
Deep learning for predicting epidermal growth factor receptor mutations of non-small cell lung cancer on PET/CT images
Introduction
Currently, the incidence and mortality rates of lung cancer remain high. Lung cancer is divided into non-small cell lung cancer (NSCLC) and small cell lung cancer. Adenocarcinoma is a common type of NSCLC (1,2). A prevalent histological form of lung cancer is lung adenocarcinoma, and the identification of the epidermal growth factor receptor (EGFR) mutation has completely changed how it is treated (3-8). Patients with lung cancer with certain EGFR gene mutations can have their prognosis improved by using EGFR tyrosine kinase inhibitors (TKIs) (9,10). Therefore, the detection of EGFR mutations is critical in first-line treatment. In EGFR mutation detection, biopsy sequencing is the most common method.
However, because of the wide heterogeneity of lung tumors, biopsies to detect EGFR mutations must locate tissue regions precisely (11,12). Also, biopsies may increase the risk of cancer metastasis (13). In addition, the difficulty of repeatedly sampling tumors, obtaining tissue samples, poor DNA quality, and higher costs limit the applicability of biopsies (14,15). In these cases, a noninvasive and easy-to-use method is needed to predict EGFR mutations.
Currently, researchers typically employ conventional radiomics, machine learning, or statistics to predict gene mutations from computed tomography (CT) images. Most researchers anticipate genetic variation using radiomics (15-19). Digumarthy et al. (16) explored the possibility of predicting EGFR mutations in NSCLC using CT images. They performed a quantitative Pearson correlation analysis on the CT images of 93 NSCLC patients. The findings demonstrated that radiomics and clinical features could effectively predict EGFR mutation. A correlation between image characteristics and gene mutations has been shown in other studies, and these investigations also looked at imaging genomics in NSCLC and evaluated the viability of predicting EGFR mutations from CT images (17,18). The relationship between radiologic characteristics and EGFR mutations in lung cancer was examined by Liu et al. (19). They concluded that combining radiomicrological characteristics with clinical data could significantly enhance EGFR mutation prediction. Rios Velazquez et al. (15) investigated the relationship between lung cancer imaging phenotypes and gene mutations. They contend that increasing prediction accuracy can be achieved by fusing imaging information with clinical models. The most effective features locate cancers with EGFR mutations.
Deep learning has a promising future in feature extraction and classification, allowing users to bypass laborious manual feature extraction (20-23). Additionally, techniques for image-based gene mutation prediction using deep learning are being explored. Wang et al. (24) proposed to train a deep learning model using CT images to predict EGFR mutations in lung adenocarcinoma. They gathered CT scans from 844 lung cancer patients and then used the CT images to build a deep learning model for EGFR gene mutation. According to several studies, deep learning models can predict gene alterations in lung cancer (25-28).
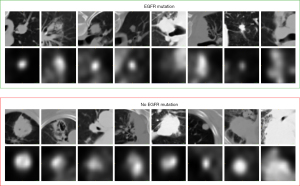
Deep learning has made great achievements in the field of robotic vision, but there are still many problems in medical image processing. First, huge datasets are necessary for high-performance deep learning models; however, obtaining large amounts of medical picture data is frequently challenging (29,30). Second, lung nodules of different mutant types have extensive changes in morphology, texture, and vision. As shown in Figure 1, a typical convolutional neural network (CNN) has difficultly extracting features from all types of lung nodules.
Researchers have carried out many studies attempting to solve the problem of medical image datasets. First, through transfer learning, it is possible to apply the image representation ability discovered in large-scale natural images to medical small-sample images (31-33). Esteva et al. (31) used Large-Scale ImageNet to pre-train GoogleNet Inception-3 CNN to identify skin cancer. Second, it is possible to extract multiple slices from 3-dimensional (3D) nodules, and 2-dimensional (2D) CNN may be expanded to handle each slice’s pictures in medical imaging (34,35). Setio et al. (35) proposed a multiview architecture based on 3D nodules and trained them with 2D CNN. The researchers studied the use of an improved deep learning model to deal with small samples of medical imaging problems (36,37). Inception-ResNet Network was used by Kang et al. (36) to categorize benign and malignant lung nodules. According to the experimental findings, lung nodule feature extraction may be greatly aided by the multiscale convolution nuclei and residual structures of Inception-ResNet. A residual attention learning network was suggested by Guan et al. (37) for the multiple categorizations of chest X-ray pictures. The feature extraction performance of CNN is significantly enhanced by this technique, which adds the attention mechanism and allows the CNN to concentrate on the lesion regions pertinent to the prediction job. The benefit of applying the attention mechanism in multiple chest X-ray image categorization tasks is shown by experimental data. Fourth, a study employed multitasking to resolve the issue of small sample size in medical imaging (38). Pellegrino et al. (39) used next-generation sequencing (NGS) to screen mutations, and the resulting data were predicted by a machine learning method.
