
Deep learning-based approach for the automatic segmentation of adult and pediatric temporal bone computed tomography images
Introduction
The temporal bone, which contains hearing and balance organs, is one of the most complex bony structures in the human body. To ensure secure temporal bone surgeries, surgeons must mentally establish a 3-dimensional (3D) interrelationship of temporal bone anatomy using 2-dimensional (2D) temporal bone computed tomography (CT) scans (1-3); this is a difficult task for resident surgeons. Image-guided technological approaches can assist surgeons in establishing and comprehending the spatial relationship between the critical structures obtained from 2D temporal bone scans and the 3D reconstruction of such scans. Image-guided techniques have been applied in various otologic surgeries, such as mastoidectomy (2), cochlear implantation (4), and acoustic neuroma resection (5,6). Sensitive structure segmentation of CT images is essential for the safety of image-guided temporal bone surgeries. However, more than 10 structures are associated with the temporal bone, and they vary widely in terms of shape, size, volume, threshold, and local contrast, among other attributes (3). Although expert manual segmentation of temporal bone CT images achieves significantly high accuracy, it is time-consuming and labor-intensive, limiting its effectiveness for image-guided temporal bone surgery (2,4-6).
Automatic segmentation is a promising approach that can address the rapidly increasing demand for the extraction of critical organs in temporal bone CT images. Over the past few years, atlas-based segmentation methods (7,8) and active shape models (ASMs) (9,10) have dominated the field of automatic segmentation of temporal bone CT images. These studies automatically extract critical structures from temporal bone CT images through a series of operations, including presegmentation, spatial registration, and label mapping (11). However, the performance of these segmentation algorithms can be compromised easily due to non-rigid registration of images, noise, and changes in the topologies of various sensitive structures, among other aspects (12,13).
In recent years, there have been significant advancements in artificial intelligence (AI). Deep learning, a subfield of machine learning, facilitates the development of novel and efficient methods to achieve the effective automatic segmentation of images generated using medical imaging techniques (14-16). Specifically, deep learning-based frameworks comprise multilayer neural networks that can learn the features of image structures, such as threshold, shape, size, volume, and texture, from the original input image (17,18) and further provide the target structures as the output. Deep learning-based computer vision and image segmentation approaches have unique capabilities (19). In recent years, some neural network models such as U-net and W-net have been proposed, which have been shown to achieve impressive results in image segmentation, compared with traditional methods (12). Applying such deep learning-based approaches in medicine has resulted in major breakthroughs in various surgical fields (20-24). However, recently published architectural designs of neural networks for temporal bone CT images are complex (1,3,25). Additionally, all the CT images used in the previously referenced studies were obtained from adults; CT images from children were not used in these studies.
This study used a previously published convolutional neural network (CNN) model (13) to segment the small and fine structures in temporal bone CT images. Automatic segmentation training of the proposed network was conducted using 11 critical structures obtained from temporal bone CT images of adults and children. Finally, the generalizability of the CNN model was evaluated to automatically segment temporal bone CT images obtained from adults and children.
The main contributions of this study are as follows:
- First, a dataset comprising 11 annotated temporal bone CT structures was developed. To our knowledge, this is the first publicly available dataset comprising annotated temporal bone CT structures. The developed dataset and the code of the CNN architecture will be shared with the GitHub website to promote future research.
- Second, because no previous studies on the automatic segmentation of pediatric temporal bone CT images were identified, CT images obtained from pediatric temporal bone imaging in the training set were included. A training model that included temporal bone CT images from adults and children was established. Therefore, the range of groups who can benefit from the application of the proposed CNN model in the medical field was expanded.
- Third, we tested the generalizability of the CNN model using adult and pediatric temporal bone CT data, thereby demonstrating the excellent segmentation performance of the CNN model in conventional temporal bone CT. These experiments showed that our method is suitable for clinical application.
The following article is presented in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-658/rc).
Methods
Dataset for deep learning
This study developed a dataset comprising 80 temporal bone CT volumes of 11 sensitive organs. Of these images, 40 were obtained from healthy adults and 40 from healthy children. Each registered patient underwent only 1 CT scan between October 2020 and March 2021. A clinical expert reviewed the developed CT image-based dataset and removed all the cases associated with otologic diseases, temporal bone surgery, and structural malformations. This study was conducted according to the Declaration of Helsinki (as revised in 2013) and approved by the Peking University Third Hospital Medical Ethics Committee (No. IRB00006761-M2019335). The requirement for written informed consent was waived due to the retrospective nature of the study.
All the temporal bone CT images were acquired using a 128-channel multidetector (Siemens/SOMATOM Definition Flash CT scan, Munich, Germany) in the Peking University Third Hospital. All patients underwent CT scanning from the top edge of the petrous part of the temporal bone to the lower edge of the external auditory canal, with a thickness of 0.625 mm, pitch of 0.30 mm, pixel of 0.412 mm, matrix size of 512×512, field of view of 220×220 mm, voltage of 120 kV, and current of 240 mAs. All temporal bone CT images were downloaded in digital imaging and communications in medicine (DICOM) format.
