
Respiratory deformation registration in 4D-CT/cone beam CT using deep learning
Introduction
Advanced techniques in cancer treatment such as volumetric modulated arc therapy (VMAT) and intensity-modulated radiation therapy (IMRT) require precise target localization to deliver the radiation to the tumor while minimizing the damage to surrounding normal tissues (1). For lung cancer treatments, patient respiratory motion induces errors in the target localization, and consequently, deviations in the dose delivery. To address this, four-dimensional imaging techniques, such as 4D-CT and 4D-CBCT, have been developed to capture the respiratory motion of the patient for treatment planning and delivery (2).
Deformable image registration (DIR) is often used to register different phases of 4D-CT or 4D-CBCT for motion modeling, contour propagation, 4D dose accumulation or target localization. DIR is a process of transforming different sets of data into one coordinate system and establishing a spatial relationship between corresponding volume elements in volumetric images via the deformable vector field (DVF). For example, by knowing the phase-to-phase DVF, dose to tumor and organ at risk (OAR) from different phases could be added to evaluate the treatment planning, and the contours of planning treatment volume (PTV) and OAR can be propagated from one phase to other phases (3,4).
Although plenty of registration methods have been proposed in recent years, it is still a challenging problem to register two images with large anatomic differences. In general, intensity-based (5) or feature-based methods (6) aimed at reducing the intensity or structure difference between the target and the deformed moving image by optimizing the deformation field iteratively. Since the DIR is a complex and ill-posed problem with excessive degrees of freedom, many constraints regularizing the DVF, such as elastic energy (7) and Laplacian term (8), have also been proposed to fit the DIR into a well-posed scenario. Despite the developments in DIR, the optimization of DVF in most conventional methods is still challenging, especially for the large anatomic difference in the moving and target images. Generally, the conventional DIR algorithm suffers from (I) long computational time due to the iterative process, (II) manual parameter tuning to achieve the optimal performance, which makes the process non-automatic and user-dependent and (III) the possibility of being trapped in a local optimum.
Recently, deep learning methods have been introduced to address some of the limitations of conventional registration methods (9). Kim et al. (10) introduced a patch-based initial deformation prediction framework to estimate the initial deformation between the moving and the target using brain images from HAMMER. The aim of this framework is to improve the registration performance of existing registration algorithms by providing an initial estimation of the DVF. Furthermore, Cao et al. (11) has investigated the direct deformable registration using different brain image datasets based on a patch-based convolutional neural network (CNN) model. It was found that despite the variable anatomical differences across the different datasets, the trained CNN model from one dataset can be transferred to another dataset. These studies demonstrated the potential of the patch-based deep learning model as a novel method in DIR.
In this paper, a patch-based CNN is proposed to directly register DVF between individual phases of 4D-CT or 4D-CBCT images.
In the training stage, image patch pairs from the moving and target images were used as input, and the network is supervised with the reference DVF generated with VelocityAI (Varian). In the prediction stage, the trained network takes image patch pairs as the input and predict the corresponding DVF. This method demonstrated the feasibility of using deep learning to register respiratory motions. Compared with conventional iterative methods, the deep learning method is fast, fully automatic, and user-independent, making it much more preferable for clinical applications.
Methods
A patch-based CNN architecture is proposed to register the 4D image pairs (e.g., phase 1, the moving image M, to phase n, the target image(s) T, of a 4D-CT volume) to obtain the corresponding deformation field ϕ. The reference deformation field is generated with VelocityAI (Varian). The aim of our network is to establish the local mapping between the input image pair. Therefore, our registration process has three parts, data preparation, training and predicting. All the coding procedures were implemented with MATLAB2018a on a PC (Intel Xeon CPU, 32GB RAM) using a single NVIDIA Tesla K40s GPU with 12GB memory.
Sample preparation
For a pair of moving image M and target image T with their deformation vector field ϕ, a patch pair {PM(u), PT(u)} and the displacement vector ϕ(u) = (dx,dy,dz) at the same position u were extracted, where PM(u) is the patch of moving image, PT(u) is the patch of the target image, and ϕ(u) is the displacement vector registered by VelocityAI at position u. A sample is represented as {PM(u), PT(u)lϕ(u)}. The characteristics of the sample were determined by the patch center and the patch size. The centers of the patch pair determine the location of the patch, and the patch size determines the range of features included in the patch pair.
