
Cartilage morphometry and magnetic susceptibility measurement for knee osteoarthritis with automatic cartilage segmentation
Introduction
Knee osteoarthritis (KOA) is the most common degenerative joint disorder leading to chronic pain and disability (1,2). Although KOA involves the whole joint, cartilage degeneration was deemed as KOA’s hallmark (3). Thus, early detection and accurate quantification of cartilage morphological and compositional changes are necessary to appropriately treat KOA. Magnetic resonance imaging (MRI) is a non-invasive method that can visualize cartilage directly and help diagnose articular cartilage disorders (4). MRI-based morphologic quantitative parameters (thickness and volume) and compositional data (e.g., relaxometry) are considered as important image biomarkers for the diagnosis and tracking of OA progression (5-7). However, this promising technique is not easy for clinical translation due to: (I) relatively long image acquisition time, and (II) time-consuming image post-processing, especially in cartilage segmentation, which is often performed manually or semi-automatically.
Quantitative susceptibility mapping (QSM) is a modern MR imaging technique that estimates the voxel-wise spatial distribution of magnetic susceptibility based on gradient recalled echo (GRE) sequence. QSM’s high sensitivity to dominant magnetic susceptibility sources in tissue (e.g., deoxyhemoglobin, iron deposit, proteins, the orientation of collagen fiber) has been shown and used in studies of knee cartilage (8-11). According to previous studies, the susceptibility value diversity in cartilage was attributed to different alignment angles of collagen relative to the B0 field. Nykänen et al. (10) showed that QSM was sensitive to the integrity of the collagen network based on bovine cartilage specimens. Wei et al. (9) demonstrated that the arrangement of collagen fibrils is the dominant source of magnetic susceptibility anisotropy, and QSM is sensitive to collagen network damage or degeneration based on knee OA patients. In contrast to another quantitative MR sequence (T2 relaxation, T1ρ, 23Na, etc.), QSM does not need to add extra image acquisition time or special sequence. The acquired cartilage GRE images could be separated into magnitude and phase images. The magnitude images were used for cartilage segmentation and morphologic measurement, whereas the raw phase images were used to obtain the susceptibility via image post-processing. Therefore, QSM is expected to be a promising clinical quantitative MR technique to simultaneously assess cartilage anatomical parameters and cartilage biochemical composition.
Although manual cartilage segmentation is currently considered the gold standard, it is laborious and time-consuming and the results obtained are subject to analyst bias and error. In recent years, deep learning has become a powerful approach and primary option in medical image segmentation (12). Unlike traditional machine learning techniques, deep learning depends on the concept of building the neural network, which is inspired by the biological neural network, comprised of a large number of identical and linked simple computational units to achieve high performance on complex tasks with powerful electronic computers. Currently, Many network structures are explored and applied in deep learning, including convolutional neural networks (CNNs) (13), fully convolutional networks (FCN) (14), recurrent neural networks (RNNs) (15), and U-Net (16,17). nnU-Net (no-new-U-Net), which is a UNet-based segmentation model, can automatically configure itself without tuning any hyperparameter. Currently, nnU-Net has achieved better performance than highly specialized solutions in the 23 public datasets used in international biomedical segmentation competitions (18). However, nnU-Net has seldom been applied to knee cartilage segmentation based on MR images. Thus, we hypothesized that nnU-Net model could automatically segment knee cartilage and quantify cartilage parameters based on three-dimensional water-selective (3D_WATS) MR cartilage images.
The two main goals of this study were as follow: (I) to develop an automatic cartilage segmentation method based on the nnU-Net model for 3D_WATS MR cartilage images; and (II) to obtain the cartilage parameters representing its morphometry and magnetic susceptibility information, such as cartilage thickness, volume, and susceptibility values, and evaluate them for KOA severity analysis. We present the following article in accordance with the STARD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1245/rc).
Methods
Subjects
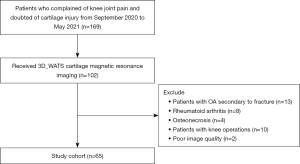
Participating subjects were individuals who had undergone health checks at our hospital from September 2020 to May 2021 and volunteered to participate in this study. All participants were recruited by one orthopedic surgeon with 10 years of experience (LW). Those who had OA secondary to fracture, rheumatoid, arthritis, and osteonecrosis or had knee operations were excluded. Flow diagram of the selecting process has been illustrated in Figure 1. A total of 65 consecutive human subjects (52.2±15.1 years, 34 males) were enrolled in this study. Identification of OA was based on an anterior-posterior weight bearing knee radiograph. The severity of OA was assessed using the X-ray based Kellgren-Lawrence (KL) scale (19). According to the KL grade and clinical symptoms, subjects were divided into three groups: normal control (n=20, KL0), mild OA (n=20, KL1-2), and severe OA (n=25, KL3-4). The heights and weights of the participants were recorded at the time of examination. All participants completed the standardized Western Ontario and McMaster Universities Arthritis Index questionnaire for pain, stiffness, and functional impairment. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The protocol was approved by the institutional review board of Shanghai Ninth People’s Hospital (No. SH9H-2020-T395-2) and written informed consent was obtained from all participants.