Experimental design
We selected 60 annotated temporal bone CT volumes (from 30 adults and 30 children), which were randomly divided into 5 groups comprising 12 annotated CT volumes (6 for adults and 6 for children) in each group. We selected 4 groups as the training set, and the remaining group was selected as the validation set. Five-fold cross-validation was performed in this study (13). It should be noted that the adult and pediatric CT volumes were not segregated and were trained together during the modeling process.
We used the remaining annotated CT volumes as the adult testing set (n=10) and the child testing set (n=10) to assess the generalizability of our CNN model. First, we compared the segmentation accuracy of target organs using our CNN model and the manual annotation method in healthy adults’ and children’s CT images. Second, we analyzed the difference in the automatic segmentation performance of the CNN model for CT images obtained from healthy adults and that for CT images obtained from healthy children. It should be noted that annotated CT volumes of the test set were not used in the training and cross-validation of the CNN model. The experiments were conducted using a workstation with a Xeon Silver 4110 CPU (16 GB memory, Intel, Santa Clara, CA, USA) and an NVIDIA RTX 2080Ti GPU (Nvidia, Santa Clara, CA, USA).
Empirical evidence
The manual segmentation of all the temporal bone CT structures was performed by 2 trained otologic surgeons and an otologist with over 20 years’ experience analyzing and reviewing annotated images generated through medical imaging techniques. We used a combination method involving increasing thresholds and regions to extract 7 small structures, including the facial nerve (FN), ossicle, inner ear (IE), cochlea, vestibule, the lateral semicircular canal (LSC), and the superior and posterior semicircular canals (SPSC). In this process, the threshold range of each structure was determined first, and then the erasure method or delineated method was used to ensure the accurate range of the presegmentation structures. Interesting structures were extracted using image analysis employing the software’s region increasing method. Finally, the surgeon manually cleaned and corrected the threshold structures. An expert with over 20 years of image analysis experience used the thresholds to review the segmented structures and provide suggestions. The same approach was also used to extract 4 large structures, including the internal auditory canal (IAC), internal carotid artery (ICA), jugular bulb (JB), and the external auditory canal (EAC). The threshold was set to −700 to 500 Hounsfield units (HU) for the FN, 226 to 3,071 HU for the ossicle, −700 to 1,011 HU for the IE, cochlea, vestibule, LSC, SPSC, IAC, IC, and JB, and −1,024 to 1,011 HU for the EAC. All structures used in this study were delineated and reconstructed using Mimics image processing software (version 20.0; Materialise NV, Leuven, Belgium).
Data augmentation technology
The new training samples were extracted through geometrical transformation approaches, such as random left-right flipping, scaling, rotating, and translating. The testing sets did not use data augmentation technology. In our study, 2,400 annotated CT volumes were generated through data augmentation.
Network architecture
We adopted a fully supervised approach to segment temporal bone images. The architecture of our CNN model, which was described in our previous study (13), is shown in Figure 1. The CNN models comprised convolutional layers, decoder-encoder units, and skip connections. The CNN model had 2 analysis (decoding) paths, 2 syntheses (encoding) paths, and 3 skip connection paths. Both analysis and synthesis paths contained two 3×3×3 convolutions in each layer. The kernel size of the transposed convolution was 2×2×2 with a stride of 2. The segmentation structures were located in the middle of the CT image data, and the surrounding edges had no segmentation target area. Therefore, the pixel padding value in the 3D convolution operation of our CNN model was set to 1. This ensured that the output dimensions of the network were similar to those of the input (64×64×80 voxels). In this study, we used the adaptive moment estimation (Adam) optimizer to optimize parameters during the iterative process (26). The Dice loss function was used to improve the performance of the CNN model during the training process.
The current CNN model is different from the standard 3D U-Net (27). First, a high-resolution feature map was used as the input of convolution blocks in each layer of the network because of the relatively small structure of the temporal bone. Second, we innovatively introduced the design of a cross skip connection layer, such that the information between the network layers could be better shared and more thoroughly trained. Then, the design of parameter sharing was used for the convolution blocks of the 2 decoding paths to increase the speed of network fitting and reduce the number of network parameters. Therefore, the CNN parameters’ total amount was less than one-tenth of that of a standard U-Net.
Evaluation metrics
In this study, the Dice coefficient (DC) and the average symmetric surface distance (ASSD) were used as the metrics for evaluating the segmentation accuracy to reflect the similarity index of automatic segmentation and manual segmentation (28-31). They are defined as follows:
where R and R0 represent the voxels annotated by clinicians and the CNN model, respectively. r and r0 represent any surface points of R and R0, respectively. d (r, r0) represents the Euclidian distance between points r and r0. NR and NR0 represent the number of surface voxels represented using R and R0, respectively. For DC values, the higher the better (maximum value of 1), whereas the reverse is true for ASSD values (minimum value of 0). A DC value of more than 0.7 indicates good agreement between automatic and manual segmentation (11,32).
Statistical analyses were carried out using SPSS 24.0 software (IBM Corp, Armonk, NY, USA). To evaluate the generalizability of the CNN model, the test set results of adults and children were compared. Two independent-sample t-tests were used, and P<0.05 was considered to indicate a statistically significant difference.
