Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network
Introduction
The analysis of in vivo cells plays a pivotal role in describing genetic expressions and surveying the movements and structures of cells in clinical medicine (1,2). Moreover, the morphological change of human skin cells in vivo is a key cutaneous property of skin investigations, the study of which could offer new perspectives into the analyses of different cutaneous components and their age-related differences and the research of skin disorders such as melasma. Assessing the cellular structure and the depth of lesion tissue is critical for skin disorder treatments. Sometimes, biopsies are infeasible due to the distribution characteristics of skin disorders, thus requiring multiple sampling locations (3). Therefore, in vivo skin imaging techniques are being researched extensively and their application value since their noninvasiveness can retain the native state of the tissue and avoid the resection of tissue and scarring (4-6). In recent years, many noninvasive diagnostic methods using optical tissue imaging in vivo have been developed, including multiphoton microscopy (MPM). Lentsch et al. (3) evaluated the melanin content, melanin volume fraction, and distribution of melasma using MPM. MPM has been proved to be an effective biological imaging technique due to its deep penetration depth and low photobleaching and toxicity (7). MPM imaging relies on non-linear optical excitation processes such as two-photon excited fluorescence (TPEF) and second harmonic generation (SHG) (8,9). The TPEF can show the cellular morphology, and the SHG is sensitive to collagen fibers (3). MPM can directly visualize cells and tissues compared with conventional ways and has been widely applied in surveying the structure and dynamic interactions of cells and tissues. Lin et al. have developed a classification method of hepatocellular carcinoma using deep learning and speculate that MPM imaging may become irreplaceable for in vivo research (10). A key step for the study of in vivo imaging techniques is using cellular image segmentation to analyze and measure cellular boundaries and outlines. The quantitative analysis of in vivo cells using MPM imaging and cellular segmentation algorithms is of tremendous clinical value since it can supply new insights into the quantitative measurements of skin structures. To our knowledge, the MPM image segmentation results are dissatisfactory in general due to the blurred cellular boundaries, inhomogeneous depths, low amounts of data, and low signal-to-noise ratio (SNR) (8,11). As a result, it is rarely exploited in clinical scenarios due to the lack of an effective automatic segmentation method for MPM images.
Conventional methods like the threshold method, region growth, edge detection, watershed-based algorithms, and level set methods attempt to find the boundaries and outlines of in vivo cells without any marking information (8). A deformable model has been devised by Ma et al. (12), which utilizes the color information to semiautomatically segment dermoscopic images. The accuracy of the segmented results is low. As the usage of deep-learning and convolutional neural networks (CNN) has grown dramatically, the accuracy of the segmented results has increased in kind, and gained attention in medical image processing. Different from conventional algorithms, the use of fully convolutional networks (FCN) is an elegant segmentation method that exports segmentation maps (13). Deconvolutional operation is executed to upsample the feature map in the last convolutional layer of FCN to achieve pixel-level semantic segmentation. An FCN architecture named U-Net, which was proposed by Olaf et al. (14) has achieved outstanding achievements in medical image segmentation. Based on two well-known public datasets, a multistage FCN model was designed for skin lesion segmentation by Bi et al. (15). The segmented ability of the FCN was improved by this work. An automatic segmentation technique based on the deep FCN that used the Jaccard distance loss function with the ISBI training datasets for skin lesion segmentation was established by Yuan et al. (16). Also, Damseh et al. established a modified FCN to segment 2-photon microscopy images (17). An end-to-end segmentation network with 97 convolutional layers was also set up. High accuracy has been achieved by these approaches with well-known public datasets or clear medical images. Xiao et al. presented the Res-UNet with a weighted mechanism to segment retinal vessels from the DRIVE and the STARE datasets (18). U-Net was outperformed by an improved model named the stacked dilated U-Net, which was developed by Reza et al. (19) that could segment inner human embryonic cells. Another improved U-Net model by Zhang et al. was developed to conduct liver segmentation by (20). Despite these innovations, there is still no customized segmented model for human skin in vivo multiphoton microscopy images. Research does exist on CNN–MPM combined imaging technology for human skin that mainly adopts 3D image processing (7,8,11) (including separating the dermis and the epidermis in 3D structures), but the details of the cellular boundary segmentation in these studies are absent.
