3]
where T and G denote the reconstructed images and ground-truth images, respectively, M and N are the number of pixels for a row and a column, respectively, and the structure similarity index matrix (SSIM), which is defined as follows:
[4]
where μT is an average of T,
Results
Layer-freezing or whole-network fine-tuning
Two methods of transfer learning were evaluated. Table 1 summarizes the evaluation data for this study. As the second column of Table 1 shows, compared to the quality of the images produced by the U-Net model alone, transfer learning effectively further augmented the quality of images for specific patients. Figure 3 shows the augmented images obtained using different methods and the ground truth images. The third column of Figure 3 shows that the difference map of a basic U-Net image and ground truth image has a higher absolute value in the lung area than the difference map between the transfer learning images and ground truth images. Further, the difference map shows that the difference is more uniform. The fourth column of Figure 3 compares two transfer learning methods. The two transfer learning methods and the basic U-Net image have approximately the same range, and the difference between the two transfer learning methods is also very slight.

Full table
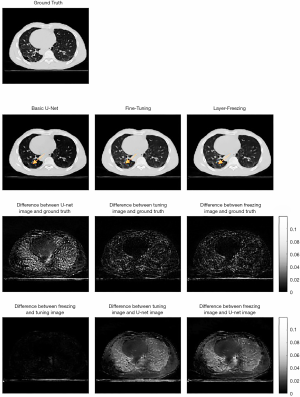
Figure 4 shows the extracted lung parts from the whole-body volume that were used to compare the augmented image quality for lung texture. The lung evaluation data in Table 1 also shows that the transfer learning methods improved the quality of the images produced. However, it should be noted that the SSIM difference between the two methods was only 0.5%. Figure 5 shows the analysis of the body area (excluding the lung area). The fourth column in Table 1 shows that the transfer-learning method only produced a slight improvement in displaying the muscle tissue in the body area.
The results outlined above show that the performance of the layer-freezing method was slightly higher than that of the fine-tuning method. However, as the difference between them was less than 0.5%, these two methods were deemed to have approximately equal competence in re-augmenting image quality from a basic machine-learning image. Notably, the training time for the layer-freezing model using a common graphic card RTX2060 was approximately 10 minutes, while fine-tuning model took four times longer to retrain the model, as it slows the learning rate. In light of these results, we used the layer-freezing method as the transfer-learning method in the next part of the study.
A comparison of sequential training and all-data training
Two methods for organizing training data were evaluated. In the first sequential training method, the first-4-day’s data were separated based on days. Each data set comprised 120 slices of limited projection CBCT images that had been paired with ground truth CT images. In the second all-data training method, all the data from the first 4 days were packed into one package containing 480 slices of two-dimensional (2D) CT-CBCT image pairs.
Figure 6 shows the results of the sequential and non-sequential image comparison for the day 5 images. The images from day 5 were selected, as to use the model to predict the data on the 5th day and later. In addition to the day 5 data evaluation, we also conducted a numerical evaluation for the data for the 3rd and the 4th days (see Table 2, Figure 7).

Full table
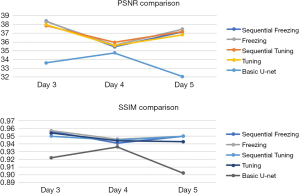
As the data in Table 2 shows, the augmentation of images as a result of all-data training was slightly higher than that for sequential training. In addition, all-data training was much easier to perform, as the first-4-day’s data were packed together to train the model, and thus there was no need to feed the model data on a day-by-day basis. In addition to comparing the data across the different days, we also examined the augmentation effect and found that this effect did not increase as the amounts of a patient’s specific data increased (see Table 2).
The effect of the number of projections
The previous images used for augmentation were reconstructed using 20% of the full number of projections. The total projection numbers were then set to 10% of the full number of projections to examine how the number of projections affects data augmentation.
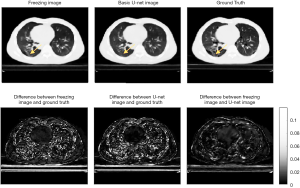
Figure 8 shows a comparison of augmented CBCT using 10% projection number and ground truth images. The SSIMs were 0.9306 and 0.9161 and the PSNRs are 35.65 and 34.65 for the transfer-learning and U-Net images, respectively. These evaluation data appear to indicate that when the projection number is very small, the machine learning method is not able to recover details as well as it can for an image that has more projections. Notably, the transfer learning method’s ability to recover details was also affected. However, the lung texture in the transfer learning images is of a slightly higher quality than that in the images that were augmented using the basic U-Net model.
