
Wide field-of-view volumetric imaging by a mesoscopic scanning oblique plane microscopy with switchable objective lenses
Introduction
Light sheet fluorescence microscopy (LSFM), or selective plane illumination microscopy (SPIM) has become an essential imaging modality in life science (1-3), enabling high resolution 3D imaging over a large volume. The typical configuration of LSFM/SPIM has two orthogonally aligned objective lenses to decouple the excitation and collection by separate optical paths. By only illuminating regions that fluorescence is collected from, LSFM prevents out-of-plane exposure and ensures low photodamage and low photo-toxicity. However, due to the orthogonal arrangement of two objective lenses, the imaging space for the sample is limited, which makes it difficult to use large samples or for integration with conventional microscopic platforms. In addition, conventional LSFM acquires volumetric datasets by mechanically translating components with nontrivial amounts of inertia, such as the primary objective lens or the sample, which can be challenging for high-speed imaging of large volumes. Inverted and ‘Open-top’ LSFM/SPIM with orthogonal dual-objective architecture (4-6) or with recently reported non-orthogonal dual-objective architecture (7,8), improve the sample mounting protocols by placing both of the objective lenses above or below the sample but still need mechanical translation. The adoption of the focus tunable lens and piezoelectric actuator makes the z-stacking process faster, yet still being limited within 1Hz volume rate when imaging zebrafish brain (9,10).
To improve the resolution, increase volumetric imaging speed, and enable flexible sample mounting/positioning protocols, different variants of LSFM/SPIM have emerged over the past two decades (11-17). Among them, oblique plane microscopy (OPM) provides an attractive optical design and provides excellent balance among the above three aspects. In contrast to conventional LSFM that has two objective lenses, OPM uses only a single objective lens at the specimen but applies an off-axis oblique light sheet excitation and a remote focusing system to capture the light sheet in a near-orthogonal angle (11-13,15,16). This “single objective lens layout” of an OPM liberates the sample placement space, and can be set up in the existing inverted, or upright microscopes. The first embodiment of OPM was developed by Dunsby, with mechanical translation of the sample (11). By moving one of the remote objective lenses in an OPM setup, near video-rate 3D fluorescence imaging of calcium dynamics has been presented (12). Swept confocally-aligned planar excitation (SCAPE) microscopy innovatively introduced scanning and descanning imaging strategy (18), enables completely translationless three-dimensional imaging. SCAPE demonstrated high-speed volumetric imaging of up to 200 volume per second (VPS) in the intact living samples (19). To increase the light acceptance angle and improve the resolution, an immersion fluid change can be introduced in the remote imaging system in the plane of the re-imaged sample illumination plane (20). A tilt-invariant scanned OPM (SoPi) was investigated (21); two-photon excitation technique was adopted (22,23); and simultaneous multimodal 3D imaging with optical coherence tomography (OCT) was demonstrated (24).
So far, the existing OPM studies have largely relied on a high numerical aperture (NA) objective lens, which typically means a high magnification lens, limiting the achievable FOV to a microscopic level typically <1×1 mm2. A diffractive OPM circumvented this constraint by redirecting the oblique image plane with a diffraction grating. As a result, it could achieve a larger FOV with the help of a low NA objective lens (25). Here, we leverage a seemingly counterintuitive phenomenon that the angle between the excitation and detection light paths does not need to be near-orthogonal (24), and the tilted angle for the intermediate image plane can be maintained with varying magnifications, simply under the Scheimpflug condition (26). As a result, our OPM setup is highly versatile in using a low NA objective lens to obtain mesoscopic FOVs up to ~5×6 mm2, the largest one reported by OPM so far. We demonstrated using 10× 0.3NA and 20× 0.5NA objective lens to imaging different biological specimens crossing a large range of length scales, from sub-millimeter C. elegans, whole zebrafish larvae, to a mouse cortex in centimeter scale.
Methods
Experimental setup
Our design derived from our previous work on oblique scanning laser ophthalmoscopy (oSLO) that essentially implements scanning OPM using natural ocular optics (27). We previously simulated the diffraction-limited 3D resolutions at varying NA of the objective lens from 0.1 to 0.9 (24), and demonstrated oSLO in rodent in vivo and in human in vitro (27-29). Here we implemented and remodel the optical design used in oSLO in a microscopic setting. The schematic layout of the experimental setup is shown in Figure 1A. The objective lens OL1 is a low NA and low magnification objective lens, which makes it possible for a wide FOV in OPM.
