
Multicenter quantitative 18F-fluorodeoxyglucose positron emission tomography performance harmonization: use of hottest voxels towards more robust quantification
Introduction
Standardized uptake value (SUV) is widely used in quantitative analysis of 18F-fluorodeoxyglucose (FDG) Positron emission tomography/computed tomography (PET/CT) imaging and quantifies metabolically active regions of interest images (1). While visual inspection is the primary method for interpreting 18F-FDG PET/CT images, the quantitative information from SUV is used to assess lesion diagnostic and prognostic factors and treatment response in clinical practice (2-6).
Although SUV is a simple image-based measure, at the same time, various biological, patient-related, technical, and physical factors can hamper its reliability and utility (7-10). In a study by Boellaard et al. (8), different factors affecting FDG PET quantification were reviewed, and a typical range and maximum effect were reported for each factor. If we neglect the technical errors (e.g., paravenous injection) and biological factors (e.g., blood glucose level), the most important physical factors in FDG PET quantifications are scan acquisition parameters, image reconstruction parameters, ROI definition, and SUV normalizations (8). These parameters can significantly affect the accuracy of 18F-FDG PET/CT quantification and lead to variability in SUV measurements (11-13). Also, partial volume effects (PVE) due to the limited spatial resolution of the PET/CT scanner, tumor size, background activity in the lesion’s surroundings, voxel size, and image sampling could affect the accuracy of quantification (14).
Even though combined PET/CT scanners have been in clinical operation for several years, the technology has undergone rapid advances in hardware, software, and acquisition methods. The current generation of PET/CT scanners are equipped with point spread function (PSF) correction, time of flight (TOF) capability, and vendor-developed image reconstruction algorithms that improve the diagnostic performance of scanners, image quality, and lesion detection, but it has led to scanner and image reconstruction-dependent variability in SUV calculations (2,15,16). Although these algorithms are valuable, they have significantly produced upward-biased recovery coefficients (RCs) in phantom studies and SUV in patient studies compared with the routine iterative reconstruction algorithm (17). RC represents the ratio of measured voxels uptake to the static ground truth case as a function of sphere size in phantom research. Therefore, variable quantitative results should be considered while comparing SUVs for therapy monitoring in multicenter clinical studies (18).
In recent years, several guidelines have been recommended for patient preparation, normalization, image acquisition and reconstruction, processing, and clinical interpretation (19,20). Harmonization strategies are needed to achieve more reproducibility in SUV measurements and assure quantitative comparability (21,22). Some groups, such as EANM/EARL, QIBA/UPICT, and SNMMI/CTN, have been able to provide harmonization programs (11). These programs are based on PET scanner calibration compliance to standard imaging guidelines using reconstruction via applying appropriate filters (7) to extract comparable quantitative metrics from all accredited centers (17). The reference SUV (SUVref) methodology was proposed in 2011 to minimize the non-biological effects influencing SUV calculation. In this method, a specific smoothing filter is used only for quantification in clinical data analysis, besides a reconstructed image for visualization with the best quality (7).
Images post-filtered for quantification can reduce SUV variability in PET images (7) but can cause blurring in images (3). Considering reconstruction-dependent variations, EARL has proposed applying a filter during reconstruction using advanced algorithms to meet the specific range of harmonizing standard reference values of RC (23). Some centers use two reconstructions, one for optimized visualization and diagnosis of lesions and another for quantitative analysis to meet harmonization (17).
Accuracy and reproducibility are essential when reporting any quantitative metrics in multicenter trials. Reproducibility refers to inter-scanner variability when the same patient is scanned across different PET/CT scanners (13,21) and indicates any uncertainty originating from acquisition, image reconstruction algorithms, and image analysis techniques in various institutes (13). High reproducibility might be more important than absolute accuracy in therapy assessment (11). Harmonization methods reduce variability, causing SUVs to be comparable regardless of the make and model of PET scanners (23).
In this study, we assessed the RC of average-pooling ten hottest voxels (RC10V), besides other quantitative metrics frequently used for harmonization. Using the average of several hottest voxels regardless of their location within the tumor might be helpful in the quantification of its aggressive portion while it does not require segmentation. We also focused on the reproducibility of quantitative parameters in different scanners by determining sets of image reconstruction parameters that yield optimal matches in quantitative performance as a function of object size. By selecting the appropriate reconstruction algorithms, we provide harmonizing criteria range with an accurate RC value and explore quantitative metrics for a comparable and robust biomarker of metabolic activity in a multicenter setting.
Methods
A schematic representation of the workflow applied in our study is shown in Figure 1.
PET/CT scanners and phantom data acquisition
Using the NEMA IEC Phantom, data acquisition was performed on six PET/CT scanners from different vendors. An experienced medical physicist was responsible for all phantom preparations and measurements. More information about vendors, the model of the scanners, and their performance parameters are presented in Table 1. Each scanner was calibrated following the corresponding manufacturer’s instructions. Cross calibration for each scanner was examined by measurement of RCmean in the background of the NEMA IEC body phantom.
Table 1
| Characteristics | Siemens Biograph6 TruePoint TrueV | Siemens Biograph6 TruePoint | Siemens Biograph-mCT | GE Discovery690 | Philips Ingenuity TF | Mediso Anyscan |
|---|---|---|---|---|---|---|
| Crystal material | LSO | LSO | LSO | LYSO | LYSO | LYSO |
| Crystal size (mm3) | 4.0×4.0×20 | 4.0×4.0×20 | 4.0×4.0×20 | 4.2×6.3×25 | 4.0×4.0×22 | 3.9×3.9×20 |
| Axial field of view (cm) | 21.6 | 16.2 | 16.2 | 15.7 | 18 | 15.2 |
| Photodetector | PMT | PMT | PMT | PMT | PMT | PMT |
| Sensitivity at the center (cps/kBq) | 7.6 | 4.2 | 5.8 | 7 | 7.4 | 5.8 |
| Axial resolution @1/10 cm | 5.5/6 | 5.5/6 | 5.5/6 | 5.6/6.3 | 4.7/5.2 | 4.9/5.7 |
| Transaxial resolution @1/10 cm | 5.9/6 | 5.9/6 | 5.9/6 | 4.9/5.5 | 4.7/5.2 | 4.8/5.6 |
| PSF algorithm | Y(TrueX) | Y(TrueX) | Y(TrueX) | Y(VUE Point FX) | Y(PSF) | Y(Tera-Tomo) |
| TOF algorithm | N/A | N/A | N/A | Y(VPF–SharpIR) | Y | N/A |
PET/CT, positron emission tomography/computer tomography; cps, count per second; PSF, point spread function; TOF, time of flight; LSO, lutetium oxyorthosilicate; PMT, photomultiplier tube; N/A, not available; LYSO, lutetium yttrium oxyorthosilicate.
