
Whole slide imaging-based deep learning to predict the treatment response of patients with non-small cell lung cancer
Introduction
According to Chinese national statistics, about 631,000 people die of lung cancer in China every year (1). The number of patients with lung cancer is much higher than that of patients with colorectal cancer, breast cancer, and prostate cancer combined (2). Among patients with lung cancer, the 5-year survival rate of those with non-small cell lung cancer (NSCLC) is less than 18% (3). The treatment strategy of NSCLC depends on the tumor subtype, clinical stage, and physical condition of the patient (4). Patients with stage I, II, and IIIA disease tend to be treated surgically (5), while those with stage IIIB and IV disease are often administered chemotherapy and radiotherapy (CRT) because of the severe late-stage lesions accompanying the lung cancer (6). In patients undergoing CRT for nonsurgical advanced lung cancer, many factors can lead to different treatment effects, such as age, comorbidity (7), existence of tumor heterogeneity (8), and treatment-induced complications, such as radiation pneumonia (9). Some patients may show signs of tumor development and distant metastasis. Therefore, it is necessary to monitor the treatment response in patients with lung cancer.
Radiologists have evaluated the treatment response of patients to CRT according to the Response Evaluation Criteria in Solid Tumors (RECIST) (10). Compared to patients with progressive disease (PD) or stable disease (SD) after CRT, patients with complete response (CR) or partial response (PR) are more likely to have a better prognosis. Both CR and PR are considered responsive states, indicating that CRT is extremely beneficial to overall survival (OS) and disease-free survival (DFS) (11,12). Some authors argue that patients with initial SD or PD after first-line chemotherapy have poorer survival outcomes and worse measures of life quality than do those with PR or CR (13). Some deep learning (DL) models have been used to predict CRT response. Xu et al. developed a model based on computed tomography (CT) images to predict the CRT response of patients with NSCLC (14). Li et al. constructed a 3-dimensional (3D) DL model based on CT images for the pretreatment evaluation of treatment response in locally advanced patients with esophageal cancer (15). However, compared with traditional medical images, histopathological images can show tumor heterogeneity more intuitively and help to visualize the subtle changes caused by the treatment.
Hematoxylin and eosin (HE) staining of biopsy tissue is widely used for the diagnosis of lung cancer (16). Whole slide imaging (WSI) enables the browsing of histopathologic sections to not be limited by space and time, and thus improves the shareability of sections and promotes quantitative and retrospective research (17,18). Unprecedented advances in DL have enabled the synergy of artificial intelligence and digital pathology (19). The convolutional neural network (CNN), which is a DL algorithm, has helped to facilitate excellent performance for image classification and segmentation (20), and it is also widely used in WSI. Coudray et al. predicted the subtypes and gene mutations of NSCLC using CNN based on WSI (21). Yang et al. used CNN to classify and predict 6 subtypes of lung cancer based on WSI (22). In predicting lymph node metastasis, Pham et al. also obtained good prediction results by combining histopathological images with the CNN algorithm (23). The combination of WSI and CNN has also been applied in the research of other systemic diseases, such as the classification of hepatocellular nodular lesions (24), the differentiation between eyelid basal cell and sebaceous carcinoma (25), the development of a gastric cancer detection system for clinical application (26), the recognition of different kidney tissues such as glomeruli and renal arteries (27), and the evaluation of hormone receptor status in breast cancer (28). In brief, CNN has shown good performance in the analysis of WSI, and it can help doctors in analyzing complex histopathological sections.
In this study, we constructed a CNN model based on WSI for predicting the treatment response in patients with NSCLC. The predicted results of the model can provide a reference for doctors to improve treatment planning and patient prognosis. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1098/rc).
Methods
Patients
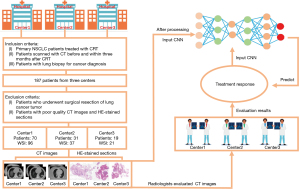
A total of 187 patients from 3 hospitals from 2016 to 2022 were included in the study (29). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Hospital of Shandong First Medical University (No. SB-KJCX2101) and the Ethics Committee of the Xiangya Hospital of Central South University (No. 202207167). The requirement for written informed consent was waived due to the retrospective nature of this investigation.