Results
Performance evaluation of the adult test set
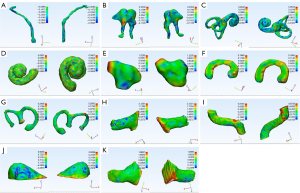
Automated segmentation over the adult test data was performed to test the generalizability of the models developed from the training set (n=10). Table 1 summarizes the automatic segmentation metrics of the adult test set using the CNN model. Figure 2 shows the masks of each structure obtained through the surgeon annotation and the CNN model automatic segmentation of temporal CT images obtained from adults. Figure 3 shows surface rendering of the temporal CT images obtained from adults through automatic segmentation and surgeon annotations.
Table 1
| Metrics | SP | FN | Ossicle | IE | Cochlea | Vestibule | LSC | SPSC | IAC | ICA | GJ | EAC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DC | Max | 0.861 | 0.915 | 0.918 | 0.930 | 0.903 | 0.878 | 0.870 | 0.893 | 0.920 | 0.884 | 0.875 |
| Min | 0.503 | 0.867 | 0.905 | 0.864 | 0.831 | 0.809 | 0.807 | 0.808 | 0.750 | 0.537 | 0.820 | |
| AVG | 0.746 | 0.891 | 0.912 | 0.897 | 0.862 | 0.843 | 0.842 | 0.854 | 0.868 | 0.714 | 0.859 | |
| SD | 0.116 | 0.016 | 0.005 | 0.022 | 0.022 | 0.026 | 0.023 | 0.025 | 0.057 | 0.102 | 0.017 | |
| ASSD (mm) | Max | 0.457 | 0.096 | 0.110 | 0.171 | 0.192 | 0.136 | 0.129 | 0.488 | 0.749 | 2.570 | 0.430 |
| Min | 0.118 | 0.061 | 0.076 | 0.054 | 0.126 | 0.094 | 0.092 | 0.199 | 0.134 | 0.375 | 0.236 | |
| AVG | 0.239 | 0.079 | 0.087 | 0.107 | 0.159 | 0.111 | 0.113 | 0.341 | 0.298 | 1.189 | 0.286 | |
| SD | 0.106 | 0.012 | 0.009 | 0.039 | 0.021 | 0.016 | 0.011 | 0.089 | 0.201 | 0.628 | 0.055 |
CNN, convolutional neural network; SP, statistical parameters; FN, facial nerve; IE, inner ear; LSC, lateral semicircular canal; SPSC, superior and posterior semicircular canal; IAC, internal auditory canal; ICA, internal carotid artery; GJ, glomus jugulare; EAC, external auditory canal; DC, Dice coefficient; ASSD, average symmetric surface distance; Max, maximum; Min, minimum; AVG, average; SD, standard deviation.
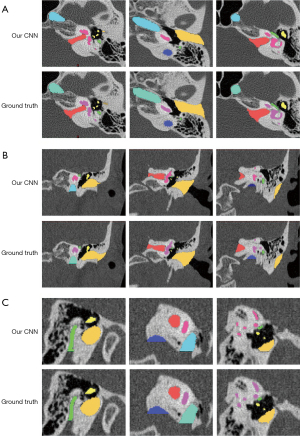
The mean DC values of all structures were over 0.7. Among them, the mean DC value of IE was the highest, reaching 0.912 (0.005). Moreover, the mean DC of individual structures that made up the IE also exceeded 0.8. The mean DC values were 0.897 (0.022) for the cochlea, 0.862 (0.022) for the vestibule, 0.843 (0.026) for LSC, and 0.842 (0.023) for SPSC. JB had the lowest mean DC value of all structures with only 0.714 (0.102). Regarding ASSD values, JB got the largest, wheres the ossicle attained the lowest, with 1.189 (0.628) mm and 0.079 (0.012) mm, respectively. The other structures had ASSD values between 0.080 and 0.400 mm.
Performance evaluation of the children test set
We further tested the generalizability of the CNN model using the temporal bone CT images obtained from children. Table 2 summarizes the CNN model’s automatic segmentation metrics of pediatric temporal bone CT images. For the thin and fine organs such as FN, ossicle, and IE structures, the mean DC values were over 0.750, whereas the mean ASSD values were below 0.200 mm. For individual structures that make up the IE, the mean accuracy of DC and ASSD was 0.886 (0.041) and 0.123 (0.046) mm, respectively, for the cochlea; 0.890 (0.016) and 0.153 (0.029) mm, respectively, for the vestibule; 0.838 (0.019) and 0.120 (0.016) mm, respectively, for LSC; and 0.849 (0.012) and 0.111 (0.013) mm, respectively, for SPSC.