This study used the NEMA IEC body phantom with the standard fillable spheres (diameters 10, 13, 17, 22, 28, and 37 mm). A standardized filling procedure was implemented to achieve low and high spheres-to-background ratios (SBR4:1 and 10:1) with a background activity concentration of approximately two kBq/mL at the scan time. As phantom preparation takes about 60 minutes, a homogenous solution of 18F-FDG was used to fill the NEMA IEC Phantom background (total activity: 40–50 MBq). The NEMA IEC Phantom was filled with two different activity concentrations for each scanner. To simulate clinical oncology situations, variably sized spheres with SBR4 were used; however, spheres with SBR10 enable a more precise measurement of the activity concentrations (24). All spheres were then filled with about 15–20 and 40–50 KBq/mL to make SBRs of 4:1 and 10:1 (with total activity of 1–1.5 and 3–4 MBq, respectively). The PET acquisition was a single bed with hot spheres placed in the center of the axial FOV. The CT-based attenuation correction (CTAC) PET image was then analyzed using vendor-provided software. Phantom data were acquired for 3 minutes at the same condition for all investigated PET/CT scanners.
Image reconstruction algorithms
PET image reconstructions were performed using vendor-provided software. To investigate the effect of reconstruction parameters on quantitative metrics, PET images were reconstructed using iterative algorithms with various numbers of iterations and subsets (i×s) and different levels of smoothing. For Ingenuity TF PET/CT scanner, PET image was reconstructed using a 3D row action maximum likelihood algorithm (3D RAMLA). Clinical vendor-recommended reconstruction parameter sets were also applied if advanced algorithms such as PSF and TOF were available. For example, in Ingenuity TF standard body image reconstruction, list-mode TOF algorithm and line-of-response TruFlight (LOR-TF) RAMLA method, so-called BLOB-OS-TF, was used with the combination of 3×33 according to the manufacturer’s recommendation. Normalization, scatter, attenuation, and decay corrections were applied during reconstructions. Image reconstruction settings for each scanner are presented in Table 2.
Table 2
| Reconstruction parameter | Siemens Biograph6 TruePoint (TrueV*) | Siemens Biograph6 TruePoint | Siemens Biograph-mCT | GE Discovery 690 | Philips Ingenuity TF | Mediso Anyscan |
|---|---|---|---|---|---|---|
| Algorithm | OSEM**-3D | OSEM-3D | OSEM-3D | OSEM-3D | BLOB-OS-TF*** | OSEM-3D |
| OSEM + PSF | OSEM + PSF | OSEM + PSF | OSEM + PSF | BLOB-OS-TF + PSF | OSEM + PSF | |
| OSEM + PSF + TOF | ||||||
| Iterations × subset | 2×8 | 2×8 | 2×8 | 2×8 | 1×6**** | 2×6 |
| 2×14 | 2×14 | 2×12 | 2×12 | 2×6 | 2×12 | |
| 4×8 | 4×8 | 4×8 | 2×18 | 2×12 | 3×12 | |
| 2×21 | 2×21 | 2×24 | 2×24 | 2×18 | 4×12 | |
| 4×14 | 4×14 | 3×24 | 3×18 | 3×10 | ||
| 3×21 | 3×21 | 2×32 | ||||
| FWHM of Gaussian filter (mm) | 2 | 2 | 2 | 2 | Normal | 2 |
| 4 | 4 | 4 | 4 | Smooth | 4 | |
| 6 | 6 | 6 | 6 | Smooth A | 6 | |
| 8 | 8 | 8 | 8 | Smooth B | ||
| 10 | 10 | 10 | ||||
| Image matrix | 168×168 | 168×168 | 200×200 | 256×256 | 144×144 | 334×334 |
| Pixel size (mm) | 4.07 | 4.07 | 4.07 | 2.73 | 4 | 2 |
*, TrueV option is increased axial field of view and higher number of image planes; **, ordered subset expectation maximization-3D; ***, Philips Ingenuity TF reconstruction: iterative reconstruction algorithm (BLOB-OS-TF) including TOF with Body (i×s=3×33) and Body-EARL mode; ****, different recovery iterations and widths of Gaussian regularization kernel for Philips Ingenuity TF (i × regularization kernel). PET/CT, positron emission tomography/computer tomography; FWHM, full width at half maximum; OSEM, ordered subset expectation maximization; 3D, 3dimentional; PSF, point spread function; TOF, time of flight.
Experienced medical physicists define a range of quantitative accurate reconstruction sets based on vendor recommendations and phantom studies in each center. Experienced physicians subjectively choose their preferred reconstructed PET images for their clinical routine across those settings, now called center standard clinical reconstruction set(s). They were compared to the proposed harmonized reconstruction sets to determine the difference between the optimized and harmonized reconstruction settings.
Quantitative metrics
One transverse slice of the reconstructed image centered on spheres was selected to visualize all spheres with the highest contrast. The same slice was used for the analysis of all spheres. Spherical volume of interest (VOI) was drawn manually on the slice mentioned above with a diameter as close as possible to the sphere diameter. Since the reconstruction setting leads to the variation of volume of interest, spherical VOI was drawn in the PET images as closely as possible to the true volume of the sphere in CT images (25-27). After that, RC was obtained by dividing the measured activity concentration by the true activity concentration. For cross-calibration, twelve spherical VOI with a 3 cm diameter within the phantom background were drawn, and the average of RCmean was calculated in each scanner. Less than a 5% difference in the RCmean is acceptable and does not need extra quality control tests (4). Quantitative analysis was performed using RCs of maximum voxel value (RCmax), the mean value of 50% isocontour of the maximum voxel value (RCISO-50), the mean value of a 1 mL spherical VOI centered on the highest uptake region (RCpeak), and mean value for ten highest uptake voxels within VOI as shown by RC10V. Interested readers are referred to Figure S1 for comparing peak and ten voxels VOIs in a transverse slice of the NEMA IEC Phantom. It has been suggested that averaging several hottest voxels, regardless of their location within the tumor, can be more accurate than SUVmax or SUVpeak (28,29). In addition, variability in SUV measurements can be further reduced by average-pooling ten hottest voxels (30). Therefore, the number of 10 voxels was selected to evaluate reproducibility in this study. In addition, the mean value of RCs for all sphere sizes (MRC) was calculated for each reconstruction setting and RC method.