The inclusion criteria of patients were as follows: (I) age older than 18 years of age; (II) with primary NSCLC; (III) treatment with CRT (the treatment dose of almost all patients was 60–66 Gy and that of a few patients was lower than or higher than 60–66 Gy); (IV) a CT scan before CRT and reexamination CT scan within 5 months after the end of the same CRT course; and (V) lung pathological biopsy for cancer diagnosis. The exclusion criteria were as follows: (I) surgical resection of lung cancer tumors; (II) poor quality CT images; and (III) poor staining quality of the HE sections. After exclusion, among the initial 187 patients, 120 patients (154 WSI) met the criteria and were finally included in the study. There were 70 patients (96 WSI) from one affiliated hospital of the Shandong First Medical University (center 1), 31 patients (37 WSI) from the Xiangya Hospital of Central South University (center 2), and 19 patients (21 WSI) from another affiliated hospital of the Shandong First Medical University (center 3). The CT images and HE-stained pathological images were collected for further analysis.
According to the RECIST, the results of the patient’s treatment response were evaluated based on the CT images before and after administration of CRT by two experienced radiologists in each center. The patients were divided into two groups: patients with CR and PR were considered responsive, and patients with SD and PD were considered nonresponsive.
The HE-stained pathological images of the patients were obtained by scanning biopsy sections with the following parameters: 0.50 µm/pixel (20× magnification) (30-32) on a slide scanner (Zhiying Medicine, Beijing, China) in center 1, 0.50 µm/pixel (20× magnification) on a slide scanner (Pannoramic MIDI, 3DHISTECH, Budapest, Hungary) in center 2, and 0.50 µm/pixel (20× magnification) on a slide scanner (Pannoramic SCAN II, 3DHISTECH, Budapest, Hungary) in center 3. The overall process of patient information collection is displayed in Figure 1.
WSI preprocessing
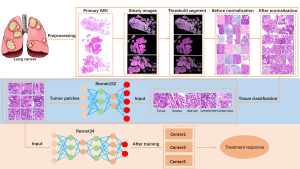
First, the WSI images were converted to binary images as red, green, and blue (RGB) images by an appropriate threshold according to the different depths of the HE staining in the three centers. Then, the binary images were cut into nonoverlapping tiles with 224×224 pixels, and tiles with tissue greater than 50% were saved. In order to improve the staining quality of the tile to better observe the tissue features in the multicenter data, 1 tile with good staining was used as a template to normalize the color of all the other tiles by software package named staintools in python (version 2.1.2, method = vahadane). Finally, tiles were obtained for the subsequent analysis.
Model construction
We constructed two CNN models to predict the treatment response of patients treated with CRT.
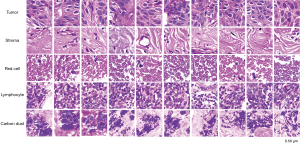
The first established CNN model was used to classify the tissue categories of the tiles. Among all normalized tiles, the pathologist selected a small number of tiles with different tissue categories. The tissue categories were tumor tissue, tumor stroma, red blood cells, lymphocyte, and carbon deposited in lung tissue (Table S1). The classified tiles were divided into training and validation sets at a ratio of 7:3 and then input into the ResNet152 network for training a model used to classify all tiles tissues. The training hyperparameters were as follows: (I) batch size, 32; (II) learning rate, 0.001; (III) optimizer, stochastic gradient descent with a momentum of 0.9; and (IV) epochs, 500. The loss function used was cross entropy. All tumor tissue tiles were identified by the trained model (33).
The second CNN model was established by using the tumor tissue tiles. The patients of center 1 were divided into training and internal validation sets at a ratio of 4:1. The patients of center 2 and center 3 were placed into external validation set 1 and external validation set 2, respectively. The training sets were used to adjust the model parameters and improve the model’s performance. The internal and external validation sets were used to test the predictive ability of the model. The labeled tiles were input into ResNet34 to perform the response evaluation task. The training hyperparameters were as follows: (I) batch size, 64; (II) learning rate, 0.001; (III) optimizer, adaptive moment estimation (Adam) with betas of 0.9 and 0.999 and a weight decay of 0.001; and (IV) epochs, 500. The loss function used was cross entropy. The second trained model predicted the treatment response of each tile. The voting method dictated that the label of tiles with the greatest quantity from one patient would be used as the label of the patient. The tile-level predictions were converted into the patient-level predictions using this method.