Real-projection data
In the previous section, the model analyses were all based on DRR images. In this section, the model is applied to the real projection data. The origin data were reconstructed using 179 (of 894) projections, a figure that represents 20% of the full number of projections.
Table 3 displays the numerical evaluation data. The first two rows of Table 3 summarize the evaluation data for the 178 projection images, and shows that the quality of the augmented image was slightly lower than of the DRR image; however, this was anticipated due to the noise in the real projection CBCT image. As the difference map in Figure 9A shows, the major improvement of the transfer-learning method was that it made the value in the image more comparable to the ground truth image; that is, it made the difference more uniform, especially in the bone areas with the most noise.

Full table
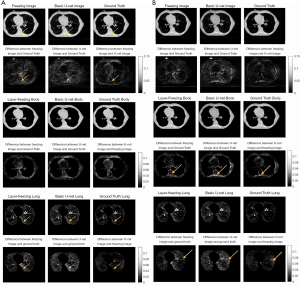
Further, as the structure-by-structure evaluation revealed, the improvement to the body area was more obvious in the difference map than it was in the DRR evaluation. However, the difference map for the lung area was not as obvious. Indeed, as can be seen, while the uniformity is better, no extra lung-texture details were recovered. Notably, the data evaluation yielded good results; the SSIM for the lung area in the transfer-learning image was 0.8574, and the SSIM for U-Net image was 0.7927. This represents an improvement of over 6%, which also supports the findings for the DRR evaluation parts.
The last two rows of Table 3 display the evaluation data for the 89-projection data. These results support the effect found in relation to projection numbers. Specifically, they indicate that the lower the projection numbers are, the lower the augmented image’s quality will be. However, while the SSIM for the 89-projection transfer learning images was still lower than that for the 179-projection transfer-learning images, the augmentation effect was better for the 89-projection images than for the 179-projection images.
Discussion
Improving the quality of 4D-CBCT images is crucial for achieving the high-precision 4D image guidance demanded by stereotactic body radiation therapy (SBRT). Previous deep learning methods developed to augment CBCT images have used a group-trained model; however, this type of model is suboptimal for individual patients. In this study, a transfer learning procedure was established to build a patient-specific model for 4D-CBCT image quality augmentation to optimize the performance of the model for individual patients. The model takes advantage of a specific patient’s prior information (gathered from planning CT images) or prior CBCT images to modify the general restoration patterns learned from various patients’ data into a specific restoration pattern for that patient. The proposed transfer learning method also demonstrated superior performance in recovering small lung textures, such as those of the bronchus and blood vessels, and in eliminating noise to make image values more accurate than those obtained by traditional deep learning methods. If high-quality 4D-CBCT images can be produced for specific patients, the inter- or intra-fraction localization in SBRT can be more accurate, which in turn should lead to more precisive dose delivery.
The SSIM measurement is a metric widely used to evaluate image quality in the medical imaging field. However, as it uses a uniform pooling method, the SSIM is highly affected by the variance in regions of high-pixel values and less affected by regions of low-pixel values (25) To address this limitation, we calculated the SSIM separately for the lung area (a region that has a low average pixel value) and the body area excluding the lung area (a region that has a high average pixel value) for the quantitative analysis. The analysis showed that the transfer-learning method produces a major improvement in the SSIM for the lung area; however, the improvement in the body area (excluding the lung area) is limited. This is because with a SSIM higher than 0.95, the augmentation of the body area using the U-Net model is already satisfactory. Conversely, the augmentation achieved by the use of the group-based U-Net model for the lung area is suboptimal due to the complexity of the lung structure and variations among patients. The transfer-learning method was capable of recovering more detailed anatomical structures than those recovered by in U-Net augmentation and indeed, could recover areas that were lost in U-Net augmentation.
