
Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy
Introduction
Accurate patient setup is critical in radiotherapy. Incorrect setup during radiotherapy may result in an unnecessary radiation dose for healthy tissue while compromising the target dose. Variation of the target position is a significant challenge in the clinical practice of external beam radiotherapy since there is a day-to-day change of tumor position caused by breathing, filling of hollow organs, and more complex changes such as weight loss and tumor regression (1). In general, cone-beam computed tomography (CBCT) is applied in image-guided radiotherapy (IGRT) for patient setup (2). The CBCT image is registered to the reference high-resolution planning CT (pCT) data set, which is acquired during the planning phase of the treatment to obtain the patient setup error (3). The outcomes are used to localize the patient. Therefore, the target during treatment is approximately at the same location during planning (3-6).
In some cases, however, distinguishing between different soft tissues on the CBCT image may prove impossible due to the limited soft-tissue contrast and scatter artifacts of CBCT images (7). Therefore, it is challenging to align CBCT with pCT accurately. Consequently, it is difficult to get a precise patient setup position and the patient must receive an added dose in the positioning procedure for intrafraction motion correction.
With the development of magnetic resonance image-guided radiotherapy (MRIgRT), an accelerator integrated with a magnetic resonance image (MRI) scanner is currently in the process of replacing the CBCT-based IGRT method (8,9). MRIgRT has several clear advantages. Firstly, the contrast of soft tissue of MRI images is much better than that of CBCT images, which can provide more distinct tumor pathology and detail of the surrounding area. Secondly, MRI is free of extra radiation and can achieve real-time imaging, which allows for rapid adaptive planning and beam delivery control based on the visualization of soft tissues (9). Previous research proposed that the use of precise MR-guided radiotherapy can further lower radiation dose to organs at risk (OARs) in pancreatic cancer (10). For MRIgRT, positioning MRI (pMRI) images, instead of CBCT images, are acquired to align with reference pCT images to calculate the target position relative to the planned reference position (8). Consequently, the coach is moved to calibrate the error.
However, cross-modality image registration between pMRI and pCT is a challenging problem due to the high variability of tissue or organ appearance caused by different imaging mechanism, which results in the lack of a general rule to compare such images (11). The pixel intensity, voxel size, image orientation, and field of view are also different in CT and MRI images, which makes multi-modality registration less straightforward than mono-modality alignment (12,13). Morrow et al. further indicated that registration between different modalities might adversely affect soft tissue-based registration for IGRT (14). For MRI/CT registration, Zachiu et al. illustrated better mono-modality registration results as compared to CT-MRI registration (15).
To tackle this challenging multi-modality image registration problem, many researchers generated CT images from MRI images (7-14). These models include the Gaussian mixture regression model (16), random forest regression model (17), segmentation-based methods (18-20), atlas-based methods (21,22), and learning-based methods (23-30). However, the loss of rich anatomical information in the CT images makes it hard to conduct precise registration, since sufficient anatomical details are also necessary for steering accurate image registration, as demonstrated in (31). So, in this paper, to achieve a more accurate patient setup, we proposed a method to decrease registration error via the synthesis of MRI images based on pCT. Thus, multi-modality image registration problem is transformed into a mono-modality alignment one. Meanwhile, rich anatomical information is enhanced, which is useful for tumor recognition during the registration process.
Much work has been carried out to estimate CT images from MRI data (7-14), but for MRI synthesis based on CT images, only a few learning-based studies have been conducted. Chartsias et al. (32) used a CycleGAN model (33) and unpaired cardiac data to generate MRI images from different patient CT images. However, this work can not directly quantitatively evaluate the generated MRI images from unpaired cardiac images, as it is challenging to obtain paired MRI and CT images from a beating heart. Jin et al. (34) improved the accuracy of CT-based target delineation in radiotherapy planning by using paired and unpaired brain CT/MRI data to generate MRI images. However, the authors did not investigate the effect of conventional supervised methods on this MRI synthesis task.