Table 2
| Metrics | SP | FN | Ossicle | IE | Cochlea | Vestibule | LSC | SPSC | IAC | ICA | GJ | EAC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DC | Max | 0.804 | 0.913 | 0.921 | 0.929 | 0.909 | 0.862 | 0.865 | 0.923 | 0.895 | 0.749 | 0.863 |
| Min | 0.671 | 0.875 | 0.907 | 0.821 | 0.858 | 0.800 | 0.821 | 0.817 | 0.847 | 0.567 | 0.796 | |
| AVG | 0.763 | 0.898 | 0.915 | 0.886 | 0.890 | 0.838 | 0.849 | 0.882 | 0.860 | 0.658 | 0.831 | |
| SD | 0.040 | 0.010 | 0.005 | 0.041 | 0.016 | 0.019 | 0.012 | 0.034 | 0.017 | 0.051 | 0.022 | |
| ASSD (mm) | Max | 0.243 | 0.116 | 0.091 | 0.194 | 0.216 | 0.154 | 0.135 | 0.408 | 0.326 | 2.366 | 0.470 |
| Min | 0.119 | 0.068 | 0.068 | 0.075 | 0.116 | 0.100 | 0.095 | 0.160 | 0.158 | 1.244 | 0.279 | |
| AVG | 0.174 | 0.080 | 0.080 | 0.123 | 0.153 | 0.120 | 0.111 | 0.239 | 0.249 | 1.619 | 0.370 | |
| SD | 0.037 | 0.014 | 0.008 | 0.046 | 0.029 | 0.016 | 0.013 | 0.075 | 0.060 | 0.398 | 0.055 |
CNN, convolutional neural network; SP, statistical parameters; FN, facial nerve; IE, inner ear; LSC, lateral semicircular canal; SPSC, superior and posterior semicircular canal; IAC, internal auditory canal; ICA, internal carotid artery; GJ, glomus jugulare; EAC, external auditory canal; DC, Dice coefficient; ASSD, average symmetric surface distance; Max, maximum; Min, minimum; AVG, average; SD, standard deviation.
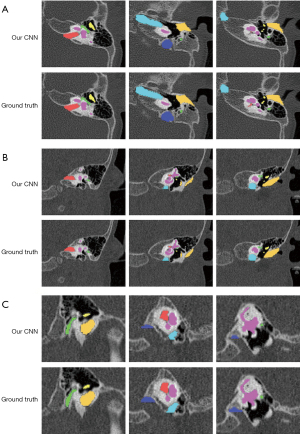
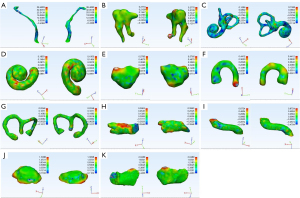
For larger structures (except JB), the mean DC metrics exceeded 0.800, and mean ASSD values were lower than 0.400 mm. JB achieved the worst result, whereby the mean DC and ASSD values were 0.658 (0.051) and 1.619 (0.398) mm, respectively. Figure 4 shows the masks of the segmented pediatric CT image structures annotated by clinicians and those annotated using the CNN model. Figure 5 shows an example of the surface rendering of automatically segmented and surgeon annotated pediatric temporal bone CT images.
Adult test accuracy versus pediatric test accuracy
Using DC as an evaluation metric, 2 independent sample t-tests showed that the value of the adult vestibule was significantly lower than that of children (0.862 vs. 0.890, respectively, P=0.004). However, there was no significant difference between the ASSD values of adult and pediatric vestibules (0.159 vs. 0.153, respectively, P=0.62). The ASSD value of pediatric IAC structures was significantly lower than that of adult IAC structures (0.341 vs. 0.239, P=0.01); however, there was no significant difference in the DC value (0.854 vs. 0.882, respectively, P=0.05). Specifically, the DC value of adult EAC structures was higher than that of pediatric EAC structures (0.859 vs. 0.831, respectively, P=0.005). In contrast, the ASSD value of adult EAC structures was lower than that of pediatric EAC structures (0.286 vs. 0.370, P=0.003, respectively,), and the differences between the DC and ASSD values were statistically significant. The values of the other adult and pediatric temporal bone CT image structures were not statistically different (Table 3).
Table 3
| Metrics | FN | Ossicle | IE | Cochlea | Vestibule | LSC | SPSC | IAC | ICA | JB | EAC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P (DC) | 0.66 | 0.28 | 0.22 | 0.47 | 0.004 | 0.58 | 0.43 | 0.05 | 0.70 | 0.14 | 0.005 |
| P (ASSD) | 0.09 | 0.86 | 0.08 | 0.41 | 0.62 | 0.23 | 0.66 | 0.01 | 0.49 | 0.09 | 0.003 |
FN, facial nerve; IE, inner ear; LSC, lateral semicircular canal; SPSC, superior and posterior semicircular canal; IAC, internal auditory canal; ICA, internal carotid artery; JB, jugular bulb; EAC, external auditory canal; DC, Dice coefficient; ASSD, average symmetric surface distance.
Discussion
Performance of atlas-based methods and ASMs
Over the past few years, atlas-based methods and ASMs have been employed as effective technologies for ensuring the automatic segmentation of temporal bone CT images (5,7,33). The principle of atlas-based methods involves using the volume extracted as an atlas for registering other volumes to be segmented (11,34). ASM-based methods employ the local texture statistical model to perform feature searching, followed by the use of a global statistical model to constrain the shape of structures (35,36). The ASM method fitted the active shape model to the image features through non-rigid registration to finish the segmentation. However, the ASM-based method only applies to cases where the target structure is similar to the active shape model, and the method is difficult to apply to a dataset with significant differences between individuals. When these methods are purely used to segment small and complex-shaped structures such as FN and IE in temporal bone CT images, the automatic segmentation performance is not entirely satisfactory (11,33,37). Therefore, atlas-based and ASMs are often combined to achieve better segmentation results. For example, Noble et al. (10) created an ASM. They placed this ASM with the target CT volume by non-rigidly registering an atlas image against the target volume and fitting the ASM to the local image features.