Data enhancement was applied to all DL models for expanding the diversity of the tiles and preventing overfitting. The specific enhancement methods were as follows: (I) random rotation within a range of 15 degrees to 15 degrees; (II) random horizontal flip; and (III) color jitter, brightness factor, contrast factor, and saturation factor varying uniformly from 0.5 to 1.5. The results are displayed in Figure S1.
The DL models were trained using PyTorch 1.9.0 (https://pytorch.org/docs/versions.html) on an Intel(R) Xeon(R) Gold 5218 CPU at 2.30 GHz CPU and one NVIDIA Quadro GV100 GPU (CUDA version 11.1).
Results
Patients
In total, 154 WSIs of 120 patients in 3 centers (Table S2) and 421,923 tiles selected after WSI cutting were included in the study. As shown in Table 1, the responsive group of center 1 comprised 17 patients, 23 WSIs, and 42,867 tiles, while the nonresponsive group of center 1 comprised 53 patients, 73 WSIs, and 132,590 tiles. The responsive group of center 2 comprised 11 patients, 11 WSIs, and 43,237 tiles, while the nonresponsive group of center 2 comprised 20 patients, 26 WSIs, and 67,189 tiles. The responsive group of center 3 comprised 6 patients, 6 WSIs, and 34,728 tiles, while the nonresponsive group of center 3 comprised 13 patients, 15 WSIs, and 139,893 tiles. These tiles were normalized and then analyzed further.
Table 1
| Subjects | Center 1 | Center 2 | Center 3 | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| R | nonR | R | nonR | R | nonR | ||||
| Patients | 17 | 53 | 11 | 20 | 6 | 13 | 120 | ||
| WSI | 23 | 73 | 11 | 26 | 6 | 15 | 154 | ||
| Tiles | 42,867 | 132,590 | 43,237 | 67,189 | 34,728 | 139,893 | 421,923 | ||
R, responsive; nonR, nonresponsive; WSI, whole slide imaging.
Model performance
We established a tissue classification model based on several tiles selected by the pathology experts. The accuracy of this model in the training set and the internal validation set was 0.966 and 0.956, respectively (Figure 2, Table S3).
Next, as shown in Table 2, we selected 181,875 tiles that were identified as tumor tissues by using the tissue classification model, consisting of 78,694 tiles in center 1, 27,209 tiles in center 2, and 75,972 tiles in center 3. There were 61,620 tiles of center 1 in the training set and 17,074 tiles of center 1 in the internal validation set. All tiles in center 2 and center 3 were used in external validation set 1 and external validation set 2, respectively. These tiles were used to train and test the final treatment response prediction model (Figure 3).
Table 2
| Subjects | Training | Internal validation | External validation 1 | External validation 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R | nonR | R | nonR | R | nonR | R | nonR | ||||
| Patients | 14 | 42 | 3 | 11 | 11 | 20 | 6 | 13 | |||
| WSI | 19 | 54 | 7 | 19 | 11 | 26 | 6 | 15 | |||
| Tiles | 16,836 | 44,784 | 3,051 | 14,023 | 8,664 | 18,545 | 18,821 | 57,151 | |||
R, responsive; nonR, nonresponsive; WSI, whole slide imaging.
Finally, we obtained a DL model with good performance for predicting the treatment response. The accuracies of patient-level prediction in the internal validation set, external validation set 1, and external validation set 2 were 0.786, 0.742, and 0.737, respectively. Table S4 shows the calculation details of the accuracy, while Table 3 displays the other evaluation indicators of the DL model.
Table 3
| Data sets | Accuracy | AUC | Precision | Recall | Specificity | F1-score |
|---|---|---|---|---|---|---|
| Internal validation set | 0.786 | 0.728 | 0.900 | 0.818 | 0.667 | 0.857 |
| External validation set 1 | 0.742 | 0.696 | 0.773 | 0.850 | 0.546 | 0.810 |
| External validation set 2 | 0.737 | 0.723 | 0.786 | 0.833 | 0.667 | 0.809 |
AUC, the area under curve.