In this work, several deep neural networks were applied to implement the synthesis task, including unsupervised generative adversarial network (GAN), such as CycleGAN (33), and supervised networks, such as Pix2Pix (35) and U-Net model (36). As well as several metrics, such as mean absolute error (MAE), mean squared error (MSE), structural similarity index (SSIM), and peak signal-to-noise ratio (PSNR), were used to quantitatively evaluate the performance of these networks. We aimed to generate synthetic MRI (sMRI) images from pCT images based on deep learning methods. Thus, the large appearance gap between pMRI and pCT images could be bridged by synthesizing MRI from pCT images, and at the same time the anatomical details could be reserved. Consequently, many conventional registration methods could be directly applied to align two similar-looking images with the same image modality, and finally achieve a precise patient positioning. To the best of our knowledge, this is the first study to generate sMRI images from pCT images for patient positioning in MRIgRT.
Methods
Neural networks for sMRI estimation
Several state-of-art deep neural networks were applied to generate sMRI from pCT, including the U-Net model (36), Pix2Pix model (35), and CycleGAN (33).
U-Net
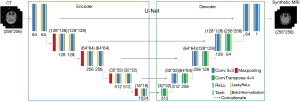
U-Net achieved great success in the segmentation task (23,37-41). The structure is illustrated in Figure 1. It is an end-to-end convolutional neural network, which consists of the mirrored encoder (left side) and decoder (right side) layers. The encoder consists of repeated 3*3 Convolutions (padded), each with a Batch-Normalization (BN) (42) layer and LeakyReLu (43) layer except input convolution. A max-pooling operation with stride two is used for downsampling. Feature channels are doubled at each downsampling step. The decoder consists of repeated 3*3 Convolutions (padded) and 4*4 ConvTranspose (padded) layers (44), each with a ReLu (45) layer and followed by a BN layer. Skip connections are used to concatenate channels from the encoder part to the corresponding decoder layers to use information from earlier layers as much as possible. At the final layer, a Tanh activation operation is used to map sMRI images.
Pix2Pix
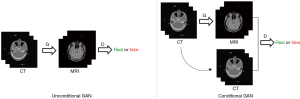
This model is a kind of conditional Generative Adversarial Networks (cGANs) (46-50). The difference between conditional and unconditional GAN is illustrated in Figure 2. Unlike unconditional GAN, the discriminator of cGANs tries to distinguish fake and real tuples, which means the input of the discriminator consists of the original input of the generator and its output as opposed to just the output of the generator. Pix2Pix model uses a “U-Net”-based generator and “PatchGAN”-based (51) discriminator. The “PatchGAN”-based discriminator penalizes structure at 2D 70*70 overlapping image patches and then the averages all responses to supply output. A smaller PatchGAN has fewer parameters and runs faster when compared to other GAN methods.
Furthermore, L1 distance is used in the Pix2Pix model to decrease blurring. Isola et al. (35) demonstrated that cGANs could produce consistent results on a variety of problems rather than focus on specific problems. In this work, we applied this network to synthesize brain MRI images.
CycleGAN
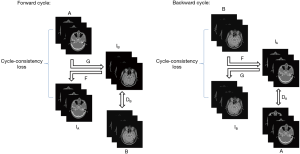
CycleGAN is a neural network that can translate an image from one domain to another using unpaired data. It alleviates the problem posed by a lack of paired datasets since there is a vast amount of unpaired image data. The schematic of this network is shown in Figure 3. The network has a forward and backward cycle. The forward cycle learns a mapping G to translate A to IB and tries to make IB as real as the ground truth of image B to fool the discriminator DB. The discriminator DB is used to identify whether the input is real or fake. The mapping of F is trained to transform IB back to IA and make IA as real as the ground truth of image A to ensure that IB can translate back to where it started. An added cycle-consistency loss is used to support this schedule. Opposite to the forward cycle, the retrograde cycle is trained to translate B to IA to improve the stability of the network. More details of CycleGAN can be seen in (33). In this work, we input brain pCT and corresponding brain MRI images as A and B, respectively.