MRI examinations
The whole knee joint (35 right, 30 left) of all participants were scanned using a 3T MRI scanner (Achieva 3.0TX; Philips Healthcare, Best, Netherlands) with an eight-channel knee coil (Philips Healthcare). The knee flexion angle was adjusted to 20–30°, and an immobilization sponge was used to increase participant comfort and reduce motion artifacts. The sagittal fat-suppressed 3D_WATS sequence [repetition time (TR)/echo time (TE) = 20/6.0, field-of-view (FOV) =14 cm, matrix =640×640, flip angle =30°, pixel size =0.35 mm × 0.35 mm, slice thickness =1.5 mm, SENSE =2, scan time =5 min 40 s] was used for imaging cartilage. The acquired images were separated into magnitude and phase images. The raw magnitude images were used for cartilage segmentation and morphologic measurement, while the raw phase images were used for QSM via image post-processing.
Automatic cartilage segmentation
Sixty-five patients’ MR magnitude images with a size of 528×528×120 were obtained. The 2D nnU-Net model was used to perform automatic cartilage segmentation. Details of the 2D nnU-Net model architecture were discussed in (20), and the segmentation model was constructed as follows: first, 21 subjects were randomly selected for manual cartilage segmentation by a radiologist (CLL, 12 years of experience), and the cartilage masks were input into the nnU-Net as ground truth for training. Second, the 3D sagittal MR magnitude images as well as the corresponding cartilage masks were broken into 2D images slice by slice and then used for training the 2D nnU-Net model. After that, the remaining subjects’ MR magnitude images were run in the well-trained U-Net model as the reference, and corresponding cartilage masks were obtained automatically. Five compartments of cartilage regions were labeled using the method of 3D connected component analysis based on Python (21) as ROIs: lateral femoral condyle (LFC), medial femoral condyle (MFC), lateral tibial plateau (LT), medial tibia plateau (MT), and patella (P).
Manual cartilage segmentation
To assess the inter-user viability of manual cartilage segmentation, 17 subjects (KL0=5, KL1–2=5, KL3–4=7) were randomly selected from all subjects to be manually segmented again by two radiologist (JLG, 5 years of experience and CLL, 12 years of experience). The operator delineated the cartilage margin of each of the five compartments of the joint slice by slice. A case of cartilage segmentation required approximately 120 slices of image processing. The manual segmentation was implemented using ITK-SNAP, which is a free open-source software tool (www.itk-snap.org). Intra-class correlation coefficient (ICC) was used to assess the consistency of quantitative cartilage parameters between the two manual segmentations obtained by two radiologists.
Cartilage morphological measurement
Cartilage thickness was determined for five distinct compartments using the Knee Segmentation and Registration Toolkit (KSRT) (https://bitbucket.org/marcniethammer/ksrt/wiki/Home), which draws a medial line in each compartment of the cartilage on the input 2D mask images and determining the cartilage thickness by calculating the minimal distance from each point on the medial line to the cartilage boundary (22). The corresponding thickness image was obtained automatically based on the cartilage mask images. The average thickness of each cartilage was determined by averaging all the non-zero pixels’ values of the thickness image. The cartilage volume of each compartment was calculated by multiplying the total number of voxels encompassing the cartilage by the volume of each voxel (23).
QSM reconstruction
The raw phase images derived from cartilage imaging were used for QSM. Firstly, phase images were unwrapped using a Laplacian operator (24). The unwrapped phase was then input to the projection onto the dipole field (PDF) algorithm (25) to remove the background phase and estimate the tissue phase data (the effect can be checked in Figure S1). Finally, a field-to-source inversion algorithm (STAR-QSM) (26) was applied to the tissue phase image to calculate the quantitative susceptibility map. During the background phase removal and susceptibility calculation, a binary mask was used to define the region of interest by thresholding the magnitude image. The mean value and standard deviation (SD) of susceptibility values derived from the QSM using the STISuite V3.0 (https://people.eecs.berkeley.edu/~chunlei.liu/software.html) for each cartilage compartment (27).