SBR impact on quantification
The difference between quantitative metrics in low and high SBRs was analyzed in all reconstructed PET images of NEMA IEC Phantom using the paired t-test and Wilcoxon signed rank test for parametric and non-parametric distribution data, respectively. Interested readers are referred to Figure S2 for additional information on the importance of SBR on quantification. In this study, statistical analyses were carried out using GraphPad PRISM software (version 8.0.2), and the Significance level was set at a P value less than 0.05 for all comparisons.
The correlation between SBR10 and SBR4 per scanner was evaluated using the spearman rank correlation coefficient. Furthermore, the Lin concordance correlation coefficient (Lin’s CCC) was used to quantify the agreement between both SBRs. Lin’s CCC was introduced as a new reproducibility index of paired variables (31). Both correlation coefficient indexes were assessed in three categories of sphere sizes: (I) all diameters; (II) diameters more than 20 mm; and (III) diameters less than 20 mm.
Optimized and harmonized reconstruction parameter sets
To find reconstruction parameter sets providing accurate quantification, the difference in the RC values between the reconstructed PET images and reference value (unity) was evaluated according to the root mean square error (RMSE) (11,32):
While the deviation between the RC values of all spheres and the RC value of the sphere with the largest diameter (37 mm) was calculated using curvature:
RCmax is independent of the observer and obtained from the single hottest voxel, though it is susceptible to variations by the noise level, which leads to uncertainty in quantification (33). Therefore, the optimization of the reconstruction task from the perspective of quantification accuracy was assessed only for RCmax as the most dependent parameter to the reconstruction setting. However, quantification of PET images in a multicenter setting is still hampered by differences in applied acquisition and reconstruction settings; therefore mean value of RCmax (MRCmax) was plotted against RMSE in two different SBRs, and reconstruction parameter sets with the lowest RMSE will be considered.
Optimized reconstruction settings per scanner were derived by considering: (I) higher RC values; (II) smaller RMSE values (excluded if the value is bigger than 0.3); (III) less upward bias due to edge artifact (overshoot at sharp intensity transitions of the object caused by PSF based image reconstruction) in larger spheres. Reconstruction parameter sets with a similar bandwidth of RCs or RC biases that fall within the range of ±10% (4,34) were taken as the appropriate reconstruction settings for harmonization.
Reproducibility
To evaluate the reproducibility of quantitation, the coefficient of variation of the MRC (CVMRC) was used to characterize agreement across various reconstruction settings in all PET/CT scanners for both SBR4 and SBR10.
Results
The initial data analysis phase was checking calibration between the dose calibrator and the PET scanner. RCmean in the background of the NEMA IEC body phantom for both SBRs was calculated for each scanner. RCmean values were 0.95, 1, 1.05, 0.97, 1, and 0.99 for Biograph6 TruePoint (TrueV), Biograph6 TruePoint, Biograph mCT, Discovery690, Ingenuity TF, and Mediso Anyscan, respectively. All calculated values fall within the range of 1±0.05.
Effects of SBR
Statistical analysis of the comparison between quantitative metrics derived from SBR10 and SBR4 is presented in Table S1. A statistically significant difference between both SBRs in all the scanners was observed (P value <0.0001), except in the smallest sphere size.
According to Table 3, the Spearman correlation coefficient was greater than 0.9 in small spheres with a diameter of less than 20 mm. Conversely, it was reduced in spheres with a diameter of more than 20 mm. The correlation between data does not mean agreement. Most scanners showed poor agreement (Lin’s CCC <0.9); thus, there is a noticeable difference between low and high SBRs. Only a few cases showed substantial agreement (Lin’s CCC >0.95), including RCmax, RCISO-50, and RCpeak obtained in the Ingenuity TF and Biograph mCT PET/CT scanners. Quantitative indices of the images acquired by Ingenuity TF demonstrated the highest agreement in both SBRs with the other PET/CT scanners. However, our results indicated a significant difference in the quantitative metrics derived from images with high and low SBRs. Accordingly, the following section assesses reconstruction settings to find separately optimized and harmonized reconstruction parameter sets for either SBR.