A novel cellular segmented model based on U-net for MPM cellular images is here proposed to address these problems. The objective of this study was to evaluate whether MPM imaging coupled with an image segmentation method based on the CNN could provide new insights into in vivo cell morphological analysis that used a small amount of data and had blurred boundaries.
Methods
U-Net model
U-Net is an advanced network that can be used to segment biomedical images (21-23). It includes a left-hand contracting path followed by an expansive right-hand path that forms a symmetrical U-Net shape (14). Some map-channels are used to connect each homologous layer of the two paths. Each map-channel functions as a bridge to convey contextual and localization information (13). The left-hand contracting path consists of 4 steps. Each step is comprised of two convolution layers (3×3) and one subsequent max-pooling layers (2×2) and uses the ReLU function as the rectified function. This path can be trained to capture the context. The expansive right-hand path is also formed of the corresponding four steps. Each step consists of an upsampling layer and two convolution layers (14), and also uses the ReLU function as the rectified function.
Proposed model: dense-UNet
U-Net ordinarily executes four downsamplings before the concatenate operation, resulting in resolution loss. The resulting resolution loss requires extensional techniques that depend on a deep network structure rather than a shallow one to improve accuracy (23). For these reasons, we employed the dense concatenated U-Net, called Dense-UNet.
Model architecture
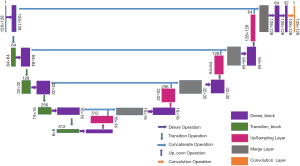
Our network is built by combining the U-net with the dense concatenation, as illustrated in Figure 1. The Dense-UNet consists of a dense downsampling (to the right) path and a dense upsampling (to the left) path, with the two paths being symmetrical. Some skip connection channels have been implemented to concatenate the two paths.
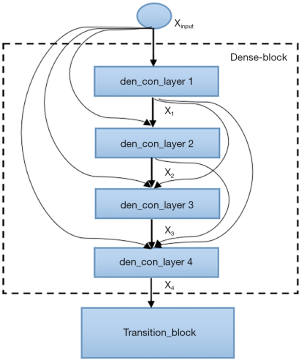
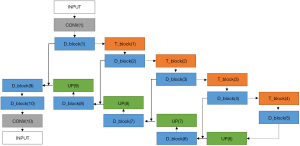
The semantic contextual features have been captured on multiple scales by the convolution operation in the dense downsampling path. It should be noted that just four downsampling layers have been applied to extract the local features, and the images have low input resolutions of 128×128×3. To address the limited depth of the U-Net architecture, we replaced the pooling and convolution operations with the dense_block operation and the transition_block operation in the downsampling path, which is named the dense downsampling path, to deepen the depth of the network. Each layer of the dense_block is linked to all its previous layers in a feed-forward mode to maximize feature reuse. Specifically, the input of each layer obtains the output feature maps from all its preceding layers (named dense concatenation). For this, the feature maps in each layer need the dense blocks to have the same feature size. The proposed Dense-UNet architecture has 10 dense_blocks: 5 dense_blocks in the dense downsampling path, and 5 dense_blocks in the dense upsampling path. Each dense_block consists of 4 densely connected layers with the same feature size, as shown in Figure 2. The transition_block is presented to achieve the layer transition. A dense_block and a transition_block form a layer, and five layers compose the dense downsampling path. We employed the upsampling layer, merger operation, and dense_block operation to reconstruct the high-resolution images in the dense upsampling path. The path is made up of five layers that localize the regions and recover the full input resolution. However, when the dense_block replaces the convolution, the dense upsampling path is substantially like the U-Net mechanism. The details of the network’s configuration are summarized in Figure 3.
As shown in Figure 2, four densely connected layers (named den_con_layers in Figure 2) have been incorporated in a dense_block. Each den_con_layer has two convolution operations. There is a total of 8 convolution operations in a dense_block. In the interest of feature reuse and compensating for the resolution loss, each layer is connected to all earlier layers to make better use of the extracted features. For example, the fourth layer of the den_con_layer receives the feature maps from layers 1, 2, and 3. The dense connections were applied in the dense_block and not in the whole architecture to avoid increasing the number of parameters.