Implementation
To generate MRI from CT images, we first rescaled the MRI and CT value of the acquired images to [0, 255], then converted these data to [0, 1] tensor and normalized each image to [−1, 1]. Instead of image patches, whole 2D images were used to train all models. The axial CT and MRI pairs were put into networks with a size of 256*256 pixels. To augment the training data, all training images were padded to the size of 286*286 pixels, and then randomly cropped to sub-images of 256*256 pixels from full images when training the networks.
For the Pix2Pix and CycleGAN models, we set the batch size as one and used Adam (52) to optimize both the generator and discriminator. Two hundred epochs were trained for each model. A fixed learning rate of 2*10−4 was applied for the first 100 epochs and linearly reduced to 0 during the training of the rest 100 epochs. The paired brain CT and MRI images were shuffled to create an unpaired dataset. The only difference between unpaired and paired datasets was that the unpaired dataset MRI was not input in the same sequence as a paired dataset. Unlike the Pix2Pix model, CycleGAN was trained with paired and unpaired CT/MRI datasets, respectively, since the synthesis result of the paired and unpaired input data form was a point of interest. To investigate the degree of misalignment of the unpaired images, we compared the randomly shuffled data and found that there were only 221/812,600 (0.27%) paired training images (each epoch with 4,063 training images, 812,600 training images for 200 epochs in total).
To contrast these models, similar parameters were applied in U-Net as used in CycleGAN and Pix2Pix model, including set batch size to 1, adopt Adam optimizer, training 200 epochs, and the learning rate was fixed at the first 100 epochs and then linearly decreased to 0 during the rest 100 epochs. Different from CycleGAN and Pix2Pix model, we used different loss functions for U-Net training. Isola et al. (35) suggested that designing sufficient losses is vital because the losses tell the CNN what to minimize. Inspired by this perspective, different losses were designed for U-Net in our experiments. We first used L2 loss to minimize the squared difference between ground truth MRI image and sMRI image. L2 loss is widely used in tasks like denoising, debugging, deblurring, and super-resolution, etc. (53). However, if there are outliers in image data, the difference between the predicted image and ground truth may be quite significant because of squaring. In this case, L1 loss may be a better choice, so we used it as a comparison. L1 loss minimizes the absolute difference between ground truth and sMRI and encourages less blurring (35). In our experiments, we found that by combining these two losses, the performance could be improved, so we used L1 + L2 as a new loss. The L1 loss can be expressed as:
|
| [1] |
L2 loss can be expressed as:
|
| [2] |
So, the new object is:
|
| [3] |
We implemented these networks in Pytorch1 and used a single NVIDIA Tesla V100 (32GB) GPU for all of the training experiments.
Evaluation
Thirty-four patient images were acquired, each with brain CT and corresponding T2-weighted MRI. The CT images were acquired with tube voltage 120 kV, current 330 mA, exposure time 500 ms, in-plane resolution 0.5*0.5 mm2, slice thickness 1 mm, image size 512*512 on SOMATOM Definition Flash (Siemens). T2-weighted MRI images [repetition time (TR): 2,500 ms, echo time (TE): 123 ms, 1*1*1 mm3, 256*256] were acquired on a 1.5T Avanto scanner (Siemens). The MRI distortion correction was applied for the acquired MRI images by the MRI data acquisition system.
To remove the unnecessary background, we first generated binary head masks through the Otsu threshold method (54). Then, we used the generated binary masks and “AND operation” to exclude unnecessary background and keep the interior brain structures. We resampled CT images of size 512*512 to 256*256 to match MRI images by bicubic interpolation (55). To align CT with the corresponding MRI images, we took CT as a fixed image, and the MRI images were registered to CT space by rigid registration. Elastix (56) toolbox was used to perform the rigid registration, and mutual information was taken as the cost function. We randomly selected 28 patients to train neural networks. The other six patients were used as test data. Overall, 4,063 training image pairs and 846 test image pairs were obtained.