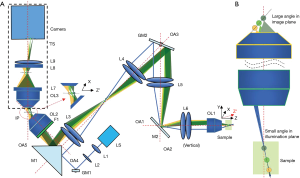
As shown in Figure 1A, excitation light is marked by blue, and emission light is marked by pale green, green, and orange. The excitation light was generated by a 488 nm laser (LS), first coupled into single-mode fiber and then collimated by lens L1 (f=10 mm). An additional focus-tunable lens L2 (Edmund: EL-3-10-VIS-26D) was used to make fine adjustments such that excitation and emission light could be confocal over a large FOV. The collimated light was first scanned by GM1 (Thorlabs: GVS201) and then redirected into a 1:1 relay lens group (L3–L4) by a right-angle prism mirror M1 (MRAK25-P01). The GM1 was placed at the focal plane of L3. The intersection point of the scanning beams was generated at the pupil plane of objective lens OL1 after two relay lens groups (L3: f=100 mm, L4: f=100 mm; L5: f=100 mm, L6: f=50 mm), and then a scanned light-sheet was created within the specimen. To maximize the angle of the oblique light-sheet, the excitation light was offset from the optical axis OA1-OA3 and incident on the edge of OL1. Another galvanometer GM2 (Nutfield: QS-12 OPD, 20 mm aperture), which was conjugated to the pupil plane of the OL1, was used to sweep the oblique light-sheet through the sample along the Y direction. As GM2 steered the excitation light-sheet and created a moving light-sheet within the sample, fluorescence emission mapped back on the same GM2 could be descanned. As a result, an intermediate stationary image plane (IP) could be created between OL2 (UPLSAPO 20×/0.75) and OL3 (UplanFL20×/0.5). The angle between the optical axis OA2 and OA3 was minimized to facilitate a more effective descanning over a large FOV. M1 was used to direct the fluorescence into OL2. An emission filter F1 (500–550 nm) was placed in the back of OL2. As the relay lens groups (L3–L6) were in the arrangement of a typical 4f-imaging system, the magnification can be calculated directly by the ratio of the focal lengths. The magnification from L4 to L3 can be calculated as follows:
[1]
Similarly, the magnification from L6 to L5 can be calculated as follows:
[2]
So, the magnification from L6-L3 can be obtained by:
[3]
The magnification of these two relay lens groups (L3–L6) was chosen to maximize the use of the numerical aperture of OL2. The lateral (perpendicular to OA1) magnification
[4]
The lateral magnification
OL1 can be switched between different low NA objective lenses, such as UplanFL10×/0.3 and UplanFL20×/0.5 from Olympus, to change FOVs and resolutions. The focal length of the collimator lens L1, as well as the two groups of relay lenses (L3–L4 and L5–L6), were carefully designed so that the Gaussian beam width and the Rayleigh range of the scanned light-sheet were ~9 µm and ~490 µm for 10× configuration. As for 20× configuration, these beam parameters were ~4.5 µm and ~122 µm, respectively. The axial magnification
[5]
It’s true that the refractive indices should be taken into consideration when calculating the magnification. However, the thickness of the mounting medium involved in imaging was <1 mm, which is small in comparison with the focal length (18 mm for 10×; 9 mm for 20×) and working distance (10 mm for 10×; 2.1 mm for 20×) of the objective. So, Eq. [5] would still hold reasonably, which is evident by our imaging results. As shown in Figure 1A, the XZ’ plane was imaged by the remote focusing system (from OL3 to L9) imaged after the optical refocusing of the sample to the intermediate image (IP) plane. As the angle of the sample plane with respect to the OA1 is small, we made the following approximation.
[6]
To ensure equal magnification on the image plane in the camera (Andor: Zyla 4.2, 2,448×2,048 pixels, 6.5 µm pixel pitch), we inserted anamorphic telescope composed of cylindrical lens L7 (Edmund: #35024, f=50 mm) and L8 (Edmund: 2 × #68-046, f=12.5 mm) between OL3 and L9 (Navitar: MVL75M1, f=75 mm), as described in (27). The magnifications of the remote focusing system in two directions are denoted as and and can be calculated as:
[7]
[8]
Then the ratio of
[9]
[10]
The magnification difference between axial and lateral magnification was corrected. Thus, the magnification of the whole optical system along all three directions (X, Y, and Z) is 2. As for 20× configuration, the later and axial magnification were 4 and 8, respectively.
Optical principle for mesoscopic OPM with low NA objective lens
Scheimpflug principle is the geometric rule that describes the orientation of the tilted object plane and image plane with respect to the lens plane. According to the Scheimpflug principle, a planar subject that is tilted with respect to the lens plane can be completely focused on a plane that is also tilted with respect to the lens plane (26). Figure 1B illustrates the main idea lies behind our design that the angle of the image plane can be increased significantly under the Scheimpflug principle to overcome the problem induced by employing a low NA objective lens in mesoscopic scanning OPM.