In this paper, we present a novel method for predicting EGFR mutation and non-EGFR mutation from both positron emission tomography/CT (PET/CT) fused images and patients’ personal information. First, we cut 32 2D views of each 3D cube of lung nodules. Then, we built an EfficientNet-V2 model to extract deep features from 32 views. Finally, we used the EfficientNet-V2 model for learning to predict both EGFR and non-EGFR mutations. In addition, we used Naive Bayes to determine the different types of patients. An adaptive weighting scheme was adopted in the training process so that the EfficientNet-V2 model could be trained end-to-end.
The main contribution of the proposed model is a fusion method for extracting 2D multiview slices. To our knowledge, this is one of the first studies to use a deep learning model to forecast multiple gene mutations concurrently. A single predicting job can benefit from multitasking due to the EfficientNet-V2 model’s enhanced ability to define nodules, which raises the predictive accuracy. Additionally, the whole model can be trained via an end-to-end methodology, negating the need for human interaction. The model’s learning process can assimilate a variety of patient data points pertinent to the predicting job. Furthermore, the suggested initial-attention-re-network model is capable of successfully extracting characteristics from different kinds of lung nodules. We present the following article in accordance with the MDAR reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-760/rc).
Methods
Our approach consists of 3 steps. First, we use torchIO (https://torchio.readthedocs.io/) to cut out the region of interest (ROI) cube of PET and CT in turn according to the imaging specialist’s label, with a cube size of 64×64×3. Second, we cut the PET cube and the CT cube into Z-score normalization, normalized the data of PET and CT per the same data area, then fused the PET cube after Z-score normalization with the CT cube to obtain a new cube of fusion. Third, we cut the cube of the ROI pulmonary nodules fused with PET/CT into 2D grayscale and merged it with the adjacent 2 pieces to form a red, green, and blue (RGB) map. Then, we input the data of PET and CT into the model of EfficientNet-V2. The output of the model was the characteristic quantity of each kind. After statistical calculation, we concluded whether the PET or EGFR cube contained a mutation.
Data preprocessing and ROI extraction
Figure 2 shows the fusion of the PET dataset and CT dataset. Due to the variable spatial resolution of different instruments in PET/CT images, we resampled them by spline interpolation and rendered them a uniform size of 1.0×1.0×1.0 mm3. Then, an experienced imaging surgeon labeled the pulmonary nodules. We calculated the coordinates of the cube center of the nodules, made a cube of 64×64×32, and cut out the ROI of the pulmonary nodules. Using this method, we obtained ROIs for PET and CT separately, and performed the first Z-score normalization on these 2 ROIs. The mean value of the processed data was 0, and the variance was 1. The main goal was to unify the data of different orders of magnitude into the same order of magnitude and to use the calculated Z-score value to measure the data, which ensured the comparability of data.
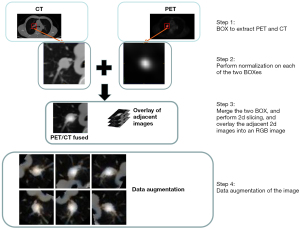
Combining PET and CT with normal distribution in the same patient, new lung nodule data were obtained that contained all the information of PET and CT. This data has many advantages over direct CT. PET was used to visualize the metabolism of tumor cells from the point of view of pathophysiology. CT was used to see the shape, which was a relatively indirect sign. We cut 3D data into 2D grayscale images sized 64×64×1 and combined them with 2 adjacent grayscale images to form an RGB image sized 64×64×3.
Data enhancement technology enhances the dataset. We used RandomGrayscale, RandomHorizontalFlip, RandomVerticalFlip, and RandomRotation. All images were then uniformly resized to 64×64×3 to accommodate the following model input. Before transferring the data to the deep neural network, we normalized the data using Z-score normalization. After normalization, the data were adjusted to a normal distribution with a mean of 0 and a variance of 1. Before activating a function, performing normalization returned an increasingly skewed distribution to a normalized distribution.