The PSNR is another metric commonly used to evaluate images. The range of PSNR is 0 to infinity, and a higher PSNR indicates a higher level of similarity between the evaluated image and the ground truth image. In our study, the PSNR measurement served as a secondary verification tool for the SSIM measurement.
The comparison of two transfer learning methods yielded similar results in terms of image quality augmentation but showed a large difference in terms of training times. All the transfer learning training processes were conducted in Python with a Keras framework, and were performed with a standard graphic card, RTX2060 (6GB VRAM). When the transfer learning training data comprised 1-day’s patient CBCT images (120 slices), the training time took 10 seconds per epoch for the layer-freezing method and 28 seconds per epoch for the whole-network fine-tuning method. The training epoch number was set to 100 for each method; however, no further drop in loss occurred after the 50th epoch for the layer-freezing method or after the 80th epoch for the fine-tuning method. Thus, the total training time was around 500 seconds for the layer-freezing method and 2,200 seconds for the fine-tuning method. The time efficiency of the layer-freezing method could be significant should the procedure be applied in a clinic to treat individual patients. Once the two transfer-learning models were trained, the testing time for augmenting the undersampled 4D-CBCT images were both approximately 5 seconds; thus, both methods would be very practical in clinical applications.
The performance of the sequential training and all-data training was comparable. Thus, the sequence of incorporating patient-specific data in the training data did not have a major effect on the accuracy of the patient-specific model. The study of the effect of the number of projections showed that the capability of transfer-learning augmentation is limited for CBCT images reconstructed from a very low number of projections, as the potential of augmentation is limited by the quality of the input image. Notably, the transfer-learning model cannot recover anatomical details that have already been lost in the input image.
In the real projection data study, the performance of transfer learning was slightly degraded due to the noises and artifacts in the fully sampled CBCT images, which were used as the ground truth images. However, the transfer-learning model performed better in relation to 10% projection CBCT images for real projection data than it did for DRR data. This might have been because the transfer learning model is better at eliminating noise and streak in the real CBCT images than is the basic U-Net model. The differences map between the U-Net images and layer-freezing images in Figure 9B shows that most differences were from the edge area of the lung and the area around bones where there is a deal of noise.
In terms of the clinical implementation of the transfer-learning method, a U-Net model should first be trained using a patient database with fully sampled CBCT images. Limited projections CBCT images should be reconstructed by extracting limited projections from the fully sampled CBCT images, and the fully sampled CBCT images should be used as ground truth images for training. Next, for every incoming new patient, fully sampled CBCT images should be acquired for the first 2 or 3 days of treatments. These fully sampled CBCT images should then be used as the training data for the transfer learning model. The trained U-Net model should then be retrained into patient-specific models for individual patients based on the transfer learning method. Finally, for the remaining treatment for each individual patient, only limited projections need to be acquired of the CBCT scans, and the reconstructed images should be augmented using the patient-specific model.
Conclusions
In conclusion, the results of the present study showed that the proposed transfer learning method can augment the image quality of under-sampled 3D/4D-CBCT images by building a patient-specific model. Compared to images produced using the traditional group-based deep-learning model, the patient-specific model further enhanced the anatomical details and reduced noise and artifacts in the images. The technique could also be used to reduce imaging dose or to improve localization accuracy using 3D/4D-CBCT images, which could be highly valuable in SBRT treatments.
Acknowledgments
Funding: This work was supported by the National Institute of Health (no. R01-CA184173 and R01-EB028324).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-655). During the conduct of the study, the authors reported that they had been awarded grants from the National Institute of Health.
Ethical Statement: This study was approved by the Ethics board of Duke (No. 00049474), and informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li T, Schreibmann E, Yang Y, Xing L. Motion correction for improved target localization with on-board cone-beam computed tomography. Phys Med Biol 2006;51:253-67. [Crossref] [PubMed]
- Sonke JJ, Zijp L, Remeijer P, van Herk M. Respiratory correlated cone beam CT. Med Phys 2005;32:1176-86. [Crossref] [PubMed]
- Sonke JJ, Rossi M, Wolthaus J, van Herk M, Damen E, Belderbos J. Frameless stereotactic body radiotherapy for lung cancer using four-dimensional cone beam CT guidance. Int J Radiat Oncol Biol Phys 2009;74:567-74. [Crossref] [PubMed]
- Kida S, Saotome N, Masutani Y, Yamashita H, Ohtomo K, Nakagawa K, Sakumi A, Haga A. 4D-CBCT reconstruction using MV portal imaging during volumetric modulated arc therapy. Radiother Oncol 2011;100:380-5. [Crossref] [PubMed]
- Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm. J Opt Soc Am A 1984;1:612-9. [Crossref]
- Milletari F, Navab N, Ahmadi SA. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV); 25-28 Oct. 2016; Stanford, CA, USA. IEEE, 2016:565-71.