The FN is the most difficult structure to identify and segment in the temporal bone. Powell et al. (11) proposed a novel atlas-based method that combined an intensity model and region-of-interest (ROI) masks, and the DC value of the FN reached 0.68 (0.09) and 0.73 (0.10), respectively. This optimized method can extract the FN more accurately. A previous study split the ossicle into malleus, incus, and stapes for the ossicular chain (11). The DC values of individual ossicles were between 0.48 (0.04) and 0.83 (0.03) (11). Therefore, we could not directly compare our results with those of individual bones based on atlas-based methods. In addition, although the ossicle was segmented as a whole in some studies, the evaluation metrics were also different (37). In the IE structures, the new method used by Powell et al. (11) can accurately locate the cochlea, semicircular canals, and vestibule with DC values of 0.7 or above 0.8. In this study, the DC values of IE and individual IE structures were all above 0.8 based on the current CNN model. Table 4 summarizes the DC results of automatic segmentation based on atlas-based and ASM methods in temporal bone images.
Table 4
| Methods | FN | Ossicle | IE | Cochlea | Vestibule | LSC | SPSC | IAC | ICA | JB | EAC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Atlas-based/ASMs (11) | 0.730 | 0.830 | – | 0.910 | 0.820 | 0.750 | 0.820 | – | – | – | – |
| ResNet (1) | 0.690 | 0.870 | 0.910 | – | – | – | – | – | – | – | – |
| U-net (1) | 0.730 | 0.860 | 0.910 | – | – | – | – | – | – | – | – |
| AH-Net (1) | 0.750 | 0.850 | 0.910 | – | – | – | – | – | – | – | – |
| 3D-DSD (3) | – | 0.822 | – | 0.836 | 0.822 | 0.700 | 0.751 | 0.811 | – | – | – |
| PWD-3D (25) | 0.720 | 0.820 | 0.900 | – | – | – | – | 0.880 | 0.820 | – | – |
| AutoCasNet (38) | – | – | 0.900 | – | – | – | – | – | – | – | – |
| CAPPU-Net (39) | 0.740 | 0.835 | 0.848 | – | – | – | – | – | – | – | – |
| Our method | 0.746 | 0.891 | 0.912 | 0.897 | 0.862 | 0.843 | 0.842 | 0.854 | 0.868 | 0.714 | 0.859 |
Some structures were not segmented in previous methods, and replaced by “–”. FN, facial nerve; IE, inner ear; LSC, lateral semicircular canal; SPSC, superior and posterior semicircular canal; IAC, internal auditory canal; ICA, internal carotid artery; JB, jugular bulb; EAC, external auditory canal; ASMs, active shape models; 3D-DSD Net, 3D deep supervised dense network; PWD-3DNet, patch-wise densely connected three-dimensional network; AutoCasNet, auto-cascaded net; CAPPU-Net, convolutional attention network with pyramid pooling U-Net.
Wang et al. (40) recently proposed a new automatic segmentation method that combines probabilistic appearance and shape models. The authors developed Bayesian inference of parametric shape models using likelihood appearance probability and prior label probability and applied it to cochlea segmentation on the clinical CT datasets. The DC values of the cochlea were between 0.85 and 0.91 using this method. Their results show that this new method’s performance is better than previously proposed unsupervised methods and comparable to supervised methods. Furthermore, Kjer et al. (41,42) proposed a statistical shape model and statistical deformation models for automatic segmentation of IE. These methods relied on rigid image registration, and the final DC metrics of the cochlea were over 0.95 in μCT images.
Currently, very few reports exist on the automatic segmentation of pediatric temporal bone CT images using atlas-based methods. Reda et al. (43) reported that the mean errors of FN segmentation were 0.23 mm using a novel atlas-based method combined with a statistical model algorithm. The ASSD value of FN images obtained from pediatric temporal CT images reached 0.174 mm in the approach proposed here. Although the evaluation indicators are not similar, our results suggest that automatically and manually segmented pediatric FN images are highly consistent.
Performance of previously published networks
Several neural networks based on deep learning have been used to segment temporal bone CT images automatically. Each neural network only realized the automatic segmentation of partial structures in the temporal bone, and the accuracy of these structures varies. Neves et al. (1) compared the performance of 3 CNN-based models (U-Net, ResNet, and AH-Net) to automatically segment the FN, ossicle, and IE in the temporal bone. The DC values of IE were over 0.9, those of the ossicle were all over 0.8, and the FN values were between 0.69 and 0.75, based on these 3 CNN models.