Statistical analysis
All statistical processing was performed using the software SPSS27 (IBM, Armonk, NY, USA). The normality of distributions for continuous variables was tested using the Kolmogorov-Smirnov (KS) test. One-way analysis of variance (ANOVA) and the post–hoc Fisher’s least significant difference (LSD) test were used to compare the difference in age, body mass index (BMI), and cartilage parameters among three groups. Categorical data were compared using the chi-square test or Fisher’s exact test. Dice scores were calculated to evaluate the consistency between manual and automatic cartilage segmentation. Pearson correlation coefficient and ICC were used to assess the consistency of quantitative cartilage parameters between the automatic and manual segmentation methods, and between two manual segmentations obtained by two radiologists. For cartilage parameters, ANOVA, with taking the significant demographic characteristics as covariates were performed to assess the differences in cartilage thickness, cartilage volume, and susceptibility values between the three groups. Support vector machine (SVM) was used to further verify the classification validity of automatically extracted cartilage parameters. Five-fold cross-validation and a grid search method (28) were used to determine the best SVM hyperparameters including the kernel, and values of C and gamma. P<0.05 denoted statistical significance.
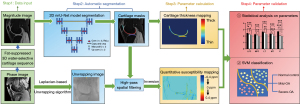
The workflow of image processing and parameters analysis is illustrated in Figure 2.
Results
Subject characteristics
The summary of the baseline characteristics of all subjects was shown in Table 1. The mild-OAs and severe-OAs had significantly higher age (P<0.001), whereas the gender, weight, height, and BMI were not significantly different between groups (P>0.05).
Table 1
| Variables | Total (n=65) | Normal (n=20) | Mild-OAs (n=20) | Severe-OAs (n=25) | P value |
|---|---|---|---|---|---|
| Age (years) | 52.2±15.1 | 37.8±11.9 | 55.1±11.9 | 61.5±10.7 | <0.001 |
| Gender | 0.11 | ||||
| Male | 34 (52.3) | 13 (65.0) | 12 (60.0) | 9 (36.0) | |
| Female | 31 (47.7) | 7 (35.0) | 8 (40.0) | 16 (64.0) | |
| Weight (kg) | 68.9±8.7 | 67.9±7.6 | 70.5±8.9 | 68.4±9.5 | 0.61 |
| Height (cm) | 167.7±7.7 | 168.9±5.2 | 168.8±6.7 | 165.9±9.8 | 0.84 |
| BMI (kg/m2) | 24.4±2.4 | 23.7±1.7 | 24.6±2.4 | 24.8±2.8 | 0.31 |
| Knee | 0.21 | ||||
| Left knee | 30 (46.2) | 6 (30.0) | 11 (55.0) | 13 (52.0) | |
| Right knee | 35 (53.8) | 14 (70.0) | 9 (45.0) | 12 (48.0) |
Continuous variables are expressed as the mean ± standard deviation. Categorical variables are presented as N (%). OA, osteoarthritis; BMI, body mass index.
Segmentation performance
As shown in Figure 3, nnU-Net provided visually accurate and acceptable cartilage segmentation, which was similar to the manual segmentation provided by the radiologist. Strong inter-observer agreements were shown in Table 2. The ICC of two radiologists ranged from 0.91–0.99 (95% CI: 0.86–0.99).
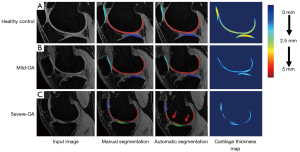
Table 2
| Compartments | Radiologist 1 | Radiologist 2 | ICC value |
|---|---|---|---|
| Average thickness (mm) | |||
| Femur | |||
| MFC | 2.30 | 2.30 | 0.99 |
| LFC | 2.11 | 2.13 | 0.99 |
| Tibia | |||
| MT | 2.48 | 2.61 | 0.91 |
| LT | 2.20 | 2.34 | 0.92 |
| Patella | 2.21 | 2.27 | 0.95 |
| Average volume (mm3) | |||
| Femur | |||
| MFC | 4486 | 4472 | 0.99 |
| LFC | 3654 | 3645 | 0.98 |
| Tibia | |||
| MT | 1750 | 1744 | 0.99 |
| LT | 1515 | 1521 | 0.98 |
| Patella | 1901 | 1910 | 0.98 |
| Average QSM mean (ppm) | |||
| Femur | |||
| MFC | −0.04 | −0.04 | 0.99 |
| LFC | −0.03 | −0.03 | 0.99 |
| Femur | |||
| MT | −0.03 | −0.03 | 0.99 |
| LT | −0.02 | −0.02 | 0.99 |
| Patella | −0.01 | −0.01 | 0.99 |
| Average QSM SD (ppm) | |||
| Femur | |||
| MFC | 0.11 | 0.11 | 0.99 |
| LFC | 0.09 | 0.09 | 0.99 |
| Femur | |||
| MT | 0.08 | 0.08 | 0.99 |
| LT | 0.09 | 0.09 | 0.99 |
| Patella | 0.08 | 0.07 | 0.98 |
QSM, quantitative susceptibility mapping; ICC, intra-class correlation coefficient; MFC, medial femoral condyle; LFC, lateral femoral condyle; MT, medial tibial plateau; LT, lateral tibial plateau; SD, standard deviation.