The patch center points were chosen on a 3-dimensional uniform grid across the lung. The density of the grid determines the number of samples. In order to ensure sufficient training samples, the density was chosen such that the number of patch pair samples is around 20,000. The center points sampling density was designed to be denser along the superior-inferior direction than the anterior-posterior and medial-lateral directions because the magnitude of the deformation field along superior-inferior direction is generally larger than the other two directions. The patch size was chosen to be large enough to cover the motion range of the anatomical structures within the patch.
CNN architecture
The workflow and CNN architecture is shown in Figure 1. There are four convolutional layers, the kernel number for each layer is 64, 128, 256, and 256, and the kernel size is (3 × 3 × z) with stride (1,1), where (x,y) is the transverse plane and z is the axial direction of the input. All the convolution operations are performed to keep the size of the output the same as the input after each convolutional layer. Each convolutional layer is followed by a batch normalization layer and a ReLU activation. The first two convolutional layers are followed by an average pooling 2-D layer with a size of 2×2 and stride of (2,2). Then, a dropout layer with 20% drop rate connects the last convolutional layer and fully connected layer. Finally, the fully connected layer with output size 3 connects the output deformation vector ϕ(u) = (dx,dy,dz). The loss function is the Mean Squared Error. In the training stage, the epochs are 60, the batch size is 128, the initial learning rate is 1e-3, and the learning rate drop is 10% every ten epochs.
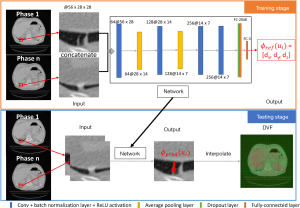
Experimental design
The goal of the network is to predict the DVF that registers the input image pair efficiently. In order to test the generalizability of our network under different circumstances, different studies combining different training and testing data sets were used, as shown in Table 1. Overall, nine 4D-CT volumes from eight patients with lung cancer from Duke University Medical Center were used for study 1, 2, 3 and 6. The study was approved by the Ethics board of Duke (No.: 00049474) and informed consent was taken from all the patients. Also, the 4D-CBCT volumes and projections from the public datasets Sparse-view Reconstruction Challenge for Four-dimensional Cone-beam CT (SPARE) (12) were used for study 4 and 5. For each dataset, there are 10 phases for the respiratory motion.

Full table
Study 1: feasibility study
The training and predicting samples are from the same 4D-CT volume. The network is trained with learning the deformation from phase 1 to phase 6, and it is applied to predict the deformation from phase 1 to phase 7. The feasibility of CNN for deformable registration was tested with this study.
Study 2: robustness study against inter-fractional anatomical differences
The training and predicting samples are from the two 4D-CT volumes from the same patient with lung cancer. The model is trained to learn the deformation from phase 1 to all the other phases from the first set of 4D-CT volume, and it is applied to predict the deformation from phase 1 to all the other phases on the second 4D-CT volume. Two sets of 4D-CT volumes were taken at different times. Therefore anatomic difference was introduced between training and predicting samples to test the network’s robustness against inter-fractional anatomical differences.
Study 3: robustness study against cone-beam sampling artifacts
Two simulated 4D-CBCT were reconstructed from digital reconstructed radiography (DRRs) generated from the double 4D-CT in study 2 based on the cone-beam geometry. The training and predicting setup is the same as trial 2. The sampling artifacts of cone-beam geometry was introduced in this test.
Study 4: robustness study against CBCT artifacts
Two 4D-CBCT volumes were reconstructed with FDK methods using the projections from SPARE. The training volume is reconstructed with 680 projections/phase for high image quality, and the predicting volume is reconstructed with 170 projections/phase to mimic the clinic 4D-CBCT acquisition. The adaptability of the network to poor image quality in predicting volume was tested.
Study 5: robustness study against cross-modality differences
A ground truth 4D-CT volume and an FDK-reconstructed 4D-CBCT volume from Sparse-view Reconstruction Challenge for Four-dimensional Cone-beam CT (SPARE) (12) were used. The ground truth 4D-CT volume is used for training, and the FDK-reconstructed 4D-CBCT is used for predicting. There is a tumor in ground truth volume but not in reconstructed 4D-CBCT volume; therefore, the adaptability of the network to the big anatomic difference between training and predicting sets was predicted. In addition, the robustness of the network against cross-modality differences was predicted.