Deep learning model
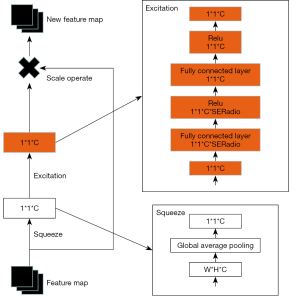
The squeeze exception (SE) module consisted of compression and excitation, and the compression module was a global average pooling. After the compression operation, the feature map was compressed to a 1×1×C vector. C is defined channel.
In order to pay more attention to the characteristics, the last step was a scale operation. After obtaining a 1×1×C vector, the original characteristic graph represented the scale operation, which was a channel weight multiplication. The original characteristic vector was width (W) × height (H) × C. Afterward, the SE module’s characteristic graph became a brand-new characteristic graph, which was the channel attention mechanism.
Figure 3 depicts the SE module, where the squeeze operation is the global average pooling. In order to simulate the correlation between channels and provide the same amount of weights as the input features, the 2 fully connected layers were combined to form a bottleneck structure. In order to ascend back via a fully connected layer following rectified linear unit (ReLu) activation, we first lowered the feature dimension to 1/16 of the input. More nonlinearity, better fitting of complicated relationships between channels, and a much lower number of parameters and computations were its main advantages compared to employing a fully connected layer. After that, a sigmoid gate was used to acquire the normalized weights in the range of 0 to 1, and a scale operation was used to weight the normalized weights according to the attributes of each channel. The formula is as follows:
where ζc is the statistic pooled by global average, and the feature graph’s dimensions are H for height, W for width, and uc for the feature map of the c filter. Next came the excitation operation, which was a gate mechanism similar to a recurrent neural network. Its operation formula is:
The channel weights were obtained by 2 fully joined operations and a sigmoid activation function. δ is at ReLU function, W1 is the first fully concatenated operation that is intended to reduce the dimension, and it has a reduction cofactor r, which is a superparameter. W2 is the parameter for the second fully joined operation, restoring the dimension to the input dimension. After 2 full connections, we used the sigmoid activation function to normalize the weights of each feature graph. The output feature map with weight was then created by multiplying the original feature map by the weight matrix of the sigmoid activation function’s output. The resulting formula is as follows:
According to Eqs. [2,3], the weight coefficients of each feature map on the channel dimension were added to make the channel features of the feature map more capable of extracting features, amplifying the effective features and reducing the invalid feature information.
The MBConv module (Figure 4) consists of an SE module, 3 convolution modules, and a dropout module. The first convolution raises the dimension of the feature and passed through Depwise Conv. Depwise Conv consists of 2 convolutions to form the first of 2 steps, then passes through the SE module and passes through 1×1 convolution reduction and dropout. Finally, it joins with the primary feature residuals to form a new feature, and all outputs can be stitched together into a depth feature map. Convolution kernels vary in size and differ in the size of the accepted domain; therefore, different types of information can be obtained from the input image. The model will be able to represent images more accurately through convolution and splicing of several output feature maps in simultaneously. The residual module fixes the gradient disappearance issue that arises during training and prevents performance loss brought on by the depth network.
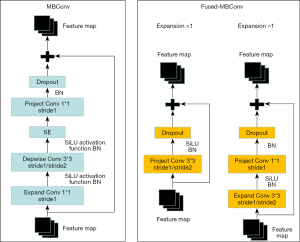
The Fused-MBConv module (Figure 4) consisted of a convolution module and dropout module, in which the specific structure was different in different cases. When the expansion ratio =1, the feature only passed through Project Conv 3×3, in which Project Conv consists of 2 convolutions, the first step is 1, the second step is 2, and then dropout was carried out. Finally, it connects with primary feature residuals, formed new features, and all outputs can be stitched together into a depth feature mapping. When the expansion ratio ≠1, the feature needs to enter Expend Conv 3×3 first, in which Expend Convolution consists of 2 parts: the first step is 1 and the second step is 2. Then, in 1×1 convolution, the dropout occurs, and finally it connects with primary feature residuals to form a new feature, and all outputs can be stitched into a depth mapping feature. The improvement of the MBConv module can make the network obtain different types of information from the output image under different circumstances and further enhance the network’s robustness. The model will be able to represent images more accurately through convolution and splicing several output feature maps simultaneously.