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. Erratum in: Nature. 2017 Jun 28;546(7660):686. [Crossref] [PubMed]
- Dong C, Loy CC, He K, Tang X. Image Super-Resolution Using Deep Convolutional Networks. IEEE Trans Pattern Anal Mach Intell 2016;38:295-307. [Crossref] [PubMed]
- Mao XJ, Shen C, Yang Y. Image restoration using very deep convolutional encoder-decoder networks with symmetric skip connections. NIPS'16: Proceedings of the 30th International Conference on Neural Information Processing Systems, 2016:2802-10.
- Chen H, Zhang Y, Kalra MK, Lin F, Chen Y, Liao P, Zhou J, Wang G, Low-Dose CT. With a Residual Encoder-Decoder Convolutional Neural Network. IEEE Trans Med Imaging 2017;36:2524-35. [Crossref] [PubMed]
- Beaudry J, Esquinas PL, Shieh CC. Learning from our neighbours: a novel approach on sinogram completion using bin-sharing and deep learning to reconstruct high quality 4DCBCT. Proc. SPIE 10948, Medical Imaging 2019 Physics of Medical Imaging 2019;1094847:1.
- Jiang Z, Chen Y, Zhang Y, Ge Y, Yin FF, Ren L. Augmentation of CBCT Reconstructed From Under-Sampled Projections Using Deep Learning. IEEE Trans Med Imaging 2019;38:2705-15. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Trans Knowl Data Eng 2010;22:1345-59. [Crossref]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform 2017;18:851-69. [PubMed]
- Shi X, Fan W, Ren J. Actively Transfer Domain Knowledge. In: Daelemans W, Goethals B, Morik K. editors. Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2008. Lecture Notes in Computer Science, vol 5212. Berlin, Heidelberg: Springer, 2008:342-57.
- Dai W, Chen Y, Xue GR, Yang Q, Yu Y. Translated Learning. Proc. 21st Ann. Conf. Neural Information Processing Systems, 2008.
- Zhuo H, Yang Q, Hu DH, Li L. Transferring Knowledge from Another Domain for Learning Action Models. In: Ho TB, Zhou ZH. editors. PRICAI 2008: Trends in Artificial Intelligence. PRICAI 2008. Lecture Notes in Computer Science, vol 5351. Berlin, Heidelberg: Springer, 2008:1110-5.
- Caruana R. Multitask Learning. Mach Learn 1997;28:41-75. [Crossref]
- Ben-David S, Schuller R. Exploiting Task Relatedness for Multiple Task Learning. In: Schölkopf B, Warmuth MK. editors. Learning Theory and Kernel Machines. Lecture Notes in Computer Science, vol 2777. Berlin, Heidelberg: Springer, 2003:567-80.
- Christodoulidis S, Anthimopoulos M, Ebner L, Christe A, Mougiakakou S. Multisource Transfer Learning With Convolutional Neural Networks for Lung Pattern Analysis. IEEE J Biomed Health Inform 2017;21:76-84. [Crossref] [PubMed]
- Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: Understanding Transfer Learning for Medical Imaging. 33rd Conference on Neural Information Processing Systems. Vancouver, Canada. 2019.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J, Wells W, Frangi A. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham, 2015:234-41.
- MathWorks. Automated lung segmentation. Available online: https://www.mathworks.com/matlabcentral/fileexchange/53114-automated-lung-segmentation
- Pambrun JF, Noumeir R. Limitations of the SSIM quality metric in the context of diagnostic imaging. 2015 IEEE International Conference on Image Processing (ICIP); 27-30 Sept. 2015; Quebec City, QC, Canada. IEEE, 2015.