Study 6: robustness study against inter-patient anatomical differences
Six 4D-CT volumes from six patients with lung cancer from Duke University Medical Center were used. Five of them were used as the training samples, and the last one was used for predicting. The interpatient anatomic difference between training and testing volumes was introduced.
Evaluation
For all studies, phase 1 and phase 7 of the 4D images were used for testing due to the large deformation between the two phases. The output of the network is the deformation vectors at control points, which are then used to generate the dense deformation vector field at individual voxels based on B-spline interpolation. Since each control points only have one deformable vector, there was no edge effect to deal with in this scheme. For qualitative evaluation, the main anatomical features, such as the diaphragm, were compared between predicted deformed image and reference deformed image. For quantitative evaluation, the cross-correlation, root-mean squared error (RMSE) and structural similarity index measure (SSIM) between predicted DVF and reference DVF obtained from Velocity was calculated. Besides the direct comparison of DVF, the deformed images by reference DVF and predicted DVF were reconstructed, and their cross-correlation was calculated in the diaphragm region. The diaphragm motion is one of the most representative features of the respiratory motion. It is, therefore, the region of interest in our analysis.
Results
Overall, our CNN network can perform decent DVF predictions, and the test results against VelocityAI (Varian) were shown below. In terms of computational time, the training of our network is around two hours, and the prediction time is within a few seconds per phase.
The result of study 1
In study 1, the training and the predicting samples were from the same 4D-CT volumes (e.g., training samples are phase 1 and phase 6 with associated reference DVF, and the application samples are phase 1 and phase 7. Note that only the result showing registering of phase 1 to phase 7 is shown in the paper, due to the most noticeable motion between these two phases). The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are both 0.99. The coefficients of cross-correlation, root mean squared error, as well as structural similarity, were shown in Table 2. The comparison of the image were shown in Figure 2.
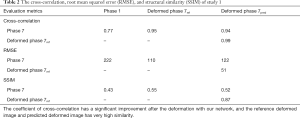
Full table
The result of study 2
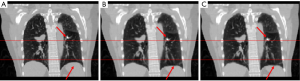
In study 2, the training and predicting samples are from different 4D-CT volumes but the same patient. The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are 0.91 and 0.99, respectively. The coefficients of cross-correlation, root mean squared error, as well as structural similarity, were shown in Table 3. The comparison of the image was shown in Figure 3.
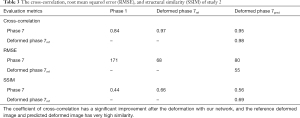
Full table
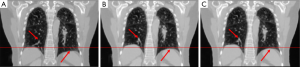
The result of study 3
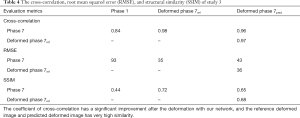
In study 3, the training and application samples are from different 4D-CBCT volumes, which are FDK-reconstructed with simulated DRRs from the double 4D-CT in study 2. The projections used for reconstruction is 340 per phase for both training and application volumes. The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are 0.77 and 0.97, respectively. The coefficients of cross-correlation, root mean squared error, as well as structural similarity, were shown in Table 4. The comparison of the image was shown in Figure 4.
Full table
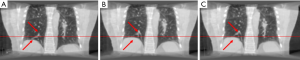
The result of study 4
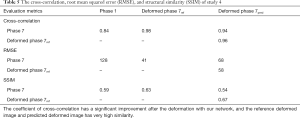
In study 4, the training and predicting samples are from different 4D-CBCT volumes but the same patient. The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are 0.90 and 0.96, respectively. The coefficients of cross-correlation, root mean squared error, as well as structural similarity, were shown in Table 5. The comparison of the image was shown in Figure 5.
Full table

The result of study 5
In study 5, the training volume is the ground truth volume of SPARE Challenge, and the application volume is the 4D-CBCT, which is FDK-reconstructed with 680 projections per phase. The training and application volumes are from the same patient. The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are 0.78 and 0.95, respectively. The coefficients of cross-correlation, root mean squared error, as well as structural similarity, were shown in Table 6. The comparison of the image was shown in Figure 6.