Each den_con_layer has a set of operations, including batch normalization, convolution, activation (ReLU), and dropout operations, as found in Table 1. The input is first fed into a batch normalization operation to alleviate the vanishing gradients. Then, the outputs are subjected to a convolution operation with a 1×1 filter to decrease the number of feature maps of the inputs. Further downsampling via a convolution operation is applied with a 3×3 filter to extract the features. The dropout layer follows to avoid overfitting.
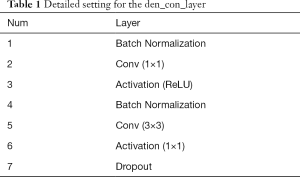
Full table
The transition_block is applied between 2 layers of the dense downsampling path to make the concatenation successful. It is composed of the batch normalization operation, the convolution operation, and max pooling with a 2×2 filter. The stride of 2 of the max pooling operations means that the filter will shift 2 pixels at a point to diminish the size of the output.
Four basic modules are illustrated in Figure 3, including the dense_block, the transition_block, the upsampling convolution layers, and the merge layers. To be specific, there are 23 layers and 89 convolution operations in total, including ten dense_blocks layers (as shown in Figure 2 and Table 1), four transition_block layers (as shown in Table 2), four upsampling layers, four merge layers, and one convolution operation.

Full table
Loss of function
The binary cross-entropy is usually employed as the loss function, which can be formulated as follows:
|
| [1] |
where f is the number of pixels, and mj and nj respectively show the predicted value and its corresponding ground-truth value. However, the resulting inefficient optimization requires the adaptive loss function due to the high susceptibility of the cross-entropy loss function to class imbalance. Therefore, Dice-loss is used as the loss function in our model as follows:
|
| [2] |
where mj is the predicted value, and nj is the corresponding ground-truth value.
Experimental framework
Imaging device
In this work, a recently labeled dataset that includes three videos that were acquired using MPM were annotated to train the neural network model. The videos were taken from the dorsal forearm of an Asian female volunteer aged 30 using a femtosecond Ti:Sa laser running at 735 nm. This RCM/MPM instrument is capable of simultaneously imaging human skin in vivo at up to 27 fps for RCM, SHG, and TPF imaging channels, and the videos of MPM signal of this instrument is integrated into 1 PET from the SHG and TPF imaging channel, while the cytoplasm is seen. The field of view of the system is 200×200 µm. The keratinocytes of the living epidermis are shown in Figure 4. In total, 15 original images with 256×256 resolution and a 24-bit depth of each pixel were obtained from different imaging depths of 3 different videos. The original pictures were divided into 60 pictures with a resolution of 128×128 to the expansion dataset (42 for training and 18 for testing). The study was approved by the University of British Columbia Research Ethics Board (no. # H96-70499). Informed consent was obtained from each volunteer subject.
Evaluation metric
Some evaluation metrics, including the Dice coefficient, accuracy, precision, and recall, were adopted to compare the performances of Dense-UNet and other methods to compare the performances of different methods.
The Dice coefficient measures the overlapping pixels between the automatic and manual segmentation of skin in vivo cells, which is calculated as follows:
|
| [3] |
where TP represents true positives and is the number of pixels that are accurately segmented as in vivo cells, TN represents true negatives and is the number of pixels that are accurately segmented as background, FP represents false positives and is the number of pixels that are wrongly segmented as in vivo cells. FN is false negatives and is the number of pixels that were missed.
Accuracy is the entire accuracy of the in vivo cell and background segmentation, which is described as the following:
|
| [4] |
Precision is the proportion of in vivo cells that are classified as true-positive pixels concerning all pixels of in vivo cells that are classified by automatic segmentation, which is delimited as follows:
|
| [5] |
The recall represents the proportion of the true positive pixels of in vivo cells that are classified by automatic segmentation versus the pixels of in vivo cells that are classified by manual segmentation, which is calculated as follows:
|
| [6] |
F1-Score is used to quantify the weighted average of in vivo cells between the precision and recall rate, with a value in [0, 1], and is calculated as follows:
|
| [7] |
Intersection over union (IOU) is a homologous metric addition determined by weighting the ratios between the overlap area and union area as:
|
| [8] |
Results
The testing was implemented on Microsoft Windows 10.0.17134 [Intel(R) Core™ i7-8700 CPU@3.20 GHz] using a GeForce GTX 1080 Ti with a memory clock rate (GHz) of 1.683. The programming is implemented using Python 3.5.2, Anaconda 4.2.0 (64-bit), TensorFlow-GPU 1.12.0, CUDA Toolkit 9.0, cuDNN v7.0.5, VS2015, and Keras 2.2.4.