Several metrics were used to compare the ground truth MRI and sMRI, including MAE, MSE, SSIM, and PSNR. These metrics were widely used in medical image evaluation (23-30,34,57,58). These metrics can be expressed as follows:
|
| [4] |
|
| [5] |
|
| [6] |
Where N is the total number of pixels inside the head region, i is the index of the aligned pixels inside the head area. MAX denotes the largest pixel value of ground truth MRI and sMRI images, here MAX =4,095.
SSIM (59) is a metrics that can be used to quantify the similarities in whole image scale, which can be expressed as
|
| [7] |
Where µMRI and µsMRI are the average of ground truth MRI image and sMRI image. σMRI and σsMRI are the variance of ground truth MRI and sMRI, respectively. σMRI·sMRI is the covariance of ground truth MRI and sMRI.
Results
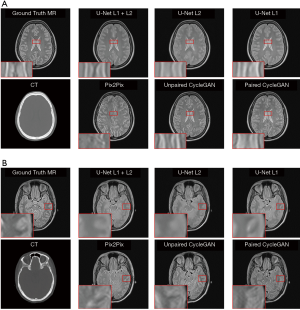
In our experiments, clinical CT images of six patients were acquired to test the proposed models, as shown. The generated sMRI images were compared with the corresponding ground truth MRI images. Figure 4 shows cross-sectional views of two representative sMRI results by using U-Net with L1 + L2 loss, U-Net with L2 loss, U-Net with L1 loss, Pix2Pix model, unpaired CycleGAN, and paired CylceGAN. For the Pix2Pix methods, the generated sMRI images were significantly different from the ground truth MRI images, which lost some anatomical information and did not predict the cerebrospinal fluid. For the CycleGAN methods, there was no clear visible difference between paired and unpaired sMRI images. Despite the images generated by CycleGAN networks all looking realistic, they tended to be noisier compared with those generated by U-Net networks for the whole image scale. As for the U-Net methods of different losses, U-Net with L2 loss generated blurry outputs, while U-Net with L1 + L2 loss was much better than other methods.
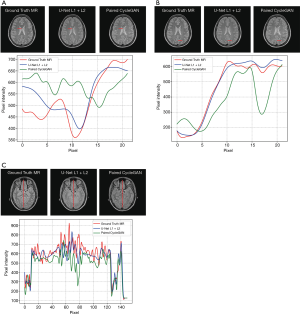
A comparison of 1D profiles passing through three different lines, which were acquired from images generated by U-Net with L1 + L2 loss and paired CycleGAN model, is shown in Figure 5A,B,C. Based on these images, the sMRI images produced by U-Net with L1 + L2 loss matched well with the ground truth MRI at pixel scale. For Figure 5A, the paired CycleGAN produced a more vibrational profile, which indicated that the CycleGAN model generated more details for sMRI images. However, these generated details may be inaccurate compared with the ground truth MRI image. We typically chose a long vertical line to compare more pixel details of these two models in Figure 5C. It was proved that at a large pixel scale, the results of the U-Net model with L1 + L2 loss were still well matched with the ground truth MRI. Figure 5A,B,C shows that even the profile tendency of the CycleGAN is dissimilar to the ground truth MRI.
Overall statistics of the four quantitative metrics over the whole brain image are shown in Table 1. The results showed that the GAN methods of paired CycleGAN, unpaired CycleGAN, and Pix2Pix model had higher MAE and lower SSIM value when compared with the U-Net model, which means that U-Net models with L1 loss, L2 loss, and L1 + L2 loss were more accurate than the GAN methods used in our experiment. Among all these methods, U-Net with L1 loss achieved much higher MSE and lowered PSNR. This showed that the pixel values of predicted sMRI images by L1 loss U-Net were more volatile. U-Net with L2 loss was more stable than that with L1 loss in MSE and PSNR. U-Net with combined L1 loss and L2 loss achieved the best quantitative results in MAE, MSE, SSIM, and PSNR out of the overall methods used in this paper. To investigate if the combination of L1 and L2 loss was statistically significant, we performed paired t-tests to compare U-Net with L1 loss and U-Net with L2 loss against that with L1 + L2 loss. The t-test results indicated that U-Net with L1 + L2 loss significantly outperformed U-Net with L1 loss in MSE and PSNR, the P values were 4.5×10−6 and 7.9×10−7 respectively, which were much less than 0.05. The results of the t-test also showed that U-Net with L1 + L2 loss was statistically better than U-Net with L2 loss in SSIM, with P<0.05 (0.017).