Discussion
In this study, we established a WSI-based DL model for predicting the treatment response of CRT in nonsurgical patients with NSCLC. This model not only accurately predicted the effect of CRT in the internal validation set [accuracy =0.786; area under the curve (AUC) =0.728] but also showed good prediction performance in the two external validation groups (set 1: accuracy =0.742; set 2: accuracy =0.737).
Most previous studies to find biomarkers to predict the treatment response of tumor patients have been based on traditional medical images, such images of CT, magnetic resonance imaging (MRI), and positron emission tomography (PET). Wang et al. constructed a DL model to predict the CRT response based on the CT images of 118 patients (34). Fave et al. established a model based on the delta features to predict the treatment response of patients with NSCLC (35). Based on the CT images, Lee et al. selected biomarkers to predict survival of patients with NSCLC (36). In traditional medical imaging, the features of tumors cannot be reflected directly due to problems such as image distortion and artifacts. Therefore, it is crucial to establish a model to predict the treatment response based on histopathological sections. Pathological biopsy has become one of the necessary examinations for lung cancer patients before treatment. Under conditions of high magnification (20× magnification), we could clearly see the morphology of tumor cells and surrounding stromal tissue, which was conducive to our DL model directly capturing the features of cellular and tissue levels of CRT-responsive and CRT-nonresponsive patients. Before treatment, knowing the predicted CRT treatment effect in advance can help doctors to adjust the treatment plan in advance. The treatment process of patients is irreversible. Previous studies established several radiomics models based on CT/MRI/PET images to predict the treatment effect. The concordance index (C-index) of the radiomic models was often between 0.60 and 0.67, which was improved to 0.72 through combining the images with clinical and genomic features (34-36). Compared with the prediction model based on traditional images, our model, which was trained with WSI, achieved better prediction performance. The accuracies of our model were 0.742 and 0.737 in external validation set 1 and set 2, respectively. Patients predicted to have poor response under the DL model after treatment can choose other treatment schemes or increase the radiotherapy dose, whereas patients with good response can use the current treatment scheme more reliably. We included data of three centers in this study, and the DL model showed excellent prediction performance and satisfactory generalization ability. The accuracies of the model were 0.742 and 0.737 in external validation set 1 and set 2, respectively. The generalizability of the model suggests that this model can also obtain good prediction results for the data of other hospitals. We also constructed a model to distinguish the types of tissue. This model can reduce the time for doctors to outline different tissue areas and thus can be used in future research.
This study also involved some limitations which should be noted. First, the amount of included data was relatively small, and thus the inclusion of more cases may improve the performance of the model. Moreover, most of the treatment response results of the patients were PR or SD, and a few treatment response results were CR or PD. We classified PR and CR patients as CRT-responsive patients, and SD and PD as CRT-nonresponsive patients. In the future, we hope to expand and balance our sample size and more carefully classify patients into the CR, PR, SD, and PD categories. Second, we established a single-mode model based on pathological images without including the clinical features, features from traditional images such as CT images, and molecular-level information such as gene and protein expression. The combination of clinical information, traditional images, histopathological images, and molecular information may improve the prediction ability of the model. In the future, we may incorporate multidimensional patient information in building the models to predict molecular expression based on images and thus assist the development of precision medicine and noninvasive examination. Finally, in the next study, we hope to include image features and pathologic information, such as tumor percentage and necrosis percentage, to increase the interpretability of the model.
In summary, we developed and validated a DL model that had good robustness for predicting the treatment response of patients with NSCLC. The model has the potential to help doctors optimize patients’ treatment plans and enhance patient’s treatment outcomes.
Conclusions
A DL model was constructed based on WSI for predicting the treatment response of patients with NSCLC. The model can assist doctors in formulating personalized CRT plans and supporting the precise treatment for patients.
Acknowledgments
The authors appreciate the academic support from the AME Oncology Collaborative Group.