The relationship of the angle of the illumination plane and IP has been derived in our previous publication (27), thus the angle
[11]
where
[12]
As the value of the numerical aperture is small for low NA objective lens, tan(NAOL1) can be approximated by ROL1/fOL1, where ROL is the radius of the pupil aperture for OL1. Thus, Eq. [12] can be reduced to:
[13]
This equation indicates that the angle of IP only depends on the radius of the pupil aperture of OL1. Therefore, the angle of IP is maintained as long as we choose objective lenses with a similar aperture size. For example, the pupil apertures of low NA Olympus objective lenses (UplanFL4x: ROL1 =5.8 mm, UplanFL10x: ROL1 =5.4 mm, and UplanFL20x: ROL1 =4.5 mm) are close to each other. As a result, the system could be switched into a different configuration with different FOV and resolution by changing one objective, slightly adjusting the position of the remote imaging system and the offset of the excitation beam. Specifically for 10× configuration (UplanFL10×/0.3) and 20× configuration (UplanFL20×/0.5) used in this study,
Imaging protocol
The synchronization of the fast scan mirror GM1, slow scan mirror GM2, and the camera trigger was modified from our previous publication (24,27). Briefly, during every synchronized period, the control system would output one incremental step signal for the slow-scan mirror (GM2), one ramping wave signal for the fast-scan mirror (GM1), and one trigger signal for the camera.
Image processing
As the acquired 2-dimensional images were oblique cross-sectional images, the affine transformation was applied to the whole volume data to recover the actual geometry of the sample (24). The geometric transformation function (imwrap) in Matlab (MathWorks) was used to implement the affine transformation. As for the 20× configuration, the magnification in the Z direction was 2 times higher than that of X or Y while the resolution in Z direction was lower than that of X or Y. So, the Z direction was oversampled. To recover the actual geometry, MATLAB function (resize) was used to scale the image along the Z direction by a factor of 0.5. The color encoded images in depth were generated by ImageJ.
The mouse cortex data set was firstly deconvoluted, and then a de-haze method using dark channel prior was used to remove the autofluorescence background (31). Both processing was performed with ImageJ toolbox (32). The patch size is set to be 15 pixels ×15 pixels to get the dark channel. And the remaining amount of haze is set to be 0.05.
Sample preparation protocols
All animal procedures were in accordance with the Institutional Animal Care and Use Committee at Boston Medical Center, Boston University, and conformed to the guidelines on the Use of Animals from the National Institutes of Health (NIH).
Fluorescent microspheres with 3.1 µm diameter were immobilized in 1% agarose gel for the resolution characterization. The agarose gel was molded in a Petri dish and covered with a coverslip (refractive index: ~1.52, thickness: ~170 µm) for imaging.
Transgenic Tg(fli1: GFP) zebrafish expressing the green fluorescent protein in the entire vasculature were used in the experiment (33). Zebrafish were cultivated at ~28 °C following standard procedures. Larvae (≥3 days post fertilization, dpf) with a length of 3–4 mm were selected for in vivo imaging. The zebrafish larvae was placed in an anesthetic water bath (tricaine, 4% w/v, commercially as MS-222) with a regular culture dish (34). The imaging experiment started after 10–30 minutes of anesthesia exposure. The refractive index of the anesthetic water is 1.33. The long working distance of the low NA objective lens allows imaging through water bath in an upright setup. After the imaging, all of the anesthetized zebrafish were released into freshwater for recovery.
Mice were perfused transcardially with phosphate-buffered saline (PBS) followed by FITC-albumin (3 mg/mL) in gelatin (10% w/v) mixture (35). The carcass was kept in crushed ice for 10 minutes for the gelatin to solidify. The brain was extracted promptly and immersed in 4% paraformaldehyde (PFA) solution for 6 h and PBS solution for 3 days on a shaker. Tissue is transferred to solutions containing 0.5% alpha-thioglycerol with increasing fructose concentrations for the defined durations (20%: 2–4 h, 40%, 60%, 80%: 12 h, 110%: 24 h). The brain is then mounted upside down inside a glass Petri dish in the last (110%, w/v) solution. The refractive index of the 110% fructose solution is ~1.5. The refractive index of the Petri dish is ~1.51, and its bottom has a thickness of 170 µm. It was imaged through the glass bottom of the dish. This protocol was modified from the method described in (36), which was technically not optical clearing but was index matching.
The transgenic C. elegans strain used in our experiment was QW1217 which pan-neuronally expresses cytoplasmic GCaMP6s. Four C. elegans were selected 5 days after the hatching stage so that the worms would have a length of ~1 mm. They were immobilized in a 13% w/v polyethylene glycol hydrogel pad, as previously described (37). The pad was then gently positioned between a glass slide and coverslip (refractive index: ~1.52, thickness: ~170 µm) in several drops of S Basal solution (100 mM NaCl, 50 mM KPO4 buffer, 5 micrograms/mL cholesterol) (38), with 5 mM tetramisole to arrest nematode movement. A 1:1 mixture of paraffin and petroleum jelly was used as a sealant.