Full table
Discussion
In this study, we proposed a sMRI generation method to help pCT align with pMRI images. This method can be used for patient positioning in the radiotherapy process. Different deep learning models were compared, including GANs methods of CycleGAN, Pix2Pix model, and traditional U-Net. To further improve the accuracy of the synthetic method, we combined L1 loss with L2 loss together as a new loss function for U-Net training. The qualitative and quantitative comparison suggests that U-Net with L1 + L2 loss achieved the best results, with the lowest overall average MAE of 74.19 and MSE of 1.035*104, respectively, and the highest SSIM of 0.9440 and PSNR of 32.44, respectively.
In our experiment, the U-Net model produced better quantitative results than the paired and unpaired CycleGAN and Pix2Pix models. This may be partially due to some hidden details of CT images being unrecognizable to the generator, since these methods belong to global optimized methods. To generate realistic-looking MRI images to deceive the discriminator in the GANs model, the generator produced some fake details to match these loss functions. However, these details are generated without foundation and are different from the ground truth MRI, which leads to worse results than U-Net.
The sMRI images generated by U-Net with L1 + L2 loss also achieved better qualitative results compared with paired CycleGAN. The difference map of U-Net with L1 + L2 loss and paired CycleGAN (the best GAN methods in our experiments) can be seen in Figure 6. According to the difference map between ground truth MRI and the corresponding sMRI, U-Net with L1 + L2 loss learns to distinguish different anatomical structures in the head area from similar CT pixel values. Errors are evenly distributed in the head region, which may be partially attributable to the non-perfect alignment between the ground truth MRI and the corresponding CT images. As Han (23) mentioned in his work, it is challenging to achieve one-to-one correspondence pixel values between training MRI and CT images by a linear registration. If there are misalignments, the errors in training data will cause inaccuracy in the model since the model will be trained to make the wrong prediction. Compared with U-Net with L1 + L2 loss, the paired CycleGAN model has a significant error, which means in our MRI synthesis task with small clinical data, a supervised learning network of U-Net is more accurate than unsupervised learning networks of CycleGAN.
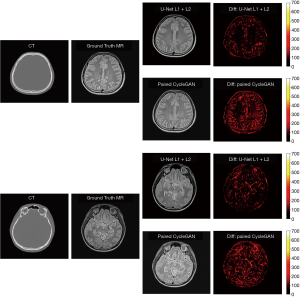
In our CT to MRI study, we generated sMRI from CT images to convert MRI-to-CT cross-modality registration problem into the single modality of MRI-to-MRI registration to facilitate the use of registration methods and further improve the registration accuracy. In this paper, we used 2D networks to achieve the MRI synthesis task, which may cause potential inter-slice discontinuity problem. In our future work, we will continue to apply 3D networks to investigate if there are any differences in MRI synthesis tasks between 2D and 3D networks.
Conclusions
In this work, we proposed a deep learning method to help patient positioning in radiotherapy progress by generating synthetic brain MRI from corresponding pCT images. Instead of aligning pCT with pMRI, it can be easier to align sMRI with pMRI. L1 loss and L2 loss can be combined as a new U-Net loss function. Several methods were compared, and U-Net with L1 + L2 loss achieved the best performance in our test set. The proposed method will be used in the increasingly popular MRIgRT scheme for future radiotherapy applications.