The EfficientNet-V2 model (Figure 5) is composed of 3 Fused-MBConv modules, 2 MBConv modules, 1 Conv module, and a classifier. First, the input image is convolved into 3×3 dimensions, then 3 Fused-MBConv 3×3 modules are processed to extract the initial features, then 2 MBConv modules are used to obtain the final features. Finally, the features are input into the classifier for EGFR and non-EGFR classification. Note that there is a shortcut when the stride =1 and input/output (I/O) Channels are equal, and there is a dropout layer only when shortcut connects. Here, the dropout layer is Stochastic Depth, which randomly drops the main branch of the block to reduce the depth of the network.
Neural architecture search (NAS)
We learned a variety of design choices to improve training speed. To find the best combination of these options, we employed a training-aware NAS. Our design space was as follows: convolutional operation type, MBConv, and Fused-MBConv; number of layers and kernel size, 3×3 and 5×5; and expansion ratio (the first expand conv 1×1 in MBConv or in Fused-MBConv the first expand conv 3×3), 1, 4, and 6. On the one hand, we did this (NAS) by removing unwanted search options, such as pooling skip operations. On the other hand, we reused the channel sizes searched in EfficientNet. Next, 1,000 models were randomly sampled in the search space, and each model was trained for 10 epochs using a smaller image scale. The search reward combined the model accuracy rate A, the standard training time S required for a step, and the model parameter size P. The reward function can be written as: A·Sω·Pν. where ω=−0.07, ν=−0.05.
We added a 3×3 kernel convolution layer after the input picture since our CT-PET images were grayscale, and the pre-trained EfficientNet-V2 network used 3 channels as its color image input.
General deep learning settings and computation environment
Using the Python package Pytorch1.7 (https://pytorch.org/), we implemented and enhanced the model. In our fully connected neural networks, a neuron’s output (yi) for neuron j is determined by:
Where F stands for the activation function, wij and bj for the weight and bias, respectively, and xj for the output of neuron i in the previous layer. Thus, the total number of neurons at a layer may be expressed as:
Weights and biases were modified during training to reduce a loss function. We used an AdamW optimizer to improve the model with a CrossEntropyLoss loss function. The sigmoid linear unit (SiLU) F was established.
All analyses were performed on a Window10 with GeForce RTX 3080 processors. The initial learning rate was 1e−3. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of Yunnan Cancer Hospital, and individual consent for this retrospective analysis was waived.
Results
Datasets
The hospital responsible for our study collected datasets from Yunnan Cancer Hospital of China that included 150 patients with EGFR mutations between 2016 and 2019. There were 59 women and 91 men. Their median age was 58, and their ages varied from 31 to 80. There were 93 wild type, 57 EGFR mutations, 64 smokers, and 86 non-smokers. The dataset included 121 patients split into training datasets and 29 patients split into testing datasets. Table 1 displays the clinical characteristics of the training and testing datasets.
Table 1
| Parameter | Training dataset (n=121) | Testing dataset (n=29) |
|---|---|---|
| Sex | ||
| Male | 76 | 15 |
| Female | 45 | 14 |
| Smoking | ||
| Yes | 57 | 7 |
| No | 64 | 22 |
| Age (year) | ||
| Minimum | 31 | 34 |
| Maximum | 80 | 78 |
| Median | 56 | 60 |
| EGFR | ||
| Mutant | 52 | 5 |
| Wild type | 69 | 24 |
EGFR, epidermal growth factor receptor.
Confusion matrix
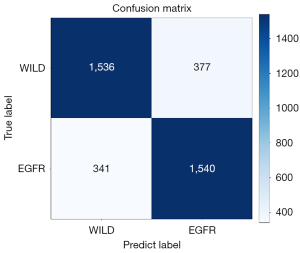
To test the performance of the model, we used 3,794 PET/CT fusion datasets after 2D slicing, where we controlled the number of wild type and EGFR datasets, of which 1,913 datasets were wild type and 1,881 datasets were EGFR. Figure 6 shows the confusion matrix of the predicted results.
Comparison with traditional methods
Earlier studies used clinical and radiological features to predict gene mutations (17-19). Age, sex, smoking status, and tumor stage were used in the clinical validation process. Radiologists with more than 8 years of experience manually segmented nodules as part of the radiomics feature extraction procedure, which also involves extracting 1,108 radiomics features using the PyRadiomics toolkit (40) and choosing 8 features via recursive feature elimination. The radiomics model’s EGFR mutation was finally predicted using a random forest with 100 trees. In order to predict EGFR, the clinical models build a random forest of 100 trees using the clinical features.
To avoid overfitting, we used cross-validation with 10 folds. In the training phase, we divided the training dataset into training (90%) and validation (10%) to train 100 epochs per fold, selected the 10 best models according to the lowest value of loss of 100 epochs per fold, tested the test set with these models, and then calculated the average. Figure 7 shows results of the experiment.