In previous studies, several neural networks for the automatic segmentation of temporal bone CT images were proposed: 3D deep supervised dense network (3D-DSD Net) (3), patch-wise densely connected three-dimensional network (PWD-3DNet) (25), auto-cascaded net (AutoCasNet) (38), and convolutional attention network with pyramid pooling U-Net (CAPPU-Net) (39). 3D-DSD Net added some designs, including a deep supervised hidden layer, densely connected block, and multipooling features fused based on a fully convolutional network, which theoretically improved the segmentation accuracy of small organs (3). Nikan et al. (25) proposed a novel fully convolutional network, and they claimed that the accuracy of PWD-3DNet surpassed that of current semiautomated segmentation techniques. PWD-3DNet has 2 architectural structures: an augmented model and a non-augmented model. The augmentation layers were introduced to the augmented model, which enlarged the input dataset (Micro-CT samples were the training sets) and generalized samples with different acquisition parameters (25). Compared with the nonaugmented model, the segmentation accuracy of the augmented model was higher for temporal bone structures (25). Hussain et al. (38) proposed a cascaded 2D U‑net architecture and used 3D-connected component refinement to segment IE in micro‑CT. The DC value of IE was 0.900 (0.007) based on this AutoCasNet. Kim et al. (39) proposed CAPPU-Net, which combined feature extraction blocks—convolutional bottleneck attention module and atrous spatial pyramid pooling. The DC values of FN, ossicle, and cochlea were 0.740 (0.214), 0.835 (0.194), and 0.848 (0.149), respectively. Table 4 shows the accuracy of automatic segmentation of temporal bones with various methods. In this study, the CNN obtained high automatic segmentation accuracy, showing that our CNN model performed very well in segmenting temporal bone CT images.
Generalizability of CNN model future applications
We attempted to use adult training models to segment pediatric temporal bone CT images automatically. However, their performance was not satisfactory. Therefore, the newly added pediatric training set can improve the recognition accuracy of pediatric CT images. In this study, our proposed network demonstrated excellent automatic segmentation performance during the test process, regardless of whether it was adult or pediatric data. Although there were statistical differences in the accuracy metrics of automatic segmentation between adult and pediatric temporal CT images for a few structures, all our approaches achieved satisfactory automatic segmentation results.
The accurate and rapid segmentation of temporal bone CT images is one of the critical techniques in the clinical application of image-guided otologic surgery. This technological approach frees clinicians from labor-intensive operations. It enables them to focus on the most critical steps of surgery, thereby reducing the fatigue and the possibility of surgical errors. The automatic segmentation of CT images also provides significant support for the application of virtual reality technology in the field of otology, and it further improves the safety of temporal bone surgery (44). In the preoperative evaluation stage, the intelligent identification of sensitive structures can prompt the surgeon to formulate an optimal surgical plan by realizing the risk warning function of temporal bone surgery. In addition, the automated segmentation of important structures will further realize the intelligent diagnosis of ear diseases based on CT images (45). The 3D reconstruction of the anatomical structure using clinical CT volumes can further establish a temporal bone surgery simulation system using a huge amount of data, strengthening the student’s understanding of anatomy, surgeons’ surgical rehearsals, and clinical education (46,47).
This study has several limitations. First, a small training dataset was used to build a neural network model. Because manual segmentation is extremely labor intensive, it was not possible to provide additional training data in a short time. This problem was also apparent in the test sets. Therefore, we are planning to expand the training and test set size further to improve the CNN model’s performance in the future. Second, all the annotated CT image structures were collected from a single institution. In our future studies, we shall include more annotated CT volumes from different institutions and scanners to further improve the generalizability of the CNN model. Third, abnormal anatomical structures were not included in this study. In a subsequent study, we shall consider the automatic segmentation of malformed structures in the temporal bone.
Conclusions
In conclusion, we trained and assessed a neural network that automatically segmented almost all temporal bone anatomy structures in this study. The proposed network can be used to accurately segment small structures and larger organs in the temporal bone. We further showed that the proposed network model has excellent generalizability for adult and pediatric CT images. The CNN model automated segmentation of temporal bone. This can help advance otologist education, intelligent imaging diagnosis, surgery simulation, application of augmented reality, and preoperative planning for image-guided otology surgery.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Funding: This research was supported by grants from the China Capital Health Development Project (No. 2016-2-4094), Key Clinical Project of Peking University Third Hospital (Nos. BYSY2017025 and BYSYZD2021015), and the National Natural Science Foundation of China (Nos. 61701014 and 61911540075).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-658/rc