Full table

The result of study 6
In study 6, the inter-patient study was conducted. The training samples were from five 4D-CT from 5 different patients with lung cancer. The application set is another 4D-CT from a new patient. The coefficients of cross-correlation of whole body DVF and image in the diaphragm region between reference and prediction are 0.5 and 0.91, respectively. The coefficients of cross-correlation and comparison of images were shown in Table 7 and Figure 7.
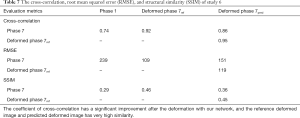
Full table
Discussion
The network predicts the deformation vectors in the center of the input patch. The coefficients of cross-correlation of the deformation vectors between the reference and the prediction are 0.99, 0.91 and 0.90 for study 1, 2 and 4 respectively and 0.77, 0.78 and 0.5 for study 3, 5 and 6 respectively. As shown by the comparison, the study 1, 2 and 4 showed a better performance than study 3, 5 and 6 in the performance of matching between the reference and the prediction. There are two reasons that contributed to the performance differences. The first reason is the image quality, which largely affects the feature extracting performance of the network, and therefore affects the learning and prediction efficiency of the network. Study 3 and study 5 both have poor image quality performance when reconstructing 4D-CBCT volumes, and it may contribute to inaccurate learning and predicting of deformation vectors. The second reason is the large anatomic difference between training and predicting image volumes. The network will extract the features and learn the respiration pattern from the training image volume. If the image volume for prediction is from a new patient or changed dramatically, which is the case of study 6 and study 5, the feature learned by the network will be different from that in the new patient’s anatomy, leading to degradation of the registration accuracy. Besides, the patient data used covered a variety of patient scenarios in terms of tumor location and motion patterns. These may affect the reconstructed phase images and, consequently, the accuracy of the registration.
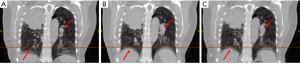
However, as shown in Figures 2-7 referenced with red lines and arrows, the prediction and reference images have a good match in the main features such as in the diaphragm. The coefficients of cross-correlation between the reference image and predicted image are all above 0.9 for all the cases in the region of the diaphragm, which also indicates the good match between the predicted image and the reference image.
Studies 3, 5 and 6 do not have high coefficients of cross-correlation in comparison of deformation vectors, but they have a good matching in the image comparison. This contrary indicates the network is able to predict accurate deformation vectors at the edge of the feature. For the smooth region such as the liver, it is difficult for the network to learn the features in the region to generate an accurate deformation vector prediction.
In order to improve the registration performance of our network in the inter-patient scenario, two things will be explored in future studies. The sample size will be increased to cover more patients, and a more sophisticated model will be adopted. However, as suggested in the latest paper (13), increasing in sample size will not further increase the performance of the network after achieving a saturation sample size. Therefore, a careful choice of sample size is of importance to balance the performance of the network and the efficiency of training.
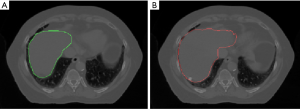
Figure 8 shows the potential application of our network, which is the contour propagation from phase 1 to all the other phases. Figure 8A shows the axial view of phase 1 with a manual contour on the liver in study 2, and Figure 8B shows the same slice but the deformed contour with our network. Our network achieved an excellent performance registering the boundary of features. In the clinic scenario, the physician only needs to contour on phase 1, and our model will perform the automatic contour propagation to other phases. Then the registration accuracy will be revised by physicians. It potentially improves contour efficiency and contour consistency, and consequently, the efficiency of the treatment flow will be increased.
A main advantage of using CNN to register phase-to-phase DVF is its fast speed. Our CNN network only takes a few seconds to predict the DVF even for large image volumes. In contrast, the VelocityAI takes around 1 min to calculate the DVF. Furthermore, our deep learning method is fully automatic without human intervention. In contrast, Velocity requires manual tuning of the parameters, which is highly dependent on the user experience and can significantly prolong the registration process.