Dice scores for the whole cartilage and five distinct compartments were shown in Table 3. Mean dice scores for manual and automatic segmentation ranged from 0.90–0.97 for five distinct compartments and the average was 0.93 for the whole cartilage. Specifically, our segmentation model based on nnU-Net achieved mean dice scores for all severe-OAs of 0.95 (LFC), 0.92 (MFC), 0.85 (LT), 0.90 (MT), and 0.85 (P); with the average value 0.89. In addition, mean dice values between two radiologists for all subjects ranged from 0.95–0.97 and for severe-OAs ranged from 0.92 to 0.96.
Table 3
| Group | LFC | MFC | LT | MT | P | Average | |
|---|---|---|---|---|---|---|---|
| Rad 1 vs. Auto | Normal control group | 0.98 | 0.97 | 0.96 | 0.97 | 0.94 | 0.96 |
| Mild-OA group | 0.93 | 0.92 | 0.94 | 0.96 | 0.95 | 0.94 | |
| Severe-OA group | 0.95 | 0.92 | 0.85 | 0.90 | 0.85 | 0.89 | |
| All subjects’ | 0.97 | 0.96 | 0.90 | 0.94 | 0.90 | 0.93 | |
| Rad 1 vs. Rad 2 | Normal control group | 0.97 | 0.97 | 0.97 | 0.95 | 0.96 | 0.96 |
| Mild-OA group | 0.95 | 0.94 | 0.98 | 0.96 | 0.93 | 0.95 | |
| Severe-OA group | 0.96 | 0.94 | 0.93 | 0.95 | 0.92 | 0.94 | |
| All subjects’ | 0.97 | 0.97 | 0.96 | 0.96 | 0.95 | 0.96 |
All subjects involve the healthy subjects, mild-OAs and severe-OAs. The Dice score value ranged between 0 and 1 with a value of 1 indicating a perfect segmentation and a value of 0 indicating no overlap at all. LFC, lateral femoral condyle; MFC, medial femoral condyle; LT, lateral tibial plateau; MT, medial tibial plateau; P, Patella; Rad, Radiologist; Auto, Automatic method using nnU-Net; OA, osteoarthritis.
Quantitative cartilage parameters
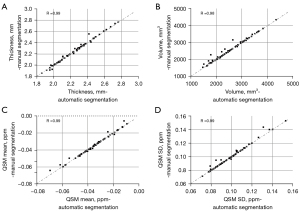
The mean and SD of cartilage thickness, cartilage volume, and QSM parameters on each compartment for the three groups from automatic segmentation are summarized in Table 4. Figure 4 shows the scatter plots for these parameters between automatic and manual segmentations, with corresponding Pearson correlation coefficients ranging from 0.98 to 0.99 (95% CI: 0.89–1.00).
Table 4
| Compartment | Parameters | Normal controls (N) (n=20) | Mild-OA (M) (n=20) | Severe-OA (S) (n=25) | Raw P value | Adjusted P value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N-M | N-S | M-S | N-M | N-S | M-S | ||||||
| LFC | Thickness (mm) | 2.41±0.20 | 2.33±0.15 | 2.15±0.16 | 0.475 | <0.001*** | 0.006** | 0.674 | 0.003** | 0.013* | |
| Volume (mm3) | 5,103±1130 | 4,521±802 | 3,925±923 | 0.225 | 0.001** | 0.185 | 0.450 | 0.041* | 0.346 | ||
| QSM mean (ppm) | −0.04±0.01 | −0.05±0.01 | −0.05±0.01 | 0.735 | 0.112 | 0.949 | 0.694 | 0.332 | 0.745 | ||
| QSM SD (ppm) | 0.11±0.01 | 0.11±0.02 | 0.127±0.03 | 1.000 | 0.337 | 0.873 | >0.99 | 0.539 | 0.783 | ||
| MFC | Thickness (mm) | 2.23±0.17 | 2.17±0.13 | 1.93±0.14 | 0.714 | <0.001*** | <0.001*** | 0.714 | 0.001** | 0.022* | |
| Volume (mm3) | 4,141±817 | 4,010±628 | 3,004±673 | 1.000 | <0.001*** | <0.001*** | >0.99 | 0.009** | 0.045* | ||
| QSM mean (ppm) | −0.04±0.01 | −0.05±0.01 | −0.07±0.03 | 0.198 | 0.033* | 0.140 | 0.410 | 0.089 | 0.470 | ||
| QSM SD (ppm) | 0.10±0.01 | 0.10±0.01 | 0.13±0.04 | 0.964 | 0.059 | 0.094 | 0.901 | 0.182 | 0.320 | ||
| LT | Thickness (mm) | 2.64±0.23 | 2.54±0.21 | 2.36±0.23 | 0.597 | 0.001** | 0.051 | 0.872 | 0.015* | 0.163 | |
| Volume (mm3) | 1,977±488 | 1,799±299 | 1,521±375 | 0.545 | 0.002** | 0.103 | 0.673 | 0.048* | 0.402 | ||