Conflicts of Interest: All authors completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-658/coif). The authors report that this research was supported by grants from the China Capital Health Development Project (No. 2016-2-4094), Key Clinical Project of Peking University Third Hospital (Nos. BYSY2017025 and BYSYZD2021015), and the National Natural Science Foundation of China (Nos. 61701014 and 61911540075). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Peking University Third Hospital Medical Ethics Committee (No. IRB00006761- M2019335). The requirement for written informed consent was waived due to the retrospective nature of our study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Neves CA, Tran ED, Kessler IM, Blevins NH. Fully automated preoperative segmentation of temporal bone structures from clinical CT scans. Sci Rep 2021;11:116. [Crossref] [PubMed]
- Zagzoog N, Yang VXD. State of Robotic Mastoidectomy: Literature Review. World Neurosurg 2018;116:347-51. [Crossref] [PubMed]
- Li X, Gong Z, Yin H, Zhang H, Wang Z, Zhuo L. A 3D deep supervised densely network for small organs of human temporal bone segmentation in CT images. Neural Netw 2020;124:75-85. [Crossref] [PubMed]
- Wang J, Liu H, Ke J, Hu L, Zhang S, Yang B, Sun S, Guo N, Ma F. Image-guided cochlear access by non-invasive registration: a cadaveric feasibility study. Sci Rep 2020;10:18318. [Crossref] [PubMed]
- McBrayer KL, Wanna GB, Dawant BM, Balachandran R, Labadie RF, Noble JH. Resection planning for robotic acoustic neuroma surgery. J Med Imaging (Bellingham) 2017;4:025002. [Crossref] [PubMed]
- Dillon NP, Balachandran R, Siebold MA, Webster RJ 3rd, Wanna GB, Labadie RF. Cadaveric Testing of Robot-Assisted Access to the Internal Auditory Canal for Vestibular Schwannoma Removal. Otol Neurotol 2017;38:441-7. [Crossref] [PubMed]
- Powell KA, Kashikar T, Hittle B, Stredney D, Kerwin T, Wiet GJ. Atlas-based segmentation of temporal bone surface structures. Int J Comput Assist Radiol Surg 2019;14:1267-73. [Crossref] [PubMed]
- Gare BM, Hudson T, Rohani SA, Allen DG, Agrawal SK, Ladak HM. Multi-atlas segmentation of the facial nerve from clinical CT for virtual reality simulators. Int J Comput Assist Radiol Surg 2020;15:259-67. [Crossref] [PubMed]
- Reda FA, McRackan TR, Labadie RF, Dawant BM, Noble JH. Automatic segmentation of intra-cochlear anatomy in post-implantation CT of unilateral cochlear implant recipients. Med Image Anal 2014;18:605-15. [Crossref] [PubMed]
- Noble JH, Labadie RF, Majdani O, Dawant BM. Automatic segmentation of intracochlear anatomy in conventional CT. IEEE Trans Biomed Eng 2011;58:2625-32. [Crossref] [PubMed]
- Powell KA, Liang T, Hittle B, Stredney D, Kerwin T, Wiet GJ. Atlas-Based Segmentation of Temporal Bone Anatomy. Int J Comput Assist Radiol Surg 2017;12:1937-44. [Crossref] [PubMed]
- Minnema J, van Eijnatten M, Kouw W, Diblen F, Mendrik A, Wolff J. CT image segmentation of bone for medical additive manufacturing using a convolutional neural network. Comput Biol Med 2018;103:130-9. [Crossref] [PubMed]
- Lv Y, Ke J, Xu Y, Shen Y, Wang J, Wang J. Automatic segmentation of temporal bone structures from clinical conventional CT using a CNN approach. Int J Med Robot 2021;17:e2229. [Crossref] [PubMed]
- Khan MA, Kwon S, Choo J, Hong SM, Kang SH, Park IH, Kim SK, Hong SJ. Automatic detection of tympanic membrane and middle ear infection from oto-endoscopic images via convolutional neural networks. Neural Netw 2020;126:384-94. [Crossref] [PubMed]
- Zhou X. Automatic Segmentation of Multiple Organs on 3D CT Images by Using Deep Learning Approaches. Adv Exp Med Biol 2020;1213:135-47. [Crossref] [PubMed]
- Lin H, Xiao H, Dong L, Teo KB, Zou W, Cai J, Li T. Deep learning for automatic target volume segmentation in radiation therapy: a review. Quant Imaging Med Surg 2021;11:4847-58. [Crossref] [PubMed]
- Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM, Giger ML. Deep learning in medical imaging and radiation therapy. Med Phys 2019;46:e1-36. [Crossref] [PubMed]
- Kong Z, Li T, Luo J, Xu S. Automatic Tissue Image Segmentation Based on Image Processing and Deep Learning. J Healthc Eng 2019;2019:2912458. [Crossref] [PubMed]
- Xia X, Kulis B. W-net: A deep model for fully unsupervised image segmentation. arXiv 2017. arXiv:1711.08506.
- van den Noort F, van der Vaart CH, Grob ATM, van de Waarsenburg MK, Slump CH, van Stralen M. Deep learning enables automatic quantitative assessment of puborectalis muscle and urogenital hiatus in plane of minimal hiatal dimensions. Ultrasound Obstet Gynecol 2019;54:270-5. [Crossref] [PubMed]
- Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, Liu T, Yang X. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys 2019;46:2157-68. [Crossref] [PubMed]
- Guo X, Schwartz LH, Zhao B. Automatic liver segmentation by integrating fully convolutional networks into active contour models. Med Phys 2019;46:4455-69. [Crossref] [PubMed]
- Chan JW, Kearney V, Haaf S, Wu S, Bogdanov M, Reddick M, Dixit N, Sudhyadhom A, Chen J, Yom SS, Solberg TD. A convolutional neural network algorithm for automatic segmentation of head and neck organs at risk using deep lifelong learning. Med Phys 2019;46:2204-13. [Crossref] [PubMed]
- Ren G, Xiao H, Lam SK, Yang D, Li T, Teng X, Qin J, Cai J. Deep learning-based bone suppression in chest radiographs using CT-derived features: a feasibility study. Quant Imaging Med Surg 2021;11:4807-19. [Crossref] [PubMed]
- Nikan S, Van Osch K, Bartling M, Allen DG, Rohani SA, Connors B, Agrawal SK, Ladak HM. PWD-3DNet: A Deep Learning-Based Fully-Automated Segmentation of Multiple Structures on Temporal Bone CT Scans. IEEE Trans Image Process 2021;30:739-53. [Crossref] [PubMed]
- Kingma DP, Ba JL. Adam: A Method for Stochastic Optimization. Available online: http://www.arxiv.org/pdf/1412.6980.pdf
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. editors. MICCAI 2015. Cham: Springer, 2015.
- Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297-302. [Crossref]
- Rashed EA, Gomez-Tames J, Hirata A. End-to-end semantic segmentation of personalized deep brain structures for non-invasive brain stimulation. Neural Netw 2020;125:233-44. [Crossref] [PubMed]
- Park J, Yun J, Kim N, Park B, Cho Y, Park HJ, Song M, Lee M, Seo JB. Fully Automated Lung Lobe Segmentation in Volumetric Chest CT with 3D U-Net: Validation with Intra- and Extra-Datasets. J Digit Imaging 2020;33:221-30. [Crossref] [PubMed]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 2004;23:903-21. [Crossref] [PubMed]
- Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imaging 1994;13:716-24. [Crossref] [PubMed]
- Noble JH, Warren FM, Labadie RF, Dawant BM. Automatic segmentation of the facial nerve and chorda tympani in CT images using spatially dependent feature values. Med Phys 2008;35:5375-84. [Crossref] [PubMed]
- Kirsch V, Nejatbakhshesfahani F, Ahmadi SA, Dieterich M, Ertl-Wagner B. A probabilistic atlas of the human inner ear's bony labyrinth enables reliable atlas-based segmentation of the total fluid space. J Neurol 2019;266:52-61. [Crossref] [PubMed]
- Bi H, Jiang Y, Tang H, Yang G, Shu H, Dillenseger JL. Fast and accurate segmentation method of active shape model with Rayleigh mixture model clustering for prostate ultrasound images. Comput Methods Programs Biomed 2020;184:105097. [Crossref] [PubMed]
- Zeng YZ, Liao SH, Tang P, Zhao YQ, Liao M, Chen Y, Liang YX. Automatic liver vessel segmentation using 3D region growing and hybrid active contour model. Comput Biol Med 2018;97:63-73. [Crossref] [PubMed]
- Noble JH, Dawant BM, Warren FM, Labadie RF. Automatic identification and 3D rendering of temporal bone anatomy. Otol Neurotol 2009;30:436-42. [Crossref] [PubMed]
- Hussain R, Lalande A, Girum KB, Guigou C, Bozorg Grayeli A. Automatic segmentation of inner ear on CT-scan using auto-context convolutional neural network. Sci Rep 2021;11:4406. [Crossref] [PubMed]
- Kim G, Jeoun BS, Yang S, Kim J, Lee SJ, Yi WJ. CAPPU-Net: A Convolutional Attention Network with Pyramid Pooling for Segmentation of Middle and Inner Ear Structures in CT Images. 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Oct 31-Nov 4, 2021. Virtual Conference.
- Wang Z, Demarcy T, Vandersteen C, Gnansia D, Raffaelli C, Guevara N, Delingette H. Bayesian logistic shape model inference: Application to cochlear image segmentation. Med Image Anal 2022;75:102268. [Crossref] [PubMed]
- Kjer HM, Paulsen RR. Modelling of the Human Inner Ear Anatomy and Variability for Cochlear Implant Applications. Kongens Lyngby: Technical University of Denmark, 2016.
- Kjer HM, Fagertun J, Wimmer W, Gerber N, Vera S, Barazzetti L, Mangado N, Ceresa M, Piella G, Stark T, Stauber M, Reyes M, Weber S, Caversaccio M, González Ballester MÁ, Paulsen RR. Patient-specific estimation of detailed cochlear shape from clinical CT images. Int J Comput Assist Radiol Surg 2018;13:389-96. [Crossref] [PubMed]
- Reda FA, Noble JH, Rivas A, McRackan TR, Labadie RF, Dawant BM. Automatic segmentation of the facial nerve and chorda tympani in pediatric CT scans. Med Phys 2011;38:5590-600. [Crossref] [PubMed]
- Hussain R, Lalande A, Marroquin R, Guigou C, Bozorg Grayeli A. Video-based augmented reality combining CT-scan and instrument position data to microscope view in middle ear surgery. Sci Rep 2020;10:6767. [Crossref] [PubMed]
- Wang YM, Li Y, Cheng YS, He ZY, Yang JM, Xu JH, Chi ZC, Chi FL, Ren DD. Deep Learning in Automated Region Proposal and Diagnosis of Chronic Otitis Media Based on Computed Tomography. Ear Hear 2020;41:669-77. [Crossref] [PubMed]
- Wijewickrema S, Piromchai P, Zhou Y, Ioannou I, Bailey J, Kennedy G, O'Leary S. Developing effective automated feedback in temporal bone surgery simulation. Otolaryngol Head Neck Surg 2015;152:1082-8. [Crossref] [PubMed]
- Chauvelot J, Laurent C, Le Coz G, Jehl JP, Tran N, Szczetynska M, Moufki A, Bonnet AS, Parietti-Winkler C. Morphological validation of a novel bi-material 3D-printed model of temporal bone for middle ear surgery education. Ann Transl Med 2020;8:304. [Crossref] [PubMed]