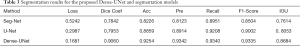
First, the Dense-UNet model is executed to evaluate the performance of U-Net. The in vivo cell segmentation performances are compared between Dense-UNet and other typical methods (Seg-Net and U-Net) on the same datasets, as is depicted in Table 3. It is demonstrated that Dense-UNet outperforms U-Net by 11.07% in the Dice coefficient, 3.95% in the accuracy, 4.28% in the precision, 1.32% in the recall, 3.33% in the F1-Score and 6.31% in the IOU. Also, Dense-UNet outperforms SegNet by 12.18% in the Dice coefficient, 10.28% in the accuracy, 12.19% in the precision, 3.89% in the recall, 8.31% in the F1-Score and 10.7% in IOU.
Full table
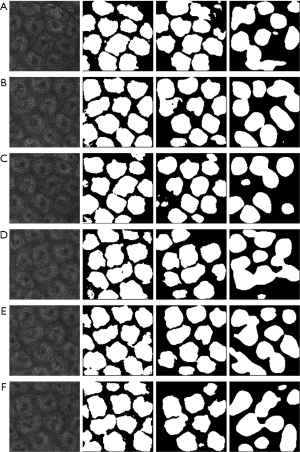
The test images were chosen from a new video to avoid the repetition of the data, as shown in Figure 5. Lower resolution MPM images are described in the first column. The results of segmentation (white areas) by the Dense-UNet, U-Net, and Seg-Net are represented in the second to fourth columns, respectively. It is clear that Seg-Net commonly commits overfitting and loses some fuzzy cells without being able to recognize and segment cells of in vivo MPM images effectively. Meanwhile, the U-Net also commits overfitting and is unable to extract the fine-detailed boundary information. The segmentation results by Dense-UNet are closer to the real boundaries than the others. In short, the segmentation results proved that the Dense-UNet could aptly manage blurry and low-resolution medical images.
Discussion
In general, judging the performance of different algorithms is done according to the accuracy of prediction results. The most recent segmentation algorithm of in vivo MPM images was proposed by Wu et al. in 2016 and is based on superpixel and watershed (8). The MPM images of human skin in vivo have some specific characteristics, including blurry boundaries, low resolutions, and inhomogeneous contrast ratios. U-Net is an ultramodern method to conduct medical image segmentation. In it, only four downsampling layers are used for feature extraction. Although the segmented results of U-Net are much better than those of Seg-Net, as shown in Table 3 and Figure 5, the U-Net is still not sensitive enough to capture the finer details. The white areas that are segmented by U-Net are still smooth by ignoring the spatial information of shallow layers. Also, the resolution of the MPM images is as low as 128×128×3, which makes it hard to extend the depth of the contracting path using the U-Net model. The Dense-UNet supplied more exact segmentation of in vivo cells than the U-Net and SegNet. The Dense-UNet enjoys the advantages both the U-net and Dense-net and uses dense concatenations to deepen the depth of the contracting path. The structural characteristics of the Dense-UNet can be summarized in the following points.
- The novel Dense-UNet model combines a dense structure with a full convolution network (FCN). By inheriting the superiority of both the FCN and deep CNN, more semantic features, and high segmentation accuracy are gained by the deeper structure.
- Feature reuse is achieved by iteratively summing all earlier feature maps for every layer of the dense_block. This way, more boundary details of blurry, in vivo skin cells can be segmented due to the feature reuse.
- The vanishing gradient and model degradation issues are relieved by the feature reuse and batch normalization.
- More efficacious optimization can be achieved as the iterative sum need not be applied between different dense_blocks, and the number of parameters can be significantly reduced due to feature reuse and bypass.
- The features from earlier layers are mapped in short paths (only 1 to 4) using the dense concatenation since there is not a dense concatenation between different dense_blocks. The resolution is recovered by using the captured features in the expansive path, which improves the discrimination capability.
As expected, the Dense-UNet achieved the most advanced segmentation of MPM images of human skin in vivo.
Conclusions
In this paper, the Dense-UNet method was proposed to solve the segmentation problems for MPM images of human skin in vivo.