Table 3
| Quantitative metrics | PET/CT scanners | All diameters | Diameters >20 mm | Diameters <20 mm | |||||
|---|---|---|---|---|---|---|---|---|---|
| Spearman/r | Lin’s CCC | Spearman/r | Lin’s CCC | Spearman/r | Lin’s CCC | ||||
| RCmax | Biograph6 TruePoint (TrueV) | 0.90 | 0.87 | 0.75 | 0.62 | 0.96 | 0.82 | ||
| Biograph6 TruePoint | 0.93 | 0.92 | 0.86 | 0.81 | 0.94 | 0.84 | |||
| Biograph mCT | 0.94 | 0.95 | 0.81 | 0.81 | 0.98 | 0.94 | |||
| Discovery690 | 0.87 | 0.85 | 0.81 | 0.79 | 0.94 | 0.77 | |||
| Ingenuity TF | 0.95 | 0.96 | 0.99 | 0.97 | 0.95 | 0.93 | |||
| Anyscan | 0.94 | 0.93 | 0.82 | 0.86 | 0.95 | 0.87 | |||
| RCISO-50 | Biograph6 TruePoint (TrueV) | 0.92 | 0.85 | 0.83 | 0.65 | 0.94 | 0.75 | ||
| Biograph6 TruePoint | 0.94 | 0.91 | 0.92 | 0.90 | 0.92 | 0.78 | |||
| Biograph mCT | 0.96 | 0.94 | 0.89 | 0.89 | 0.97 | 0.87 | |||
| Discovery690 | 0.93 | 0.87 | 0.89 | 0.70 | 0.95 | 0.79 | |||
| Ingenuity TF | 0.96 | 0.96 | 0.99 | 0.98 | 0.95 | 0.93 | |||
| Anyscan | 0.92 | 0.88 | 0.68 | 0.73 | 0.95 | 0.85 | |||
| RCpeak | Biograph6 TruePoint (TrueV) | 0.96 | 0.93 | 0.75 | 0.57 | 0.98 | 0.90 | ||
| Biograph6 TruePoint | 0.98 | 0.97 | 0.87 | 0.90 | 0.97 | 0.91 | |||
| Biograph mCT | 0.97 | 0.97 | 0.87 | 0.89 | 0.98 | 0.89 | |||
| Discovery690 | 0.96 | 0.94 | 0.74 | 0.59 | 0.98 | 0.87 | |||
| Ingenuity TF | 0.98 | 0.97 | 0.94 | 0.89 | 0.96 | 0.96 | |||
| Anyscan | 0.96 | 0.93 | 0.77 | 0.72 | 0.96 | 0.89 | |||
Pairwise statistical differences were carried out by sign test (data was not shown in the table). P value <0.0001 was observed in all cases. SBR, spheres-to-background ratio; Lin’s CCC, Lin’s concordance correlation coefficient; RC, recovery coefficient; PET/CT, positron emission tomography/computer tomography; TF, time of flight.
Proposed harmonized reconstruction settings
The mean value of RCmax for all sphere sizes (MRCmax) against RMSE for SBR10 and SBR4 is shown in Figure 2. The distribution of reconstruction parameter sets based on MRCmax versus RMSE was plotted. On these graphs, the colony of points indicates harmonized reconstructions among the six PET/CT scanners. Ideally, the RC value should be 1; thus, with a 10% bias, the range of acceptable RCmax was selected between 0.9 and 1.1 to determine the optimal reconstruction setting with high accuracy. Although several optimal reconstructions exist considering the RC value, we chose the settings with the smallest RMSE value. Mediso Anyscan PET/CT scanner was excluded because the RMSE of its reconstructions was too high. The red box in Figure 2 is the expanded view of the reconstruction parameter sets with low RMSE and accurate RCmax value to identify the reconstruction candidate for both optimization and harmonization.
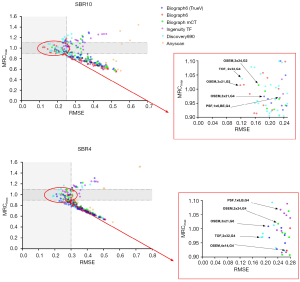
The analysis mentioned above of all the reconstruction setups resulted in several reconstruction parameter sets appropriate for harmonization between the scanners. To accommodate unavoidable inter-scanner variability and errors due to system calibration, all the RC ranges were defined to bandwidth, taking into account decreased variability among the investigated scanners. RCs as a function of sphere size for the suggested reconstruction settings are plotted (Figure 3). These plots depict the range of RC values proposed for harmonization. RC specifications based on the recommended reconstruction settings for harmonization are presented in Table 4.
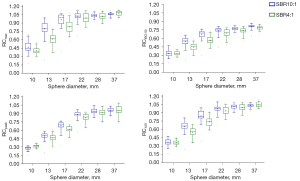
Table 4
| Diameter (mm) | SBR10 | SBR4 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RCmax | RCISO-50 | RCpeak | RC10V | RCmax | RCISO-50 | RCpeak | RC10V | ||
| 10 | 0.4–0.54 | 0.29–0.37 | 0.25–0.29 | 0.33–0.41 | 0.36–0.44 | 0.3–0.36 | 0.29–0.33 | 0.33–0.39 | |
| 13 | 0.74–0.89 | 0.5–0.6 | 0.47–0.53 | 0.61–0.71 | 0.54–0.67 | 0.39–0.49 | 0.42–0.5 | 0.5–0.61 | |
| 17 | 0.88–1.05 | 0.62–0.71 | 0.65–0.73 | 0.8–0.93 | 0.74–0.91 | 0.52–0.61 | 0.58–0.66 | 0.66–0.79 | |
| 22 | 0.99–1.07 | 0.72–0.77 | 0.85–0.91 | 0.94–1 | 0.92–1.07 | 0.65–0.75 | 0.79–0.88 | 0.88–0.99 | |
| 28 | 1.01–1.06 | 0.74–0.79 | 0.91–0.98 | 0.97–1.04 | 0.98–1.08 | 0.71–0.77 | 0.88–0.96 | 0.95–1.04 | |
| 37 | 1.04–1.08 | 0.8–0.83 | 0.94–0.99 | 1.01–1.05 | 1.04–1.11 | 0.76–0.8 | 0.91–1.03 | 1.01–1.09 | |
RC, recovery coefficient; SBR, spheres-to-background ratio.
A summary of the standard clinical reconstruction parameters and proposed parameter sets for harmonization are presented in Table 5. The reconstruction algorithm, the number of iterations × subsets, and full width at half maximum (FWHM) of the Gaussian filter are reported in Table 5. Remarkably similar reconstruction sets are recommended for high and low SBRs.