System calibration
The magnification was determined by imaging a rule and fluorescent microspheres immobilized in agarose gel. By measuring the corresponding displacement in the image after moving the beads sample in the Z direction by a caliper, we could convert the pixel coordinates to world coordinates. To calibrate the magnification in the X direction, we removed the fluorescent filter and imaged a rule. The distance can be directly read in the rule image in the acquired images. Similarly, the magnification in the Y direction was also calibrated by imaging the rule. The GM2 had to be scanned in two different positions so that the displacement could be read from the rule. The corresponding pixel number was the total steps included in the scanning range along the Y direction. The magnifications in X, Y, and Z were calibrated to be 2.02, 2.06, and 2.05, respectively, which agree well with our optical design described above.
Results
Characterization of FOV and resolution
To characterize the resolution and the FOV, phantom measurements were performed. Fluorescent microspheres with a diameter of 3.1 µm were diluted and immobilized in 1% agarose gel. Image acquisition experiments were carried out with two different dry objectives, a 10× (0.3NA) and a 20× (0.5NA). The acquired volume data sets were processed with the affine transformation as described in the Method section such that the coordinates could be transformed from image coordinates to world coordinates (XYZ). The results of the 10× (0.3NA) objective lens are shown in Figure 2 while the data from the 20× can be found in supplementary materials (Figure S1). Figure 2A,B,C are the maximum intensity projections (MIP) of the reconstructed data on three orthogonal facets, respectively. The dashed red lines in Figure 2B were used to indicate where the cross-sections were taken from to generate Figure 2A,C. Given the magnification and the camera’s pitch size (6.5 µm), the FOV along X, Y, and Z direction were measured to be 5.8, 4.9, and 0.7 mm, respectively. Figure 2D,E,F are zoom-in views of the area indicated by the square in Figure 2A,B,C. Three representative beads, which were labeled in Figure 2F, were randomly selected to measure the resolutions. The MIPs over each dimension of the three beads are shown in Figure 2G,H. Figure 2I is the intensity distribution through the center of each bead along three dimensions. Gaussian curves were fitted to the intensity data to quantify the full width at half maximum (FWHM) along each dimension, which was 5.4 µm (X), 6.9 µm (Y), and 34.5 µm (Z), respectively. To evaluate the resolution variation over the full FOV, we quantitatively analyzed the beads in different locations. The mean value and variance of the resolutions are 6.5±1.1 µm (X), 7.1±1.2 µm (Y), and 34±6.5 µm (Z) as shown in Figure 3. The results of that of the 20× configuration are shown in Figure S2, which are 1.9±0.6 µm (X), 2.7±0.5 µm (Y), and 11.5±2.4 µm (Z).
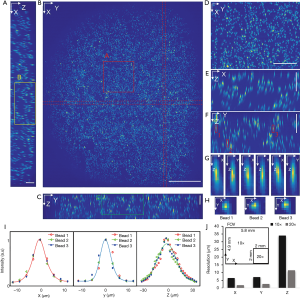
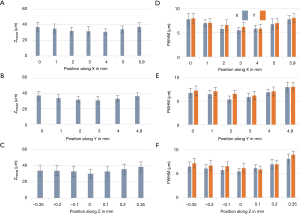
Figure 2J is a comparison of both FOV and resolution of 10× and 20× configuration, which show the FOV of 10× configurations is at least 7 times larger than that of 20× configuration while the resolution (mean value) of the 20× objective is two times better.
To demonstrate the productivity of switching between 10× and 20× configurations, we acquired dataset sequentially with different configurations. The results are shown in Figures S3,S4. The image quality under the same configuration is very similar.
Raw datasets without affine transformation for both 10× and 20× configurations are shown in Figures S5,S6, respectively. By comparing Figure 2F with Figure S5F, or Figure S1E with Figure S6E, the difference of the orientation of the beads can be observed.