Funding: This work was supported by China National Key Research and Development (No. 2021YFE0204600), the Academic Promotion Program of Shandong First Medical University (No. 2019QL009), and the Taishan Scholars Program of Shandong Province (No. TS201712065).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-1098/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1098/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Hospital of Shandong First Medical University (No. SB-KJCX2101) and the Ethics Committee of the Xiangya Hospital of Central South University (No. 202207167). The requirement for written informed consent was waived due to the retrospective nature of this investigation.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol 2021;33:40-6. [Crossref] [PubMed]
- Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. Am Fam Physician 2007;75:56-63. [PubMed]
- Ettinger DS, Akerley W, Bepler G, Chang A, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2008;6:228-69. [Crossref] [PubMed]
- Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: Biology and treatment options. Biochim Biophys Acta 2015;1856:189-210. [PubMed]
- Howington JA, Blum MG, Chang AC, Balekian AA, Murthy SC. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e278S-313S.
- Paraschiv B, Diaconu CC, Dumitrache-Rujinski S, Belaconi IN, Constantinescu T, Stroescu CC, Dantes E, Fildan AP, Bogdan MA, Toma CL. Treatment options in stage III non-small cell lung cancer. Pneumologia 2016;65:67-70. [PubMed]
- Dutkowska AE, Antczak A. Comorbidities in lung cancer. Pneumonol Alergol Pol 2016;84:186-92. [PubMed]
- de Sousa VML, Carvalho L. Heterogeneity in Lung Cancer. Pathobiology 2018;85:96-107. [Crossref] [PubMed]
- Arroyo-Hernández M, Maldonado F, Lozano-Ruiz F, Muñoz-Montaño W, Nuñez-Baez M, Arrieta O. Radiation-induced lung injury: current evidence. BMC Pulm Med 2021;21:9. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Maquilan G, Grover S, Xanthopoulos E, Evans TL, Aggarwal C, Langer CJ, Cohen RB, Stevenson JP, Simone CB 2nd, Rengan R. Analysis of the Relationship Between Response to Chemotherapy and Response to Radiation Therapy in Patients With Non-Small Cell Lung Cancer Receiving Sequential Treatment. Am J Clin Oncol 2018;41:391-5. [Crossref] [PubMed]
- Zhou X, Li C, Zhang Z, Li DY, Du J, Ding P, Meng H, Xu H, Li R, Ho E, Zhang A, Okunieff P, Lu J, Sha MY. Kinetics of plasma cfDNA predicts clinical response in non-small cell lung cancer patients. Sci Rep 2021;11:7633. [Crossref] [PubMed]
- Grossi F. Management of non-small cell lung in cancer patients with stable disease. Drugs 2012;72:20-7. [Crossref] [PubMed]
- Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, Mak RH, Aerts HJWL. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin Cancer Res 2019;25:3266-75. [Crossref] [PubMed]
- Li X, Gao H, Zhu J, Huang Y, Zhu Y, Huang W, Li Z, Sun K, Liu Z, Tian J, Li B. 3D Deep Learning Model for the Pretreatment Evaluation of Treatment Response in Esophageal Carcinoma: A Prospective Study (ChiCTR2000039279). Int J Radiat Oncol Biol Phys 2021;111:926-35. [Crossref] [PubMed]
- Nooreldeen R, Bach H. Current and Future Development in Lung Cancer Diagnosis. Int J Mol Sci 2021; [Crossref] [PubMed]
- Kumar N, Gupta R, Gupta S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J Digit Imaging 2020;33:1034-40. [Crossref] [PubMed]
- Webster JD, Dunstan RW. Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol 2014;51:211-23. [Crossref] [PubMed]
- Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20:e253-61. [Crossref] [PubMed]
- Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical Image Analysis using Convolutional Neural Networks: A Review. J Med Syst 2018;42:226. [Crossref] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Yang H, Chen L, Cheng Z, Yang M, Wang J, Lin C, Wang Y, Huang L, Chen Y, Peng S, Ke Z, Li W. Deep learning-based six-type classifier for lung cancer and mimics from histopathological whole slide images: a retrospective study. BMC Med 2021;19:80. [Crossref] [PubMed]
- Pham HHN, Futakuchi M, Bychkov A, Furukawa T, Kuroda K, Fukuoka J. Detection of Lung Cancer Lymph Node Metastases from Whole-Slide Histopathologic Images Using a Two-Step Deep Learning Approach. Am J Pathol 2019;189:2428-39. [Crossref] [PubMed]
- Cheng N, Ren Y, Zhou J, Zhang Y, Wang D, Zhang X, Chen B, Liu F, Lv J, Cao Q, Chen S, Du H, Hui D, Weng Z, Liang Q, Su B, Tang L, Han L, Chen J, Shao C. Deep Learning-Based Classification of Hepatocellular Nodular Lesions on Whole-Slide Histopathologic Images. Gastroenterology 2022;162:1948-1961.e7. [Crossref] [PubMed]
- Luo Y, Zhang J, Yang Y, Rao Y, Chen X, Shi T, Xu S, Jia R, Gao X. Deep learning-based fully automated differential diagnosis of eyelid basal cell and sebaceous carcinoma using whole slide images. Quant Imaging Med Surg 2022;12:4166-75. [Crossref] [PubMed]
- Song Z, Zou S, Zhou W, Huang Y, Shao L, Yuan J, et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun 2020;11:4294. [Crossref] [PubMed]
- Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, Roelofs JJTH, Stegall MD, Alexander MP, Smith BH, Smeets B, Hilbrands LB, van der Laak JAWM. Deep Learning-Based Histopathologic Assessment of Kidney Tissue. J Am Soc Nephrol 2019;30:1968-79. [Crossref] [PubMed]
- Naik N, Madani A, Esteva A, Keskar NS, Press MF, Ruderman D, Agus DB, Socher R. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun 2020;11:5727. [Crossref] [PubMed]
- Vanguri RS, Luo J, Aukerman AT, Egger JV, Fong CJ, Horvat N, Pagano A, Araujo-Filho JAB, Geneslaw L, Rizvi H, Sosa R, Boehm KM, Yang SR, Bodd FM, Ventura K, Hollmann TJ, Ginsberg MS, Gao JMSK MIND Consortium. Hellmann MD, Sauter JL, Shah SP. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer 2022;3:1151-64. [Crossref] [PubMed]
- Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, Parwani A, Bui M, Hartman DJ. A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association. Arch Pathol Lab Med 2019;143:222-34. [Crossref] [PubMed]
- Cao R, Yang F, Ma SC, Liu L, Zhao Y, Li Y, Wu DH, Wang T, Lu WJ, Cai WJ, Zhu HB, Guo XJ, Lu YW, Kuang JJ, Huan WJ, Tang WM, Huang K, Huang J, Yao J, Dong ZY. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020;10:11080-91. [Crossref] [PubMed]
- Shao W, Wang T, Huang Z, Han Z, Zhang J, Huang K. Weakly Supervised Deep Ordinal Cox Model for Survival Prediction From Whole-Slide Pathological Images. IEEE Trans Med Imaging 2021;40:3739-47. [Crossref] [PubMed]
- Shao L, Liu Z, Feng L, Lou X, Li Z, Zhang XY, Wan X, Zhou X, Sun K, Zhang DF, Wu L, Yang G, Sun YS, Xu R, Fan X, Tian J. Multiparametric MRI and Whole Slide Image-Based Pretreatment Prediction of Pathological Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer: A Multicenter Radiopathomic Study. Ann Surg Oncol 2020;27:4296-306. [Crossref] [PubMed]
- Wang L, Dong T, Xin B, Xu C, Guo M, Zhang H, Feng D, Wang X, Yu J. Integrative nomogram of CT imaging, clinical, and hematological features for survival prediction of patients with locally advanced non-small cell lung cancer. Eur Radiol 2019;29:2958-67. [Crossref] [PubMed]
- Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, Followill D, Jones AK, Stingo F, Liao Z, Mohan R, Court L. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 2017;7:588. [Crossref] [PubMed]
- Lee J, Li B, Cui Y, Sun X, Wu J, Zhu H, Yu J, Gensheimer MF, Loo BW Jr, Diehn M, Li R. A Quantitative CT Imaging Signature Predicts Survival and Complements Established Prognosticators in Stage I Non-Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2018;102:1098-106. [Crossref] [PubMed]