Acknowledgments
Funding: This work is supported in part by grants from the National Key Research and Develop Program of China (2016YFC0105102), the Leading Talent of Special Support Project in Guangdong (2016TX03R139), Shenzhen matching project (GJHS20170314155751703), the Science Foundation of Guangdong (2017B020229002, 2015B020233011) and CAS Key Laboratory of Health Informatics, Shenzhen Institutes of Advanced Technology, and the National Natural Science Foundation of China (61871374, 81871433, 61901463, 90209030, C03050309), Shenzhen Science and Technology Program of China grant (JCYJ20170818160306270), Fundamental Research Program of Shenzhen (JCYJ20170413162458312), the Special research project slot son of basic research of the Ministry of Science and Technology (No. 22001CCA00700), the Guangdong Provincial Administration of Traditional Chinese Medicine (20202159).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-19-885). JX is an employee of Shenzhen University General Hospital. The other authors have no conflicts of interest to declare.
Ethical Statement: The approval for this study was obtained from the Human Research Ethics Committee of the Shenzhen Second People’s Hospital and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Guckenberger M. Image-guided radiotherapy based on kilovoltage cone-beam computed tomography – a review of technology and clinical outcome. Eur Oncol Haematol 2011;7:121-24. [Crossref]
- Jaffray DA, Siewerdsen JH. Cone‐beam computed tomography with a flat‐panel imager: initial performance characterization. Med Phys 2000;27:1311-23. [Crossref] [PubMed]
- Thilmann C, Nill S, Tücking T, Höss A, Hesse B, Dietrich L, Bendl R, Rhein B, Häring P, Thieke C, Oelfke U, Debus J, Huber P. Correction of patient positioning errors based on in-line cone beam CTs: clinical implementation and first experiences. Radiat Oncol 2006;1:16. [Crossref] [PubMed]
- Lehmann J, Perks J, Semon S, Harse R, Purdy JA. Commissioning experience with cone‐beam computed tomography for image‐guided radiation therapy. J Appl Clin Med Phys 2007;8:2354. [Crossref] [PubMed]
- Oldham M, Létourneau D, Watt L, Hugo G, Yan D, Lockman D, Kim LH, Chen PY, Martinez A, Wong JW. Cone-beam-CT guided radiation therapy: A model for on-line application. Radiother Oncol 2005;75:271-8. [Crossref] [PubMed]
- Zachiu C, de Senneville BD, Tijssen RH, Kotte AN, Houweling AC, Kerkmeijer LG, Lagendijk JJ, Moonen CT, Ries M. Non-rigid CT/CBCT to CBCT registration for online external beam radiotherapy guidance. Phys Med Biol 2017;63:015027. [Crossref] [PubMed]
- Angelopoulos C, Scarfe WC, Farman AG. A comparison of maxillofacial CBCT and medical CT. Atlas Oral Maxillofac Surg Clin North Am 2012;20:1-17. [Crossref] [PubMed]
- Acharya S, Fischer-Valuck BW, Kashani R, Parikh P, Yang D, Zhao T, Green O, Wooten O, Li HH, Hu Y, Rodriguez V, Olsen L, Robinson C, Michalski J, Mutic S, Olsen J. Online magnetic resonance image guided adaptive radiation therapy: first clinical applications. Int J Radiat Oncol Biol Phys 2016;94:394-403. [Crossref] [PubMed]
- Mutic S, Dempsey JF. The ViewRay system: magnetic resonance–guided and controlled radiotherapy. Semin Radiat Oncol 2014;24:196-99. [Crossref] [PubMed]
- Bohoudi O, Bruynzeel A, Senan S, Cuijpers JP, Slotman BJ, Lagerwaard FJ, Palacious MA. Fast and robust online adaptive planning in stereotactic MR-guided adaptive radiation therapy (SMART) for pancreatic cancer. Radiother Oncol 2017;125:439-44. [Crossref] [PubMed]
- Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N. A deep metric for multimodal registration. 19th International Conference on Medical Image Computing and Computer-Assisted Intervention, 2016:10-8.