Table 5
| Scanner | Clinical reconstruction settings | Voxel size (cm3)*** | Proposed reconstruction settings for harmonization | ||||
|---|---|---|---|---|---|---|---|
| Algorithm | i×s | Gaussian filter | Algorithm | i×s | Gaussian filter | ||
| Biograph mCT | OSEM + PSF | 2×24 | 4 mm | 0.049 | A: OSEM + PSF | 3×24 | 6 mm |
| B: OSEM-3D | 3×24 | 4 mm | |||||
| Biograph6 TruePoint | OSEM + PSF | 2×21 | 5 mm | 0.05 | A: OSEM + PSF | 3×21 | 6 mm |
| B: OSEM-3D | 2×21 | 2 mm | |||||
| Biograph6 TruePoint (TrueV) | OSEM + PSF | 2×21 | 3 mm | 0.034 | A: OSEM + PSF | 3×21 | 6 mm |
| B: OSEM-3D | 3×21 | 6 mm | |||||
| Anyscan | OSEM + PSF | 2×6 | 0 | 0.008 | – | – | – |
| Ingenuity TF-A | BLOB-OS-TF | Body | Smooth A (6 mm) | 0.064 | BLOB-OS-TF + PSF | 1×6** | Smooth (4 mm) |
| Ingenuity TF-B | BLOB-OS-TF + PSF | 1×6* | Smooth A (6 mm) | 0.064 | BLOB-OS-TF | Body | Normal (4 mm) |
| Discovery 690-A | OSEM + PSF | 3×18 | 6.4 mm | 0.025 | OSEM + PSF | 3×18 | 6 mm |
| Discovery 690-B | OSEM + PSF + TOF | 2×18 | 6.4 mm | 0.025 | OSEM + PSF + TOF | 3×18 | 6 mm |
*, image reconstruction in Ingenuity TF was performed using PSF-based MLEM deconvolution resolution recovery with a combination of PSF iteration × regularization in Body mode defined in the scanner; **, combination of PSF iteration × regularization in Body-EARL mode, defined in Ingenuity TF scanner; ***, the voxel size for each scanner is the same in both clinical reconstruction and proposed harmonized reconstruction settings. OSEM, ordered subset expectation maximization; PSF, point spread function; TOF, time of flight; 3D, 3 dimentional; MLEM, Maximum-Likelihood Expectation-Maximization.
The RMSE and curvature values for all RC metrics in SBR4 and SBR10 for clinical and harmonized reconstruction settings are presented in Table S2. Accordingly, RMSE values for routine clinical reconstructions in SBR10 were within the range of 0.18–0.46, 0.27–0.55, 0.32–0.53, and 0.24–0.47 for RCmax, RCISO-50, RCpeak, and RC10V, respectively. The proposed harmonized reconstructions reduced RMSE in the ranges of 0.14–0.26, 0.3–0.42, 0.34–0.41, and 0.22–0.34 for RCmax, RCISO-50, RCpeak, and RC10V, respectively. The curvature values of RCmax, RCISO-50, RCpeak, and RC10V in the standard clinical reconstructions were within the ranges of 0.2–0.54, 0.2–0.38, 0.3–0.51, and 0.26–0.54, respectively. Curvatures for the proposed harmonized reconstructions were decreased in the ranges of 0.18–0.3, 0.16–0.25, 0.3–0.39, and 0.24–0.35 for RCmax, RCISO-50, RCpeak, and RC10V, respectively.
Furthermore, RMSE values for the routine clinical reconstructions for SBR4 were in the ranges of 0.25–0.41, 0.3–0.54, 0.32–0.5, and 0.28–0.42 for RCmax, RCISO-50, RCpeak, and RC10V, respectively. The RMSE values for RCmax, RCISO-50, RCpeak, and RC10V were reduced by 23–33%, 35–46%, 32–41%, and 27–37%, respectively, using the proposed harmonized reconstructions. For the standard clinical reconstruction setting, curvatures of RCmax, RCISO-50, RCpeak, and RC10V were in the ranges of 0.26–0.51, 0.22–0.32, 0.3–0.39, and 0.29–0.46, respectively. For the proposed harmonized reconstructions, curvature values of RCmax, RCISO-50, RCpeak, and RC10V ranged between 0.23–0.36, 0.19–0.25, 0.22–0.4, and 0.26–0.38, respectively.
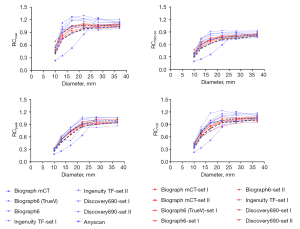
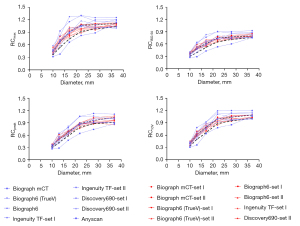
The RC curves derived from clinical and harmonized reconstruction settings are shown in Figures 4,5. RCs are plotted as a function of sphere size. Significant variations were noticeable between various makes and models of PET/CT scanners when standard clinical reconstruction modes were utilized. The variability of quantitative indices was reduced after applying the proposed harmonized reconstruction sets.
Reproducibility
As shown in Figure 6, CVMRC values for both clinical and harmonized reconstruction settings were calculated. The CVMRC shows a remarkable reduction of about 28%, 26%, 32%, and 19% in harmonized reconstruction settings for MRCmax, MRCISO-50, MRCpeak, and MRC10V, respectively. CVMRC for MRC10V in the harmonized reconstruction setting has the smallest value of 5.9% in SBR4. Moreover, the smallest value in SBR10 belongs to MRCpeak, with a value of 5.8% in the harmonized reconstruction setting.

Discussion
The main goal of this study was to determine a harmonized reconstruction setting in multiple PET/CT scanners, some equipped with different hardware and software technology. We sought the most robust and reproducible quantitative metric and proposed a novel quantitative metric averaging the ten hottest voxels regardless of location. Our experiments established reconstruction parameter sets across six scanners that would minimize inter and intra-scanner variability in SUVs obtained and their outcomes.
A statistically significant difference was observed between quantitative metrics in SBR10 and SBR4. Our study demonstrated a high agreement in SBR (Lin’s CCC >0.95) regarding the size of spheres in some of the investigated PET/CT scanners. The Spearman correlation coefficient was higher in smaller spheres (d<20 mm) than in larger ones (d>20 mm). One reason for this phenomenon may be related to edge artifact, which produces an overshoot of the measured activity at the edge of spheres.
Uptake overestimation was observed in all the PET/CT scanners equipped with advanced reconstruction algorithms applying a higher number of iteration × subsets without any or with the lowest FWHM of the Gaussian filter. Overestimation was produced in increased noise levels in smaller or single voxels (VOImax) using the PSF reconstruction algorithm (35).