| QSM mean (ppm) | −0.03±0.01 | −0.03±0.01 | −0.03±0.01 | 0.759 | >0.99 | 0.781 | 0.801 | >0.99 | 0.671 | ||
| QSM SD (ppm) | 0.08±0.01 | 0.08±0.01 | 0.09±0.01 | 0.922 | 0.404 | 0.582 | 0.712 | 0.602 | 0.781 | ||
| MT | Thickness (mm) | 2.36±0.17 | 2.33±0.16 | 2.03±0.21 | 1.000 | <0.001*** | <0.001*** | >0.99 | 0.029* | 0.039* | |
| Volume (mm3) | 1,786±334 | 1,761±356 | 1,257±449 | 1.000 | <0.001*** | 0.001** | >0.99 | 0.006** | 0.013* | ||
| QSM mean (ppm) | −0.02±0.01 | −0.03±0.01 | −0.04±0.03 | 1.000 | 0.038* | 0.055 | >0.99 | 0.130 | 0.028 | ||
| QSM SD (ppm) | 0.09±0.01 | 0.09±0.01 | 0.01±0.02 | 0.999 | 0.274 | 0.249 | >0.99 | 0.583 | 0.491 | ||
| P | Thickness (mm) | 2.75±0.29 | 2.10±0.37 | 1.78±0.40 | 0.000*** | <0.001*** | 0.025* | 0.015* | 0.003** | 0.192 | |
| Volume (mm3) | 2,468±608 | 1,713±569 | 1,205±626 | 0.001** | <0.001*** | 0.036* | 0.041* | 0.009** | 0.083 | ||
| QSM mean (ppm) | −0.03±0.01 | −0.03±0.02 | −0.039±0.07 | 1.000 | 0.938 | 0.941 | >0.99 | 0.993 | 0.910 | ||
| QSM SD (ppm) | 0.06±0.01 | 0.08±0.02 | 0.09±0.03 | 0.116 | 0.002** | 0.310 | 0.311 | 0.008** | 0.493 | ||
Data are shown as mean ± SD. P values below 0.05 were considered statistically significant. *, P<0.05; **, P<0.01; ***, P<0.001. SD, standard deviation; ANOVA, one-way analysis of variance; OA, osteoarthritis; LFC, lateral femoral condyle; MFC, medial femoral condyle; LT, lateral tibial plateau; MT, medial tibial plateau; P, patella; QSM, quantitative susceptibility mapping.
Performance in OA evaluation
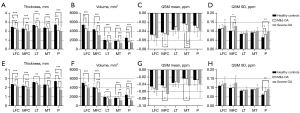
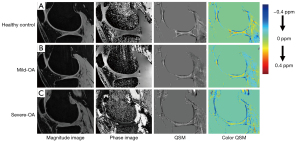
Comparisons between the three groups in terms of cartilage thickness, cartilage volume, and QSM parameters are illustrated in Figure 5. Highly significant decreases in cartilage thickness as well as in cartilage volume were observed as the OA severity increased (P<0.05). Few significant differences were found in the mean and SD of susceptibility value, with an increasing trend respectively [overall average QSM mean (ppm): −0.03±0.01, −0.03±0.01, −0.04±0.02 for normal controls, mild-OAs, and severe-OAs; overall average QSM SD (ppm): 0.09±0.01, 0.09±0.01, 0.11±0.02 for normal controls, mild-OAs, and severe-OAs]. Examples of magnitude image, phase image and QSM mapping for each OA class are illustrated in Figure 6.
Prediction performance evaluation
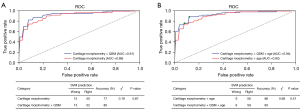
To further verify the validity of automatically extracted cartilage parameters, multi-class SVM was used to construct the predictive model for the classification of three OA classes. After evaluating the diagnostic performance of SVM models with the five-fold cross-validation, the SVM hyperparameters were set as kernel = ‘RBF’, C = 1 and gamma = 0.1. The classification results were shown in Figure 7. The cut-off points of the optimal prediction model have been determined by maximizing the Youden index and circled (green) in the Figure 7B. The constructed model based on cartilage morphological parameters (thickness and volume) obtained AUC was 0.89 (95% CI: 0.85–0.92), whereas combined with QSM a relatively higher AUC score can be achieved, with a value of 0.91 (95% CI: 0.87–0.93). Taking age as covariate, the constructed model achieved AUC 0.92 (95% CI: 0.88–0.94) and 0.94 (95% CI: 0.89–0.96), correspondingly.