- Woods RP, Mazziotta JC, Cherry SR. MRI-PET registration with automated algorithm. J Comput Assist Tomogr 1993;17:536-46. [Crossref] [PubMed]
- Al-Saleh MA, Alsufyani NA, Saltaji H, Jaremko JL, Major PW. MRI and CBCT image registration of temporomandibular joint: a systematic review. J Otolaryngol Head Neck Surg 2016;45:30. [Crossref] [PubMed]
- Morrow NV, Lawton CA, Qi XS, Li XA. Impact of computed tomography image quality on image-guided radiation therapy based on soft tissue registration. Int J Radiat Oncol Biol Phys 2012;82:e733-8. [Crossref] [PubMed]
- Zachiu C, De Senneville BD, Moonen CT, Raaymakers BW, Ries M. Anatomically plausible models and quality assurance criteria for online mono-and multi-modal medical image registration. Phys Med Biol 2018;63:155016. [Crossref] [PubMed]
- Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Med Phys 2011;38:2708-14. [Crossref] [PubMed]
- Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, Shen D. Alzheimer's Disease Neuroimaging Initiative. Estimating CT Image From MRI Data Using Structured Random Forest and Auto-Context Model. IEEE Trans Med Imaging 2016;35:174-83. [Crossref] [PubMed]
- Hsu SH, Cao Y, Huang K, Feng M, Balter JM. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Phys Med Biol 2013;58:8419. [Crossref] [PubMed]
- Zaidi H, Montandon ML, Slosman DO. Magnetic resonance imaging‐guided attenuation and scatter corrections in three‐dimensional brain positron emission tomography. Med Phys 2003;30:937-48. [Crossref] [PubMed]
- Berker Y, Franke J, Salomon A, Palmowski M, Donker HC, Temur Y, Mottaghy FM, Kuhl C, Izquierdo-Garcia D, Fayad ZA, Kiessling F, Schulz V. MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/Dixon MRI sequence. J Nucl Med 2012;53:796-804. [Crossref] [PubMed]
- Kops ER, Herzog H. Alternative methods for attenuation correction for PET images in MR-PET scanners. IEEE Nucl Sci Symp Conf Rec 2007;6:4327-30.
- Hofmann M, Steinke F, Scheel V, Charpiat G, Farquhar J, Aschoff P, Brady M, Schölkopf B, Pichler BJ. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. J Nucl Med 2008;49:1875-83. [Crossref] [PubMed]
- Han X. MR‐based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408-19. [Crossref] [PubMed]
- Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, Shen D. Estimating CT image from MRI data using structured random forest and auto-context model. IEEE Trans Med Imaging 2016;35:174-83. [Crossref] [PubMed]
- Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT Image from MRI Data Using 3D Fully Convolutional Networks. In: Carneiro G, Mateus D, Peter L, Bradley A, Tavares JMRS, Belagiannis V, Papa JP, Nascimento JC, Loog M, Lu Z, Cardoso JS, Cornebise J. editors. Deep Learning and Data Labeling for Medical Applications. DLMIA 2016, LABELS 2016. Lecture Notes in Computer Science, vol 10008. Springer, Cham, 2016: 170-8.
- Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, Shen D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins D, Duchesne S. editors. Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science, vol 10435. Springer, Cham, 2017:417-25.
- Roy S, Butman JA, Pham DL. Synthesizing CT from Ultrashort Echo-Time MR Images via Convolutional Neural Networks. Simulation and Synthesis in Medical Imaging: Second International Workshop, SASHIMI 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 10, 2017:24-32.
- Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT Synthesis Using Unpaired Data. In: Tsaftaris S, Gooya A, Frangi A, Prince J. editors. Simulation and Synthesis in Medical Imaging. SASHIMI 2017. Lecture Notes in Computer Science, vol 10557. Springer, Cham, 2017:14-23.