In a clinical study, Rogasch et al. (36) evaluated the impact of advanced reconstruction algorithms on SUVmax in lesions with low (<4.8) and high (>4.8) tumor-to-background ratio (TBR). They demonstrated that TOF and PSF integration substantially increased SUVmax in low and high TBRs. Our phantom experiments showed that this positive bias usually occurs in spheres with a diameter of 17, 22, and 28 mm. Also, it is higher in SBR10 than in SBR4, which is in line with the results obtained by Rogasch et al. (36). As FDG PET/CT also measures the response to therapy, we expect an overestimation in the uptake, which is challenging for small tumors shrinking after treatment. The difference in RCs’ upward bias in high and low SBRs in large spheres reduces the correlation between both SBRs; however, good correlation and suitable concordance were observed in small spheres. We found a significant difference and substantial/moderate concordance between both SBRs. Thus, quantitative analysis of the phantom study may be mainly affected by SBR.
A high degree of variation in quantitative metrics was observed with investigated reconstruction parameter sets in inter- and intra-scanner performance. We found that higher RC values lead to lower RMSE and more accuracy. However, harmonized reconstruction sets do not necessarily yield the highest RC values across all spheres, as shown in Figure 2. The selection of harmonized reconstruction setting demonstrates that variability can be reduced to acceptable limits. The trend of RCs against sphere diameters is almost in agreement with the results published in EANM/EARL (11,19) for SBR10 and JSNM (37) for SBR4, introducing the harmonized bandwidth of RC values. The EANM strategy is based on RCmax and RCmean analysis, whereas JSNM only recommends applying SUVmax for harmonization.
In the proposed harmonized reconstruction setting, higher RCs in small spheres were considered, as large spheres have almost accurate RC values when edge artifact is eliminated. For all the quantitative metrics, harmonized reconstruction sets yielded promising results. The proposed harmonized reconstruction settings improved the CVMRC of all the RCs across the scanners, leading to higher reproducibility. Even though our multi-scanner assessment was limited to only six scanners with different technologies, our results could predict that harmonizing RC bandwidth is feasible for the next generation of PET/CT scanners. Therefore, it is possible to harmonize the PET quantitative metrics produced by advanced reconstruction algorithms by applying post-smoothing filters during or after image reconstruction. Kelly and Declerck recommended this method to reduce reconstruction-dependent variation, called SUVref (7). Sunderland and Christian (38) showed that scanner-specific reconstruction parameters increased quantification variability, and the smoothing filter was the most influential parameter. However, considering EANM guidelines, spatial filters should not exceed a FWHM of 7 mm (20). In the current work, several FWHMs of Gaussian post-smoothing filter (low to high) available on the workstation of each system was assessed for this purpose. Gaussian filters with 4 to 6 mm FWHM could result in a more accurate and harmonized quantitation. Also, they may reduce image noise and Gibbs artifact.
Results of the present study demonstrated that effects in RCmax, RCISO-50, RCpeak, and RC10V due to changes in reconstruction setting are most noticeable for routine clinical reconstruction modes, while within the proposed reconstruction sets for harmonization, this variation was reduced. SUVmax represents a small portion of VOI, which may raise the question of how well it reflects the biology of the entire lesion. Moreover, SUVmax is less affected by PVE, while it is sensitive to image noise (39). Makris et al. (40) showed that SUVpeak is significantly less susceptible to change in the reconstruction setting. In a study by Brendle et al. (41), SUVpeak exhibited the highest reproducibility and less variation with different reconstruction protocols than SUVmax.
However, due to the VOIpeak definition, two problems exist. Firstly, inaccurate metabolic active volume; as the hottest voxels in FDG avid lesions are not necessarily close to each other, a 1 mL sphere unavoidably includes some voxels that are not the hottest ones. The second is not being able to apply VOIpeak for lesion volumes less than 1 mL. This study used the average value of 10 voxels with maximum uptake to offset these problems, producing higher and harmonized recoveries. The average SUVmax of several voxels accounts for the heterogeneity of the lesion uptake (42). Laffon et al. (28) demonstrated that SUV from 40 hottest voxels might be superior to SUVpeak to assess the most metabolically active portions of tumors and improve SUV variability performance. The SUVmax-40 index can be easily implemented in clinical practice with low intra- or inter-observer variability. The variability of the average SUV from several hottest voxels is significantly lower than those of SUVmax and SUVpeak (42). Nevertheless, the number of hottest voxels should be optimized depending on the clinical situation to provide small-size lesions on specific image reconstruction sets in specific PET scanners (28,33).
Burger et al. (30) determined the repeatability of SUVs as a function of the number of voxels. They concluded that repeatability is noticeably increased by deriving SUVs from the ten hottest voxels. According to Figure 6, the reproducibility of RCpeak and RC10V is almost the same, but regarding the voxel sizes reported in Table 5, RC10V grants a privilege for quantification of lesion volumes less than 1 mL. Moreover, the 1 mL sphere in RCpeak is drawn in the highest uptake region of the tumor, where it may not include all the hottest voxels in the entire VOI. This may affect delineation and quantification, especially in the aggressive portion of the tumor. As averaging several hottest voxels regardless of their location within the tumor can be more accurate than SUVpeak (29,30), and it does not require segmentation, RC10V is suggested for quantitative harmonization as the most robust and reproducible quantitative measure. It should be noted that RC10V, like other quantitative metrics, is applied to the same reconstructed PET image. Thus its use may not be affected by the reconstruction matrix (i.e., voxel size).
The current study had some limitations. First, the range of harmonized RCs may change with the development of new systems. Second, we only evaluated reproducibility, while evaluating repeatability and reproducibility simultaneously can yield better results. Third, reconstruction protocols may depend on voxel size differences, which were disregarded in this study. Fourth, while harmonization using phantom scans has been shown, its consistency when applied to patient studies has yet to be demonstrated. Fifth, we investigated what has been “clinically demonstrated” in tumor quantification (31), i.e., averaging the ten hottest voxels in phantoms ideally considered homogenous, so quantifications may be affected by noise or PVE, although optimized reconstruction settings were applied. Finally, these results cannot be directly transferred to scanner harmonization involving other PET isotopes, such as 68Ga. There are quite a few reasons for that, including; (I) the lower administered dose and shorter half-life, with nearly the same uptake time for 68Ga, results in lower count statistics at the time of imaging; (II) a higher positron range in 68Ga results in PET images with degraded spatial resolution.