Despite the network showing decent registration performance in the main features of respiration, there are some limitations to this method. First, the control points were chosen uniformly. The result of this is that some patches containing no features will be included in the training samples, and the network cannot learn anything from the patch pair without corresponding features. Therefore, useless patches will affect the learning efficiency of the network. Second, the reference DVF used for supervision of the network training has inaccuracy itself. The Velocity uses a smoothing and regularization function derived from B-spline to optimize the free-form deformation, but such regularization limits the ability to deform small localized warping and therefore affects the accuracy (14).
In our work, the network is to mimic the deformation procedures of Velocity and the accuracy of prediction is limited by the accuracy of the Velocity DVF. In order to get rid of this limitation, an unsupervised network directly learns the deformation from the moving image to the target image can be considered.
Conclusions
The feasibility and generalizability of using deep learning to register phase-to-phase respiratory DVF from 4D images were demonstrated using 4D-CT/CBCT images. CNN based deep learning achieved comparable deformable registration accuracy as Velocity. Compared to Velocity registration, the deep learning method is faster and fully automatic without user dependency, which makes it more preferable in clinical applications.
Acknowledgments
Funding: This work was supported by the National Institutes of Health under Grant No. R01-CA184173 and R01-EB028324.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-19-1058). The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics board of Duke (No.: 00049474), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Macchia G, Deodato F, Cilla S, Cammelli S, Guido A, Ferioli M, Siepe G, Valentini V, Morganti AG, Ferrandina G. Volumetric modulated arc therapy for treatment of solid tumors: current insights. Onco Targets Ther 2017;10:3755-72. [Crossref] [PubMed]
- Vedam SS, Keall PJ, Kini VR, Mostafavi H, Shukla HP, Mohan R. Acquiring a four-dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol 2003;48:45-62. [Crossref] [PubMed]
- Kadoya N. Use of deformable image registration for radiotherapy applications. J Radiol Radiat Ther 2014;2:1042.
- Rosu M, Hugo GD. Advances in 4D radiation therapy for managing respiration: part II - 4D treatment planning. Z Med Phys 2012;22:272-80. [Crossref] [PubMed]
- Johnson HJ, Christensen GE. Consistent landmark and intensity-based image registration. IEEE Trans Med Imaging 2002;21:450-61. [Crossref] [PubMed]
- Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 2011;15:622-39. [Crossref] [PubMed]
- Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vis Image Underst 1997;66:207-22. [Crossref] [PubMed]
- Shen D, Davatzikos C. Very high-resolution morphometry using mass-preserving deformations and HAMMER elastic registration. Neuroimage 2003;18:28-41. [Crossref] [PubMed]
- Chen Y, Yin FF, Jiang Z, Ren L. Daily edge deformation prediction using an unsupervised convolutional neural network model for low dose prior contour based total variation CBCT reconstruction (PCTV-CNN). Biomed Phys Eng Express 2019;5:065013. [Crossref] [PubMed]
- Kim M, Wu G, Wang Q, Lee SW, Shen D. Improved image registration by sparse patch-based deformation estimation. Neuroimage 2015;105:257-68. [Crossref] [PubMed]
- Cao X, Yang J, Zhang J, Nie D, Kim M, Wang Q, Shen D. Deformable Image Registration Based on Similarity-Steered CNN Regression. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2017 (Internet). Cham: Springer International Publishing, 2017:300-8. Available online: http://link.springer.com/10.1007/978-3-319-66182-7_35
- Shieh C, Gonzalez Y, Li B, Jia X, Rit S, Mory C, Riblett M, Hugo G, Zhang Y, Jiang Z, Liu X, Ren L, Keall P. SPARE: Sparse‐view reconstruction challenge for 4D cone‐beam CT from a 1‐min scan. Medical Physics (Internet). 2019 Jul 19 (cited 2019 Aug 12); Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/mp.13687
- Li Z, Zhou S, Huang J, Yu L, Jin M. Investigation of Low-Dose CT Image Denoising Using Unpaired Deep Learning Methods. IEEE Trans Radiat Plasma Med Sci 2020.1-1. [Crossref]
- Shekhar R, Lei P, Castro-Pareja CR, Plishker WL, D'Souza WD. Automatic segmentation of phase-correlated CT scans through nonrigid image registration using geometrically regularized free-form deformation. Med Phys 2007;34:3054-66. [Crossref] [PubMed]