- Xiang L, Wang Q, Nie D, Zhang L, Jin X, Qiao Y, Shen D. Deep embedding convolutional neural network for synthesizing CT image from T1-Weighted MR image. Med Image Anal 2018;47:31-44. [Crossref] [PubMed]
- Zhao C, Carass A, Lee J, Jog A, Prince JL. A supervoxel based random forest synthesis framework for bidirectional MR/CT synthesis. Simul Synth Med Imaging 2017;10557:33-40. [Crossref] [PubMed]
- Cao X, Yang J, Gao Y, Wang Q, Shen D. Region-adaptive deformable registration of CT/MRI pelvic images via learning-based image synthesis. IEEE Trans Image Process 2018;27:3500-12. [Crossref] [PubMed]
- Chartsias A, Joyce T, Dharmakumar R, Tsaftaris SA. Adversarial Image Synthesis for Unpaired Multi-modal Cardiac Data. In: Tsaftaris S, Gooya A, Frangi A, Prince J. editors. Simulation and Synthesis in Medical Imaging. SASHIMI 2017. Lecture Notes in Computer Science, vol 10557. Springer, Cham, 2017:3-13.
- Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. Proceedings of the IEEE International Conference on Computer Vision 2017:2223-32.
- Jin CB, Jung W, Joo S, Park E, Saem AY, Han IH, Lee JI, Cui X. Deep CT to MR Synthesis using Paired and Unpaired Data. Sensors 2019;19:2361. [Crossref] [PubMed]
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017:1125-34.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention 2015:234-41.
- Lin G, Milan A, Shen C, Reid I. Refinenet: Multi-path refinement networks for high-resolution semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017:1925-34.
- Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y. The one hundred layers tiramisu: Fully convolutional densenets for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2017:11-9.
- Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:2481-95. [Crossref] [PubMed]
- Milletari F, Navab N, Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. Fourth International Conference on 3D Vision (3DV) 2016:565-71.
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention 2016:424-32.
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015.
- Xu B, Wang N, Chen T, Li M. Empirical evaluation of rectified activations in convolutional network. arXiv preprint arXiv:1505.00853, 2015.
- Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434, 2015.
- Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th International Conference on Machine Learning (ICML-10) 2010:807-14.
- Reed S, Akata Z, Yan X, Logeswaran L, Schiele B, Lee H. Generative adversarial text to image synthesis. arXiv preprint arXiv:1605.05396, 2016.
- Denton E, Chintala S, Szlam A, Fergus R. Deep generative image models using a laplacian pyramid of adversarial networks. NIPS'15: Proceedings of the 28th International Conference on Neural Information Processing Systems - Volume 1, December 2015:1486-94.
- Mirza M, Osindero S. Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784, 2014.
- Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. NIPS'14: Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2, December 2014:2672-80.
- Gauthier J. Conditional generative adversarial nets for convolutional face generation. Class Project for Stanford CS231N: Convolutional Neural Networks for Visual Recognition Winter Semester 2014;2014:2.
- Li C, Wand M. Precomputed real-time texture synthesis with markovian generative adversarial networks. Comput Vis ECCV 2016:702-16.
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- Zhao H, Gallo O, Frosio I, Kautz J. Loss functions for image restoration with neural networks. IEEE Trans Comput Imaging 2016;3:47-57. [Crossref]
- Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979;9:62-6. [Crossref]
- Prashanth HS, Shashidhara HL, KN BM. Image Scaling Comparison Using Universal Image Quality Index. International Conference on Advances in Computing, Control, and Telecommunication Technologies 2009: 859-63.
- Ben-Cohen A, Klang E, Raskin SP, Soffer S, Ben-Haim S, Konen E, Amitai MM, Greenspan H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng Appl Artif Intell 2019;78:186-94. [Crossref]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Bi L, Kim J, Kumar A, Feng D, Fulham M. Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs). In: Cardoso MJ, Arbel T, Gao F, Kainz B, van Walsum T, Shi K, Bhatia KK, Peter R, Vercauteren T, Reyes M, Dalca A, Wiest R, Niessen W, Emmer BJ. editors. Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment. RAMBO 2017, CMMI 2017, SWITCH 2017. Lecture Notes in Computer Science, vol 10555. Springer, Cham, 2017:43-51.
- Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13:600-12. [Crossref] [PubMed]