Volumetric imaging of in vivo zebrafish larvae and ex vivo mouse cortex at 10× configuration
Zebrafish is a popular model in biomedical research due to its optical transparency from embryo to larval stages. As the length of the zebrafish larvae is usually longer than 3 mm (≥3 dpf), the whole-body volumetric imaging without stitching is difficult by microscopic FOV. Here, transgenic zebrafish expressing green fluorescent protein (≥3 dpf) were imaged in vivo. The longitudinal body axis of the zebrafish was aligned to be parallel to the light sheet to maximize imaging efficiency. The exposure time of the camera was 14 ms resulting in a frame rate of ~71 Hz. The whole imaging process lasted for ~3 seconds under 1.2 mW laser power with 220 frames captured using a 3.15 µm slow-scanning step. The size of the acquired volume is 3.2×0.63×0.69 mm3 with a pixel dimension 995×218×200. Figure 4A,B,C are MIPs of the whole volume along each dimension. The vascular network can be visualized in a depth-encoded en face MIP in Figure 4B. The dorsal longitudinal anastomotic vessels (DLAVs), as well as intersegmental arteries (ISA) and intersegmental veins (ISV) in zebrafish trunk vasculature, is clearly present, each with vessel diameter of ~10 µm (39,40). By plotting a line profile through a pair of ISA and ISV along the Z direction as shown in Figure 4C, the space between the paired ISA and ISV is calculated to be ~45 µm. This paired feature can be well resolved in 3D as shown in Figure 4B (lateral) as well as in Figure 4C (axial view/dorsal view). The discrimination of the contra-lateral parachordal vessels (ISA/ISV pair) served as a good demonstration of the axis resolution in our low NA mesoscopic scanning OPM. Figure 4A is a cross-section in the area of the common cardinal vein and the sinus venosus. This feature can also be observed in Figure 4D which are color rendered images in different depth. Each picture in Figure 4D was a projection of 10 planes center at a particular depth. The structural features presented in each depth is distinct through a total depth of 500 µm.
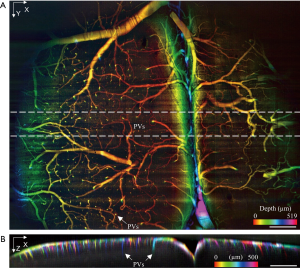
To demonstrate the extended mesoscopic FOV of the proposed setup, the vasculature of an ex vivo mouse cortex with FITC-infusion and optical clearing, as described in the Method section, was imaged with a 10× objective lens. The whole volume was acquired under 0.9 mW laser power with 1,550 frames captured by a 3.15 µm slow-scanning step. The resulting volume size is 5.8×4.9×0.7 mm3 with a pixel dimension of 1,800×1,550×220 [Visualization 1]. The depth-encoded en face image is shown in Figure 5A in which serval major blood vessels span across the whole FOV as well as their smaller branches. Figure 5A also clearly reveals that the penetrating vessels (PVs) in the mouse cortex, plunging vertically into a deeper layer. Penetrating vessel structure is one of the main vascular topologies in the mouse cortex that supports the blood flow to the neocortex (41). The vertical stripes shown in Figure 5A were caused by the synchronization of the rolling shutter and the scanning laser (fast scanning direction). Figure 5B displays the cross-section with color-encoded projection along Y-axis. Penetrating vessels with different colors, which represent the different locations in the Y direction, can be clearly observed. The resolution and image quality is well maintained over the curvature across the FOV (~0.5 mm in depth), thanks to the extended depth of view by the low NA objective lens and the weakly focused excitation. By converting the pixel coordinates in the acquired volume data to world coordinates, the penetration depth into the highly scattering cortex is estimated to around 0.15–0.25 mm. The original mouse cortex data set and the deconvoluted results are shown in Figures S7,S8.
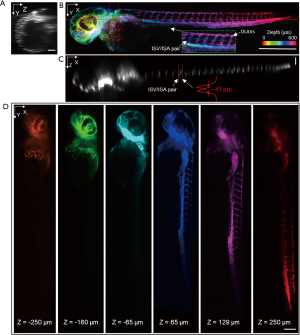
In vivo volumetric imaging of a whole zebrafish larvae [expressing Tg(fli1: GFP)]. (A, C) Maximum intensity projection (MIP) of the selected layers between the dash lines in panel (B). Both vertical and horizontal scale bars are 100 µm. (B) Depth encoded en face view of the whole volume. The scale bar is 500 µm. (D) Depth encoded en face view in deferent depth. The scale bar is 200 µm. DLAVs, the dorsal longitudinal anastomotic vessels; ISV, intersegmental veins; ISA, intersegmental arteries.
Volumetric imaging of the vasculature in a fixed mouse brain [labeled by Fluorescein IsoThioCyanate (FITC)]. (A) Depth encoded en face view of the mouse brain; (B) X-Z cross-section with color encoded along Y axis. The scale bar is 500 µm. PVs, penetrating vessels.