Dense-UNet adds a dense concatenation to U-Net with 89 convolutional layers, which allows it to combine the advantages of the method with the superiority of the deep CNN. The adaptability of Dense-UNet for MPM images of skin in vivo is better than that of U-Net under low resolutions and inhomogeneous contrast ratios. This is the first time that the U-Net has been combined with the deep CNN and used for in vivo cell image segmentation. The quantitative analysis of MPM images of human skin in vivo based on this deep-learning model can be used to research the variations of cellular features more effectively with pathologies and improve clinical diagnosis.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (U1805262, 61701118, 61901117, 61871131, 61571128), by the United Fujian Provincial Health and Education Project for Tackling the Key Research of China (2019-WJ-03), by the Natural Science Foundation of Fujian Province (2019J01272), by the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R10), by the Special Funds of the Central Government Guiding Local Science and Technology Development (2017L3009),by the research program of Fujian Province (2018H6007), by the Special Fund for Marine Economic Development of Fujian Province (ZHHY-2020-3), by the Scientific Research Innovation Team Construction Program of Fujian Normal University (IRTL1702), by the Canadian Institutes of Health Research (MOP130548), and by the Canadian Dermatology Foundation, the VGH & UBC Hospital Foundation, and the BC Hydro Employees Community Services Fund.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-19-1090). HL reports grants received during the conduct of the study from the Canadian Institutes of Health Research, grants from the Canadian Dermatology Foundation, the VGH & UBC Hospital Foundation, and the BC Hydro Employees Community Services Fund; also, HL has a patent with Zeng H, Lui H, McLean D, Lee A, Wang H, and Tang S: Apparatus and Methods for Multiphoton Microscopy. US patent # 9687152B2, June 27, 2017 Canadian Patent # 2832162; issued May 14, 2019. HZ reports grants received during the conduct of the study from the Canadian Institutes of Health Research, the Canadian Dermatology Foundation, the VGH & UBC Hospital Foundation, and from the BC Hydro Employees Community Services Fund; also, HZ has a patent with Zeng H, Lui H, McLean D, Lee A, Wang H, and Tang S: Apparatus and Methods for Multiphoton Microscopy. US patent # 9687152B2, June 27, 2017 Canadian Patent # 2832162; issued May 14, 2019. YW reports grants received during the conduct of the study from the National Natural Science Foundation of China. GC reports grants received outside the submitted work from the United Fujian Provincial Health and Education Project for Tackling the Key Research of China, the Natural Science Foundation of Fujian Province, the Program for Changjiang Scholars and Innovative Research Team in University, the Special Funds of the Central Government Guiding Local Science and Technology Development, and the Scientific Research Innovation Team Construction Program of Fujian Normal University. The other authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the University of British Columbia Research Ethics Board (no. H96-70499). Informed consent was obtained from each volunteer subject.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fereidouni F, Bader AN, Colonna A, Gerritsen HC. Phasor analysis of multiphoton spectral images distinguishes autofluorescence components of in vivo human skin. J Biophotonics 2014;7:589-96. [Crossref] [PubMed]
- Chen G, Lui H, Zeng H. Image segmentation for integrated multiphoton microscopy and reflectance confocal microscopy imaging of human skin in vivo. Quant Imaging Med Surg 2015;5:17-22. [PubMed]
- Lentsch G, Balu M, Williams J, Lee S, Harris RM, König K, Ganesan A, Tromberg BJ, Nair N, Santhanam U, Misra M. In vivo multiphoton microscopy of melasma. Pigment Cell Melanoma Res 2019;32:403-11. [Crossref] [PubMed]
- Wang H, Lee AM, Frehlick Z, Lui H, McLean DI, Tang S, Zeng H. Perfectly registered multiphoton and reflectance confocal video rate imaging of in vivo human skin. J Biophotonics 2013;6:305-9. [Crossref] [PubMed]