Discussion
In this study, we performed a fully automated cartilage segmentation and quantification method based on the nnU-Net model and extracted quantitative parameters including cartilage morphometry and QSM at 3T. The results showed that nnU-Net model provided high mean dice scores (>0.90) for cartilage segmentation and obtained high Pearson correlation coefficients for cartilage thickness, cartilage volume, and susceptibility values compared with manual segmentation; indicating that this model was able to segment cartilage accurately. Significant decreases in cartilage thickness and volume, as well as increases in mean and SD of susceptibility values, were observed in mild OA and severe OA compared with normal control. Additionally, The model achieved higher classification validity by integrating QSM. These results suggest the potential of QSM as an important complementary image biomarker for the comprehensive evaluation of OA in clinical practice.
The performance of nnU-Net model for cartilage segmentation was evaluated using manually generated labels by the experienced radiologist as ground truth. Two radiologists segmented the knee cartilage independently and we obtained high ICC values between the two manual segmentations, ranging from 0.91 to 0.99, and high Dice scores with an average value of 0.96. This indicates the high accuracy and consistency of manual segmentation. Dice scores were calculated to assess the accuracy of automatic cartilage segmentation. Our results showed that nnU-Net model provided high mean dice scores (>0.90) for cartilage segmentation in different knee compartments. In normal and mild OA, our automatic segmentation method has achieved dice scores of 0.96 and 0.93, respectively. Xue et al. (29) designed a transfer learning-based U-Net CNN model and performed automatic segmentation on knee cartilage. The average dice score was 0.82±0.08. Zhou et al. (30) used a segmentation pipeline based on a semantic segmentation CNN and 3D fully connected CRF, and achieved dice coefficients between 0.8 and 0.9 including the femoral cartilage (0.806±0.062), tibial cartilage (0.801±0.052), and patellar cartilage (0.807±0.101). In the 2019 International Workshop on Osteoarthritis Imaging (IWOAI) knee segmentation challenge (31), six networks from different teams were evaluated for their segmentation performance on a dataset consisting of 3D knee MRI from 88 subjects. The best Dice scores among these networks ranged from 0.81 to 0.90, for femoral cartilage (using 2D DeeplabV3+ Densenet, Dice =0.90±0.02), tibial cartilage (using 2D multi-planar U-Net, Dice =0.89±0.03) and patellar cartilage (using 3D V-Net, Dice =0.86±0.07). In comparison to these methods, our method for automatic knee cartilage segmentation achieved better dice scores indicating higher accuracy. In severe-OAs, however, our method’s dice scores were lower with average values of 0.89, and 0.85, 0.90, and 0.85 in LT, MT, and P respectively. This may be attributed to cartilage thinness or local loss due to cartilage attrition in these areas with a high incidence of lesions. Even though degenerative cartilage compartments segmentation in some of the sever-OAs was relatively inaccurate, all dice scores were higher than 0.85 and the segmentation result did not have a significant impact on subsequent parameter calculations.
In our study, cartilage thickness and cartilage volume significantly decrease in all knee compartments compared with normal controls, especially in the medial knee joint, which may be attributed to the prevalence of varus knee. More loading forces concentrated in the medial joint lead to medial cartilage degeneration. This result was consistent with previous research (5,32). In terms of QSM, significant decreases in mean susceptibility values were found in MFC and MT compared with normal controls, and significant increases in SD of susceptibility values were found within MFC and P in OA patients, respectively. Variation of QSM was attributed to the loss of organization in the collagen fibrils, cartilage attrition, or degeneration that affects QSM contrast (33). In addition, we calculated simple global mean values in different knee compartments, which may lead to collagen fibril heterogeneity due to different degrees of cartilage attraction. In the future, the sub-regions of the knee joint compartment or different cartilage layers should be assessed to ensure QSM accuracy. Few studies were published for the evaluation of OA using QSM. Wei et al. (9,33) have demonstrated that magnetic susceptibility has the potential to demonstrate a visible contrast change of degenerative cartilage in patients with OA and may provide information about the change in collagen that may exist before structural changes in cartilage thickness. Our results indicated that the increasing variance of magnetic susceptibility value is proportional to the thinning of cartilage. This means that quantitative susceptibility mapping is a useful biomarker for the evaluation of OA progression.
To further verify the classification validity of automatically extracted cartilage parameters, a linear SVM model was used to construct the predictive model for OA classification. Our results showed that automatically extracted cartilage parameters using nnU-Net model had high validity for the OA classification. The constructed model based on cartilage morphological parameters (thickness and volume) and age obtained AUC was 0.92 (95% CI: 0.88–0.94); whereas, by integrating QSM, we achieved a relatively higher AUC with a value of 0.94 (95% CI: 0.89–0.96). These results suggest that QSM has the potential for clinical translation for the comprehensive evaluation of OA.