Conclusions
We demonstrated that it is feasible to harmonize the performance of PET/CT systems with different makes and models, some equipped with advanced reconstruction algorithms. Among several reconstruction sets assessed in this study, sub-iterations between 42 and 72 with Gaussian post-smoothing filters with 4 to 6 mm FWHM resulted in more accurate and harmonized quantitation across all scanners. Our introduced RC10V metric was found to be superior to other quantitative indices for higher recovery in smaller spheres and improved reproducibility.
Acknowledgments
We are grateful to the staff and management of these PET/CT centers for their cooperation; Nuclear Medicine Department of Razavi Hospital, Nuclear Medicine Department of Shariati Hospital, Khatam PT/CT Center, Ferdous PET CT Center in Masih Daneshvari Hospital, PET/CT Center of Kowsar Hospital, and Payam PET/CT Center.
Funding: This study was part of a Ph.D. dissertation supported under grant number 961625 by Mashhad University of Medical Sciences, Mashhad, Iran.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-443/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. As a phantom research, this study was exempt from ethical approval and informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Devriese J, Beels L, Maes A, Van de Wiele C, Pottel H. Impact of PET reconstruction protocols on quantification of lesions that fulfil the PERCIST lesion inclusion criteria. EJNMMI Phys 2018;5:35. [Crossref] [PubMed]
- Ferretti A, Chondrogiannis S, Rampin L, Bellan E, Marzola MC, Grassetto G, Gusella S, Maffione AM, Gava M, Rubello D. How to harmonize SUVs obtained by hybrid PET/CT scanners with and without point spread function correction. Phys Med Biol 2018;63:235010. [Crossref] [PubMed]
- Iizuka H, Daisaki H, Ogawa M, Yoshida K, Kaneta T. Harmonization of standardized uptake values between two scanners, considering repeatability and magnitude of the values in clinical fluorine-18-fluorodeoxyglucose PET settings: a phantom study. Nucl Med Commun 2019;40:857-64. [Crossref] [PubMed]
- Boellaard R, Oyen WJ, Hoekstra CJ, Hoekstra OS, Visser EP, Willemsen AT, Arends B, Verzijlbergen FJ, Zijlstra J, Paans AM, Comans EF, Pruim J. The Netherlands protocol for standardisation and quantification of FDG whole body PET studies in multi-centre trials. Eur J Nucl Med Mol Imaging 2008;35:2320-33. [Crossref] [PubMed]
- Liu S, Feng Z, Wen H, Jiang Z, Pan H, Deng Y, Zhang L, Ju X, Chen X, Wu X. (18)F-FDG PET/CT can predict chemosensitivity and proliferation of epithelial ovarian cancer via SUVmax value. Jpn J Radiol 2018;36:544-50. [Crossref] [PubMed]
- Kitajima K, Watabe T, Nakajo M, Ishibashi M, Daisaki H, Soeda F, Tanemura A, Kanekura T, Yamazaki N, Ito K. Tumor response evaluation in patients with malignant melanoma undergoing immune checkpoint inhibitor therapy and prognosis prediction using (18)F-FDG PET/CT: multicenter study for comparison of EORTC, PERCIST, and imPERCIST. Jpn J Radiol 2022;40:75-85. [Crossref] [PubMed]
- Kelly MD, Declerck JM. SUVref: reducing reconstruction-dependent variation in PET SUV. EJNMMI Res 2011;1:16. [Crossref] [PubMed]
- Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med 2009;50:11S-20S. [Crossref] [PubMed]
- Adams MC, Turkington TG, Wilson JM, Wong TZ. A systematic review of the factors affecting accuracy of SUV measurements. AJR Am J Roentgenol 2010;195:310-20. [Crossref] [PubMed]
- Aide N, Lasnon C, Veit-Haibach P, Sera T, Sattler B, Boellaard R. EANM/EARL harmonization strategies in PET quantification: from daily practice to multicentre oncological studies. Eur J Nucl Med Mol Imaging 2017;44:17-31. [Crossref] [PubMed]
- Kaalep A, Sera T, Rijnsdorp S, Yaqub M, Talsma A, Lodge MA, Boellaard R. Feasibility of state of the art PET/CT systems performance harmonisation. Eur J Nucl Med Mol Imaging 2018;45:1344-61. [Crossref] [PubMed]
- Panetta JV, Daube-Witherspoon ME, Karp JS. Validation of phantom-based harmonization for patient harmonization. Med Phys 2017;44:3534-44. [Crossref] [PubMed]
- Lammertsma AA, Boellaard R. The need for quantitative PET in multicentre studies. Springer; 2014.
- Srinivas SM, Dhurairaj T, Basu S, Bural G, Surti S, Alavi A. A recovery coefficient method for partial volume correction of PET images. Ann Nucl Med 2009;23:341-8. [Crossref] [PubMed]
- Lodge MA. Repeatability of SUV in Oncologic (18)F-FDG PET. J Nucl Med 2017;58:523-32. [Crossref] [PubMed]
- Bae H, Tsuchiya J, Okamoto T, Ito I, Sonehara Y, Nagahama F, Kubota K, Tateishi U. Standardization of [F-18]FDG PET/CT for response evaluation by the Radiologic Society of North America-Quantitative Imaging Biomarker Alliance (RSNA-QIBA) profile: preliminary results from the Japan-QIBA (J-QIBA) activities for Asian international multicenter phase II trial. Jpn J Radiol 2018;36:686-90. [Crossref] [PubMed]
- Houdu B, Lasnon C, Licaj I, Thomas G, Do P, Guizard AV, Desmonts C, Aide N. Why harmonization is needed when using FDG PET/CT as a prognosticator: demonstration with EARL-compliant SUV as an independent prognostic factor in lung cancer. Eur J Nucl Med Mol Imaging 2019;46:421-8. [Crossref] [PubMed]
- Rubello D, Colletti PM. SUV Harmonization Between Different Hybrid PET/CT Systems. Clin Nucl Med 2018;43:811-4. [Crossref] [PubMed]
- Boellaard R, O'Doherty MJ, Weber WA, Mottaghy FM, Lonsdale MN, Stroobants SG, et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging 2010;37:181-200. [Crossref] [PubMed]
- Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging 2015;42:328-54. [Crossref] [PubMed]
- Tsutsui Y, Daisaki H, Akamatsu G, Umeda T, Ogawa M, Kajiwara H, Kawase S, Sakurai M, Nishida H, Magota K, Mori K, Sasaki M. Multicentre analysis of PET SUV using vendor-neutral software: the Japanese Harmonization Technology (J-Hart) study. EJNMMI Res 2018;8:83. [Crossref] [PubMed]
- Panetta JV, Scheuermann J, Karp JS, Daube-Witherspoon ME. editors. Do phantom harmonization efforts translate into harmonized patient images? IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC): IEEE; 2015.