- Sdobnov AY, Darvin ME, Genina EA, Bashkatov AN, Lademann J, Tuchin VV. Recent progress in tissue optical clearing for spectroscopic application. Spectrochim Acta A Mol Biomol Spectrosc 2018;197:216-29. [Crossref] [PubMed]
- Chung HY, Greinert R, Kärtner FX, Chang G. Multimodal imaging platform for optical virtual skin biopsy enabled by a fiber-based two-color ultrafast laser source. Biomed Opt Express 2019;10:514-25. [Crossref] [PubMed]
- Decencière E, Tancrède-Bohin E, Dokládal P, Koudoro S, Pena AM, Baldeweck T. Automatic 3D segmentation of multiphoton images: a key step for the quantification of human skin. Skin Res Technol 2013;19:115-24. [Crossref] [PubMed]
- Wu W, Lin J, Wang S, Li Y, Liu M, Liu G, Cai J, Chen G, Chen R. A novel multiphoton microscopy images segmentation method based on superpixel and watershed. J Biophotonics 2017;10:532-41. [Crossref] [PubMed]
- Liu Y, Tu H, You S, Chaney EJ, Marjanovic M, Boppart SA. Label-free molecular profiling for identification of biomarkers in carcinogenesis using multimodal multiphoton imaging. Quant Imaging Med Surg 2019;9:742-56. [Crossref] [PubMed]
- Lin H, Wei C, Wang G, Chen H, Lin L, Ni M, Chen J, Zhuo S. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophotonics 2019;12:e201800435. [Crossref] [PubMed]
- Decencière E, Tancrède-Bohin E, Dokládal P, Koudoro S, Pena AM, Baldeweck T. Automatic 3D segmentation of multiphoton images: a key step for the quantification of human skin. Skin Res Technol 2013;19:115-24. [Crossref] [PubMed]
- Ma Z, Tavares JM. A Novel Approach to Segment Skin Lesions in Dermoscopic Images Based on a Deformable Model. IEEE J Biomed Health Inform 2016;20:615-23. [Crossref] [PubMed]
- Stiaan W, Steve K, Hendrik DV. Unsupervised pre-Training for fully convolutional neural networks. 2016 Pattern Recognition Association of South Africa and Robotics and Mechatronics International Conference, PRASA-RobMech 2016, January 10, 2017.
- Olaf R, Philipp F, Brox T. U-net: Convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), v 9351, p 234-241, 2015, Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 - 18th International Conference, Proceedings.
- Bi L, Kim J, Ahn E, Kumar A, Fulham M, Feng D. Dermoscopic Image Segmentation via Multistage Fully Convolutional Networks. IEEE Trans Biomed Eng 2017;64:2065-74. [Crossref] [PubMed]
- Yuan Y, Chao M, Lo YC. Automatic Skin Lesion Segmentation Using Deep Fully Convolutional Networks With Jaccard Distance. IEEE Trans Med Imaging 2017;36:1876-86. [Crossref] [PubMed]
- Damseh R, Cheriet F, Lesage F. Fully Convolutional DenseNets for Segmentation of Microvessels in Two-photon Microscopy. Conf Proc IEEE Eng Med Biol Soc 2018;2018:661-5. [PubMed]
- Xiao X, Lian S, Luo Z, Li S. Weighted Res-UNet for High-Quality Retina Vessel Segmentation. Proceedings - 9th International Conference on Information Technology in Medicine and Education, ITME 2018, p 327-331, December 26, 2018.
- Reza RM, Saeedi P, Au J, Havelock J. Multi-resolutional ensemble of stacked dilated U-net for inner cell mass segmentation in human embryonic images. Proceedings - International Conference on Image Processing, ICIP, p 3518-3522, August 29, 2018.
- Zhang L, Xu L. An Automatic Liver Segmentation Algorithm for CT Images U-Net with Separated Paths of Feature Extraction. 2018 3rd IEEE International Conference on Image, Vision and Computing, ICIVC 2018, p 294-298, October 15, 2018.
- Krapac J, Segvic S. Ladder-Style DenseNets for Semantic Segmentation of Large Natural Images. Proceedings - 2017 IEEE International Conference on Computer Vision Workshops, ICCVW 2017, v 2018-January, p 238-245, January 19, 2018.
- Jégou S, Drozdzal M, Vazquez D. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, CVPRW 2017, v 2017-July, p 1175-1183, August 22, 2017.
- Wu ZX, Chen FJ, Wu DY. A Novel Framework Called HDU for Segmentation of Brain Tumor. 2018 15th International Computer Conference on Wavelet Active Media Technology and Information Processing, ICCWAMTIP 2018, p 81-84, January 31, 2019.