Volumetric imaging of in vivo C. elegans at 20× configuration
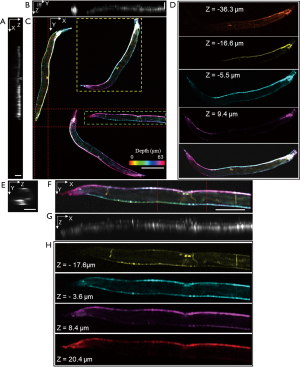
By switching to a 20× objective lens and minor refocusing of the remote focusing system, we can flexibly zoom-in a smaller FOV with twice higher 3D resolutions. Figure 6 demonstrates in vivo volumetric imaging of four C. elegans expressing GCaMP6s appearing within the same FOV. Figure 6A,B,C are MIPs along each dimension in which Figure 6C is depth-encoded. Neurons located in various ganglia in the head and tail, along the ventral cord, and inside the body are visible. The excitation laser power was 0.6 mW. One thousand frames were acquired at a slow-scanning step of 1.17 µm. The size of the acquired volume is 1.5×1.2×0.07 mm3 with a pixel dimension of 1,350×1,000×88. Figure 6D is the magnified view in different depths of the selected worm in Figure 6C, revealing distinct anatomical features in different depths across the length of the body. Details of an individual nematode indicated by the square in Figure 6C are shown in Figure 6E,F,G,H. MIPs of an individual nematode are displayed in Figure 6E,F,G, in which the profiles of the nematode are visible. Color rendered en face views in depth are shown in Figure 6H. The individual neurons along the ventral cord and inside the body, and even the thin motor commissures can be observed in different depths.
Discussion
We present a scanning OPM design that can achieve mesoscopic volumetric imaging up to ~5×6×0.7 mm3 FOV. The proposed method maintains the angle of the intermediate image under Scheimpflug condition by demagnification, and thus allows the use of low NA objective lenses. As compared to the previously reported results (9-22), the achievable FOV (5.8 mm ×4.9 mm ×0.7 mm) is an order of magnitude higher, the largest FOV in OPM so far to the best of our knowledge. Depth discrimination of vascular structure in zebrafish larvae, mouse cortex, as well as the neurons in C. elegans, were demonstrated. Our optical design is versatile that different FOV with varying resolutions can be easily switched by simply changing one objective lens. Thanks to the single objective lens layout, most common sample formats can be accepted in the proposed method. The new capability of mesoscopic volumetric imaging can provide rich structural information that can be further quantified for pathological understanding, diseases modeling and phenotyping, all of which would produce clinical impact.
One major issue of OPM is the light loss in the remote imaging system. Improvements in light acceptance angle can be made by applying the liquid immersion method as described in (20). As every pixel represents 3.17 µm in the axial direction of the 10× configuration which is a tenth of the resolution in this dimension, the sampling is redundant in the Z direction in the current setup. By implementing a more suitable sampling scheme, the imaging speed will improve significantly. Due to the highly scattering nature of the mouse brain, we only demonstrated ~0.25 mm depth penetration. By switching the Gaussian beam to the Bessel beam or two-photon excitation, the deeper cortex layer could be better revealed. Optical aberrations such as field curvature due to large FOV can be observed in Figure 2A, which can be improved and corrected by employing adaptive optics (42). To evaluate the aberration caused by the mounting medium and demagnification design, we did a simulation by Zemax as shown in Figures S9,S10. The aberration caused by the mounting mediums (thickness: <1 mm, refractive index: ~1.33 to 1.52) was negligible as shown in the comparison in Figure S11A,B. As can be seen from Figure S11 that the aberrations caused by the demagnification design were mainly field curvature and astigmatism. As for the remote focusing system, the aberrations were mainly spherical and field curvature as shown in Figure S12. Future improvements can be made by choosing objective lenses with the option of liquid immersion and applying adaptive optics. The measured lateral resolution under 20× configuration is comparable to the size of the fluorescent microspheres. In order to accurately characterize the imaging resolution, we performed a numerical simulation (as shown in Figures S13,S14) to convolute the microsphere with varying resolutions and created a lookup table to associate the FWHM measurement and the actual resolution. We then used the lookup table (Figure S15) to retrieve the actual resolution from the measured FWHM.
With the mesoscopic FOV volumetric imaging capability, simple sample mounting protocol, and the versatility of changeable FOVs/resolutions, our system will be ready for the future applications requiring in vivo volumetric imaging over a large length scale, such as neural dynamics and vasculature development.
Acknowledgments
Funding: This study is supported by the National Institute of Health (R01CA224911, R01CA232015, R01NS108464, andR21EY029412) and Bright Focus Foundation (G2017077and M2018132).
Footnote
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-806). The special issue “Advanced Optical Imaging in Biomedicine” was commissioned by the editorial office without any funding or sponsorship. JY has a patent 10337995 issued. The authors have no other conflicts of interest to declare.