- Lasnon C, Salomon T, Desmonts C, Dô P, Oulkhouir Y, Madelaine J, Aide N. Generating harmonized SUV within the EANM EARL accreditation program: software approach versus EARL-compliant reconstruction. Ann Nucl Med 2017;31:125-34. [Crossref] [PubMed]
- Senda M. Standardization of PET imaging and site qualification program by JSNM: collaboration with EANM/EARL. Ann Nucl Med 2020;34:873-4. [Crossref] [PubMed]
- Ketabi A, Ghafarian P, Mosleh-Shirazi MA, Mahdavi SR, Rahmim A, Ay MR. Impact of image reconstruction methods on quantitative accuracy and variability of FDG-PET volumetric and textural measures in solid tumors. Eur Radiol 2019;29:2146-56. [Crossref] [PubMed]
- Balcerzyk M, Fernández-López R, Parrado-Gallego Á, Pachón-Garrudo VM, Chavero-Royan J, Hevilla J, Jiménez-Ortega E, Leal A. Application of EARL (ResEARch 4 Life®) protocols for [18F]FDG-PET/CT clinical and research studies. A roadmap towards exact recovery coefficient. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 2017;873:39-42. [Crossref]
- Vosoughi H, Hajizadeh M, Emami F, Momennezhad M, Geramifar P. PET NEMA IQ Phantom dataset: image reconstruction settings for quantitative PET imaging. Data Brief 2021;37:107231. [Crossref] [PubMed]
- Laffon E, Burger IA, Lamare F, de Clermont H, Marthan R. SUVpeak Performance in Lung Cancer: Comparison to Average SUV from the 40 Hottest Voxels. J Nucl Med 2016;57:85-8. [Crossref] [PubMed]
- Mosleh-Shirazi MA, Nasiri-Feshani Z, Ghafarian P, Alavi M, Haddadi G, Ketabi A. Tumor volume-adapted SUV(N) as an alternative to SUV(peak) for quantification of small lesions in PET/CT imaging: a proof-of-concept study. Jpn J Radiol 2021;39:811-23. [Crossref] [PubMed]
- Burger IA, Huser DM, Burger C, von Schulthess GK, Buck A. Repeatability of FDG quantification in tumor imaging: averaged SUVs are superior to SUVmax. Nucl Med Biol 2012;39:666-70. [Crossref] [PubMed]
- Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989;45:255-68. [Crossref] [PubMed]
- Lasnon C, Desmonts C, Quak E, Gervais R, Do P, Dubos-Arvis C, Aide N. Harmonizing SUVs in multicentre trials when using different generation PET systems: prospective validation in non-small cell lung cancer patients. Eur J Nucl Med Mol Imaging 2013;40:985-96. [Crossref] [PubMed]
- Vanderhoek M, Perlman SB, Jeraj R. Impact of the definition of peak standardized uptake value on quantification of treatment response. J Nucl Med 2012;53:4-11. [Crossref] [PubMed]
- Kaalep A, Sera T, Oyen W, Krause BJ, Chiti A, Liu Y, Boellaard R. EANM/EARL FDG-PET/CT accreditation - summary results from the first 200 accredited imaging systems. Eur J Nucl Med Mol Imaging 2018;45:412-22. [Crossref] [PubMed]
- Prieto E, Domínguez-Prado I, García-Velloso MJ, Peñuelas I, Richter JÁ, Martí-Climent JM. Impact of time-of-flight and point-spread-function in SUV quantification for oncological PET. Clin Nucl Med 2013;38:103-9. [Crossref] [PubMed]
- Rogasch JM, Steffen IG, Hofheinz F, Großer OS, Furth C, Mohnike K, Hass P, Walke M, Apostolova I, Amthauer H. The association of tumor-to-background ratios and SUVmax deviations related to point spread function and time-of-flight F18-FDG-PET/CT reconstruction in colorectal liver metastases. EJNMMI Res 2015;5:31. [Crossref] [PubMed]
- Standard P. Imaging protocols and phantom test procedures and criteria: executive summary. Japanese Society of Nuclear Medicine website; 2016.
- Sunderland JJ, Christian PE. Quantitative PET/CT scanner performance characterization based upon the society of nuclear medicine and molecular imaging clinical trials network oncology clinical simulator phantom. J Nucl Med 2015;56:145-52. [Crossref] [PubMed]
- Lindholm H, Staaf J, Jacobsson H, Brolin F, Hatherly R, Sânchez-Crespo A. Repeatability of the Maximum Standard Uptake Value (SUVmax) in FDG PET. Mol Imaging Radionucl Ther 2014;23:16-20. [Crossref] [PubMed]
- Makris NE, Huisman MC, Kinahan PE, Lammertsma AA, Boellaard R. Evaluation of strategies towards harmonization of FDG PET/CT studies in multicentre trials: comparison of scanner validation phantoms and data analysis procedures. Eur J Nucl Med Mol Imaging 2013;40:1507-15. [Crossref] [PubMed]
- Brendle C, Kupferschläger J, Nikolaou K, la Fougère C, Gatidis S, Pfannenberg C. Is the standard uptake value (SUV) appropriate for quantification in clinical PET imaging? - Variability induced by different SUV measurements and varying reconstruction methods. Eur J Radiol 2015;84:158-62. [Crossref] [PubMed]
- Laffon E, Lamare F, de Clermont H, Burger IA, Marthan R. Variability of average SUV from several hottest voxels is lower than that of SUVmax and SUVpeak. Eur Radiol 2014;24:1964-70. [Crossref] [PubMed]