While our initial results are promising, several potential limitations exist in our study. First, the single center and small sample size may result in biased conclusions. Further validation in a larger sample size is needed. Second, we used the 2D nnU-Net, instead of 3D nnU-Net, to train the decomposing labeled slices from the 3D MR images. Although the nnU-Net model for cartilage segmentation results was successful, there is still a need to investigate the optimal sample size to train the network for robust segmentation, especially in severe OA. Third, there was a lack of real ground truth. We used manual segmentation as the reference standard which may change because of user variability. Fourth, the cartilage MR imaging was not isotropic. Thus, the relatively poor slice resolution, and the curved cartilage surface may result in partial volume artifacts; affecting cartilage segmentation and quantitative cartilage parameters. Finally, we only calculated a simple global mean value of cartilage parameters in different knee compartments. In the future, the sub-regions of the knee joint compartment or different cartilage layers should be assessed to ensure QSM accuracy.
Conclusions
The proposed cartilage segmentation and quantification method for 3D_WATS cartilage images based on nnU-Net can produce reliable segmentation of five major compartments in cartilage regions. The automatic extraction of QSM and cartilage morphometry parameters showed high validity for OA classification. This suggests that 3D_WATS cartilage imaging has the potential for clinical translation for the comprehensive evaluation of OA.
Acknowledgments
We are really grateful to Ahmed Aburas for his help in the language polishing.
Funding: This study was sponsored by the Interdisciplinary Program of Shanghai Jiao Tong University (No.YG2019QNA17) and Shanghai Municipal Commission of Health and Family Planning. They have no role in study design, data collection, analysis, interpretation or in writing the manuscript.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-1245/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1245/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fang L, Xia C, Xu H, Ge Q, Shi Z, Kong L, Zhang P, Xu R, Zou Z, Wang P, Jin H, Tong P. Defining disease progression in Chinese mainland people: Association between bone mineral density and knee osteoarthritis. J Orthop Translat 2020;26:39-44. [Crossref] [PubMed]
- Wen C, Xiao G. Advances in osteoarthritis research in 2021 and beyond. J Orthop Translat 2022;32:A1-2. [Crossref] [PubMed]
- Yusup A, Kaneko H, Liu L, Ning L, Sadatsuki R, Hada S, Kamagata K, Kinoshita M, Futami I, Shimura Y, Tsuchiya M, Saita Y, Takazawa Y, Ikeda H, Aoki S, Kaneko K, Ishijima M. Bone marrow lesions, subchondral bone cysts and subchondral bone attrition are associated with histological synovitis in patients with end-stage knee osteoarthritis: a cross-sectional study. Osteoarthritis Cartilage 2015;23:1858-64. [Crossref] [PubMed]
- Wang YJ, Wang J, Deng M, Liu G, Qin L. In vivo three-dimensional magnetic resonance imaging of rat knee osteoarthritis model induced using meniscal transection. J Orthop Translat 2015;3:134-41. [Crossref] [PubMed]
- Peterfy CG, Gold G, Eckstein F, Cicuttini F, Dardzinski B, Stevens R. MRI protocols for whole-organ assessment of the knee in osteoarthritis. Osteoarthritis Cartilage 2006;14 Suppl A:A95-111.
- Zhou F, Han X, Wang L, Zhang W, Cui J, He Z, Xie K, Jiang X, Du J, Ai S, Sun Q, Wu H, Yu Z, Yan M. Associations of osteoclastogenesis and nerve growth in subchondral bone marrow lesions with clinical symptoms in knee osteoarthritis. J Orthop Translat 2021;32:69-76. [Crossref] [PubMed]
- Yu K, Ying J, Zhao T, Lei L, Zhong L, Hu J, Zhou JW, Huang C, Zhang X. Prediction model for knee osteoarthritis using magnetic resonance-based radiomic features from the infrapatellar fat pad: data from the osteoarthritis initiative. Quant Imaging Med Surg 2023;13:352-69. [Crossref] [PubMed]
- Yuen J, Hung J, Wiggermann V, Robinson SD, McCormack R, d'Entremont AG, Rauscher A. Multi-echo GRE imaging of knee cartilage. J Magn Reson Imaging 2017;45:1502-13. [Crossref] [PubMed]
- Wei H, Dibb R, Decker K, Wang N, Zhang Y, Zong X, Lin W, Nissman DB, Liu C. Investigating magnetic susceptibility of human knee joint at 7 Tesla. Magn Reson Med 2017;78:1933-43. [Crossref] [PubMed]
- Nykänen O, Sarin JK, Ketola JH, Leskinen H, Te Moller NCR, Tiitu V, Mancini IAD, Visser J, Brommer H, van Weeren PR, Malda J, Töyräs J, Nissi MJ. T2* and quantitative susceptibility mapping in an equine model of post-traumatic osteoarthritis: assessment of mechanical and structural properties of articular cartilage. Osteoarthritis Cartilage 2019;27:1481-90. [Crossref] [PubMed]
- Wang F, Zhang M, Li Y, Li Y, Gong H, Li J, Zhang Y, Zhang C, Yan F, Sun B, He N, Wei H. Alterations in brain iron deposition with progression of late-life depression measured by magnetic resonance imaging (MRI)-based quantitative susceptibility mapping. Quant Imaging Med Surg 2022;12:3873-88. [Crossref] [PubMed]
- Hesamian MH, Jia W, He X, Kennedy P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging 2019;32:582-96. [Crossref] [PubMed]
- Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 2013;16:246-53.
- Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys 2017;44:5221-33. [Crossref] [PubMed]
- Chen J, Yang L, Zhang Y, Alber M, Chen DZ. Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. Advances in Neural Information Processing Systems 29 (NIPS 2016).
- Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K, Bendszus M, Biller A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage 2016;129:460-9. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science. Cham: Springer; 2015.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Liu C, Liu C, Si L, Shen H, Wang Q, Yao W. Relationship between subchondral bone microstructure and articular cartilage in the osteoarthritic knee using 3T MRI. J Magn Reson Imaging 2018; Epub ahead of print. [Crossref] [PubMed]
- Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger PF, Kohl S, Wasserthal J, Koehler G, Norajitra T, Wirkert S, Maier-Hein KH. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv:180910486.
10.1007/978-3-658-25326-4_7 10.1007/978-3-658-25326-4_7 - Zuendorf G, Kerrouche N, Herholz K, Baron JC. Efficient principal component analysis for multivariate 3D voxel-based mapping of brain functional imaging data sets as applied to FDG-PET and normal aging. Hum Brain Mapp 2003;18:13-21. [Crossref] [PubMed]
- Solloway S, Hutchinson CE, Waterton JC, Taylor CJ. The use of active shape models for making thickness measurements of articular cartilage from MR images. Magn Reson Med 1997;37:943-52. [Crossref] [PubMed]
- Li X, Benjamin Ma C, Link TM, Castillo DD, Blumenkrantz G, Lozano J, Carballido-Gamio J, Ries M, Majumdar S. In vivo T(1rho) and T(2) mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage 2007;15:789-97. [Crossref] [PubMed]
- Schofield MA, Zhu Y. Fast phase unwrapping algorithm for interferometric applications. Opt Lett 2003;28:1194-6. [Crossref] [PubMed]
- Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed 2011;24:1129-36. [Crossref] [PubMed]
- Wei H, Dibb R, Zhou Y, Sun Y, Xu J, Wang N, Liu C. Streaking artifact reduction for quantitative susceptibility mapping of sources with large dynamic range. NMR Biomed 2015;28:1294-303. [Crossref] [PubMed]
- Li W, Wu B, Liu C. STI Suite: A software package for quantitative susceptibility imaging. Proc Intl Soc Mg Reson Med 2014;22:0819.
- Lameski P, Zdravevski E, Mingov R, Kulakov A. SVM parameter tuning with grid search and its impact on reduction of model over-fitting. In: Yao Y, Hu Q, Yu H, Grzymala-Busse J. editors. Rough Sets, Fuzzy Sets, Data Mining, and Granular Computing. Lecture Notes in Computer Science. Cham: Springer; 2015:464-74.
- Xue YP, Jang H, Byra M, Cai ZY, Wu M, Chang EY, Ma YJ, Du J. Automated cartilage segmentation and quantification using 3D ultrashort echo time (UTE) cones MR imaging with deep convolutional neural networks. Eur Radiol 2021;31:7653-63. [Crossref] [PubMed]
- Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80:2759-70. [Crossref] [PubMed]
- Desai AD, Caliva F, Iriondo C, Mortazi A, Jambawalikar S, Bagci U, Perslev M, Igel C, Dam EB, Gaj S, Yang M, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari ASIWOAI Segmentation Challenge Writing Group. The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset. Radiol Artif Intell 2021;3:e200078. [Crossref] [PubMed]
- Welsch GH, Mamisch TC, Hughes T, Domayer S, Marlovits S, Trattnig S. Advanced morphological and biochemical magnetic resonance imaging of cartilage repair procedures in the knee joint at 3 Tesla. Semin Musculoskelet Radiol 2008;12:196-211. [Crossref] [PubMed]
- Wei H, Lin H, Qin L, Cao S, Zhang Y, He N, Chen W, Yan F, Liu C. Quantitative susceptibility mapping of articular cartilage in patients with osteoarthritis at 3T. J Magn Reson Imaging 2019;49:1665-75. [Crossref] [PubMed]