Ethical Statement: All animal procedures were in accordance with the Institutional Animal Care and Use Committee at Boston Medical Center, Boston University, and conformed to the guidelines on the Use of Animals from the National Institutes of Health (NIH).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siedentopf H, Zsigmondy R. Uber Sichtbarmachung und Größenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Ann Phys 1902;315:1-39. [Crossref]
- Huisken J, Swoger J, Bene FD, Wittbrodt J, Stelzer EHK. Optical Sectioning Deep Inside Live Embryos by Selective Plane Illumination Microscopy. Science 2004;305:1007-9. [Crossref] [PubMed]
- Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Methods 2017;14:360-73. [Crossref] [PubMed]
- Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Bao Z, Colón-Ramos D, Shroff H. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci 2011;108:17708-13. [Crossref] [PubMed]
- McGorty R, Liu H, Kamiyama D, Dong Z, Guo S, Huang B. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt Express 2015;23:16142-53. [Crossref] [PubMed]
- Mcgorty R, Xie D, Huang B. High-NA open-top selective-plane illumination microscopy for biological imaging. Opt Express 2017;25:17798-810. [Crossref] [PubMed]
- Bishop KW, Glaser AK, Liu JTC. Performance trade-offs for single- and dual-objective light-sheet microscope designs. bioRxiv 2020. doi: 10.1101/2020.05.10.087171. [Crossref]
- Glaser AK, Bishop KW, Barner LA, Serafin RB, Liu JTC. A hybrid open-top light-sheet microscope for multi-scale imaging of cleared tissues. Bioengineering 2020. [Crossref]
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 2013;10:413-20. [Crossref] [PubMed]
- Fahrbach FO, Voigt FF, Schmid B, Helmchen F, Huisken J. Rapid 3D light-sheet microscopy with a tunable lens. Opt Express 2013;21:21010-26. [Crossref] [PubMed]
- Dunsby C. Optically sectioned imaging by oblique plane microscopy. Opt Express 2008;16:20306-16. [Crossref] [PubMed]
- Kumar S, Wilding D, Sikkel MB, Lyon AR, MacLeod KT, Dunsby C. High-speed 2D and 3D fluorescence microscopy of cardiac myocytes. Opt Express 2011;19:13839-47. [Crossref] [PubMed]
- Cutrale F, Gratton E. Inclined selective plane illumination microscopy adaptor for conventional microscopes. Microsc Res Tech 2012;75:1461-6. [Crossref] [PubMed]
- Li T, Ota S, Kim J, Wong ZJ, Wang Y, Yin X, Zhang X. Axial Plane Optical Microscopy. Sci Rep 2014;4:7253. [Crossref] [PubMed]
- Sikkel MB, Kumar S, Maioli V, Rowlands C, Gordon F, Harding SE, Lyon AR, MacLeod KT, Dunsby C. High speed sCMOS-based oblique plane microscopy applied to the study of calcium dynamics in cardiac myocytes. J Biophotonics 2016;9:311-23. [Crossref] [PubMed]
- Kim J, Wojcik M, Wang Y, Moon S, Zin EA, Marnani N, Newman ZL, Flannery JG, Xu K, Zhang X. Oblique-plane single-molecule localization microscopy for tissues and small intact animals. Nat Methods 2019;16:853-7. [Crossref] [PubMed]
- An S, Ziegler KF, Zhang P, Wang Y, Kwok T, Xu F, Bi C, Matosevic S, Yin P, Li T, Huang F. Axial plane single-molecule super-resolution microscopy of whole cells. Biomed Opt Express 2019;11:461-79. [Crossref] [PubMed]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, Hillman EMC. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat Photonics 2015;9:113-9. [Crossref] [PubMed]
- Voleti V, Patel KB, Li W, Campos CP, Bharadwaj S, Yu H, Ford C, Casper MJ, Yan RW, Liang W, Wen C, Kimura KD, Targoff KL, Hillman EMC. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat Methods 2019;16:1054-62. [Crossref] [PubMed]
- Yang B, Chen X, Wang Y, Feng S, Pessino V, Stuurman N, Cho NH, Cheng KW, Lord SJ, Xu L, Xie D, Mullins RD, Leonetti MD, Huang B. Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat Methods 2019;16:501-4. [Crossref] [PubMed]
- Kumar M, Kozorovitskiy Y. Tilt-invariant scanned oblique plane illumination microscopy for large-scale volumetric imaging. Opt Lett 2019;44:1706-9. [Crossref] [PubMed]
- Kumar M, Kishore S, Nasenbeny J, McLean DL, Kozorovitskiy Y. Integrated one- and two-photon scanned oblique plane illumination (SOPi) microscopy for rapid volumetric imaging. Opt Express 2018;26:13027-41. [Crossref] [PubMed]
- Shin Y, Kim D, Kwon H-S. Oblique scanning 2-photon light-sheet fluorescence microscopy for rapid volumetric imaging. J Biophotonics 2018;11:e201700270. [Crossref] [PubMed]
- Zhang L, Capilla A, Song W, Mostoslavsky G, Yi J. Oblique scanning laser microscopy for simultaneously volumetric structural and molecular imaging using only one raster scan. Sci Rep 2017;7:8591. [Crossref] [PubMed]
- Hoffmann M, Judkewitz B. Diffractive oblique plane microscopy. Optica 2019;6:1166-70. [Crossref]
- Scheimpflug T. Improved Method and Apparatus for the Systematic Alteration or Distortion of Plane Pictures and Images by Means of Lenses and Mirrors for Photograpy and for other purposes. GB190401196A, 1904 [cited 2020]. Available online: https://patents.google.com/patent/GB190401196A/en
- Shao W, Song W, Yi J. Is oblique scanning laser ophthalmoscope applicable to human ocular optics? A feasibility study using an eye model for volumetric imaging. J Biophotonics 2020;13:e201960174. [Crossref] [PubMed]
- Zhang L, Song W, Shao D, Zhang S, Desai M, Ness S, Roy S, Yi J. Volumetric fluorescence retinal imaging in vivo over a 30-degree field of view by oblique scanning laser ophthalmoscopy (oSLO). Biomed Opt Express 2017;9:25-40. [Crossref] [PubMed]
- Song W, Zhou L, Yi J, Yi J. Volumetric fluorescein angiography (vFA) by oblique scanning laser ophthalmoscopy in mouse retina at 200 B-scans per second. Biomed Opt Express 2019;10:4907-18. [Crossref] [PubMed]
- Greivenkamp JE. Field Guide to Geometrical Optics. SPIE press; 2004 [cited 2019]. Available online: https://spie.org/publications/book/547461?webSyncID=38130918-6d12-2052-aea1-eae0191454cd&sessionGUID=c86b71ae-3a42-1bcb-b935-2c7a36979445&SSO=1
- He K, Sun J, Tang X. Single Image Haze Removal Using Dark Channel Prior. IEEE Trans Pattern Anal Mach Intell 2011;33:2341-53. [Crossref] [PubMed]
- ImageJ. [cited 2020]. Available online: https://imagej.net/Welcome
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 2002;248:307-18. [Crossref] [PubMed]
- Wilson JM, Bunte RM, Carty AJ. Evaluation of Rapid Cooling and Tricaine Methanesulfonate (MS222) as Methods of Euthanasia in Zebrafish (Danio rerio). J Am Assoc Lab Anim Sci JAALAS 2009;48:785-9. [PubMed]
- Kiliç K, Erdener E, Ning S, Sunil S, Chen A, Boas DA. Ex-vivo angiography for deeper imaging in chronic stroke models. Society for Neuroscience; 2018 [cited 2020]; San Diego. Available online: https://www.abstractsonline.com/pp8/#!/4649/presentation/9102
- Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, Lyden PD, Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci 2009;29:14553-70. [Crossref] [PubMed]
- Burnett K, Edsinger E, Albrecht DR. Rapid and gentle hydrogel encapsulation of living organisms enables long-term microscopy over multiple hours. Commun Biol 2018;1:73. [Crossref] [PubMed]
- Stiernagle T. Maintenance of C. elegans. WormBook: The Online Review of C. elegans Biology. WormBook; 2006 [cited 2020]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK19649/
- Zygmunt T, Trzaska S, Edelstein L, Walls J, Rajamani S, Gale N, Daroles L, Ramírez C, Ulrich F, Torres-Vázquez J. ‘In parallel’ interconnectivity of the dorsal longitudinal anastomotic vessels requires both VEGF signaling and circulatory flow. J Cell Sci 2012;125:5159-67. [Crossref] [PubMed]
- Geudens I, Coxam B, Alt S, Gebala V, Vion AC, Meier K, Rosa A, Gerhardt H. Artery-vein specification in the zebrafish trunk is pre-patterned by heterogeneous Notch activity and balanced by flow-mediated fine-tuning. Development 2019;146:dev181024. [Crossref] [PubMed]
- Blinder P, Shih AY, Rafie C, Kleinfeld D. Topological basis for the robust distribution of blood to rodent neocortex. Proc Natl Acad Sci 2010;107:12670-5. [Crossref] [PubMed]
- Potsaid B, Bellouard Y, Wen JT. Adaptive Scanning Optical Microscope (ASOM): A multidisciplinary optical microscope design for large field of view and high resolution imaging. Opt Express 2005;13:6504-18. [Crossref] [PubMed]