The area under the curve (AUC), accuracy (ACC), and F1 indices were our main metrics for comparison with conventional radiomics and clinical models. Table 2 demonstrates that, in terms of predicting, the model we used was superior than the conventional methods. The validation dataset’s AUC value for the anticipated EGFR mutation was 83.64%, and the ACC value was 81.92%. The outcomes predicted by the radiomics and clinical models were then compared. The validation data’s ACC was 80.62%. This occurred because a radiomics model extracts more effective features from PET/CT fusion images. However, radiomics models require radiologists to artificially segment tumor regions, requiring more labor and time.
Table 2
| Methods | AUC (%) | ACC (%) | F1 (%) |
|---|---|---|---|
| Clinical model | 79.27±2.87 | 76.35±3.36 | 71.22±2.74 |
| Radiomics model | 82.41±2.34 | 80.62±2.51 | 76.35±2.61 |
| Our model | 83.64±2.41 | 81.92±2.86 | 81.90±2.27 |
AUC, area under the curve; ACC, accuracy; F1, weighted harmonic average of precision rate and recall rate.
Model visualization
Our model seeks to investigate the associations between EGFR mutations and tumor imaging characteristics. To illustrate the prediction process and determine the tumor locations most useful for finding EGFR mutations, we employed deep learning visualization. Since our model is an end-to-end model, physicians may concentrate on the tumor regions that require care by using tumor region visualization.
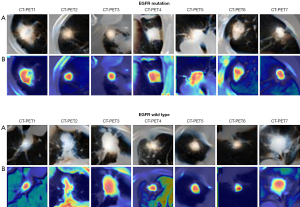
We visualized the intermediate layer of our model in order to assess the feature extraction performance of our model and comprehend why the PET/CT fusion picture has outstanding feature extraction performance. A total of 7 nodules from the EGFR mutation/wild type were chosen at random. The class activation map (CAM) of the stem layer is visualized, where CAM is a thermodynamic diagram that reveals regions associated with prediction (41). Figure 8 shows the results of EGFR and wild type activation. Rows (A) and (B) represent tumor regions and CAM, respectively. When used correctly, the model activates the corresponding nodules of EGFR.
Comparison with different types of datasets
In order to evaluate the advantage of PET/CT fusion image on the model, we compared the performance of CT, PET, and PET/CT image datasets on the model, respectively. The results are shown in Table 3. Compared with PET, the CT image had a higher resolution and could extract the relevant features better. However, the CT image mainly observed the shape of tumor cells, and the tumor cells sometimes appeared as a smaller block, resulting in the characteristic information of the tumor being lost. By fusing CT and PET images, we obtained the characteristic information of tumor morphology and tumor cell metabolism, which was the advantage of molecular image and anatomical image.
Table 3
| Dataset | AUC (%) | ACC (%) | F1 (%) |
|---|---|---|---|
| PET | 77.38±2.61 | 76.34±3.25 | 74.62±2.68 |
| CT | 81.40±2.17 | 79.21±2.51 | 77.36±1.94 |
| PET/CT | 83.64±2.41 | 81.92±2.86 | 81.90±2.27 |
AUC, area under the curve; ACC, accuracy; F1, weighted harmonic average of precision rate and recall rate; PET, positron emission tomography; CT, computed tomography.
Comparison with different training strategies
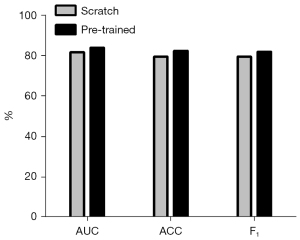
In order to demonstrate that pre-training could improve the predictive performance of the model, we evaluated the model’s performance under pre-training conditions. We trained the EfficientNet-V2 model in 25 epochs using cat versus dog. Table 4 and Figure 9 show that the pre-trained EfficientNet-V2 model performs better than the scratch (un pre-trained) EfficientNet-V2 model.
Table 4
| Training strategy | AUC (%) | ACC (%) | F1 (%) |
|---|---|---|---|
| Scratch | 80.64±2.64 | 78.67±2.71 | 79.46±2.04 |
| Pre-trained | 83.64±2.41 | 81.92±2.86 | 81.90±2.27 |
EGFR, epidermal growth factor receptor; AUC, area under the curve; ACC, accuracy; F1, weighted harmonic average of precision rate and recall rate.
Discussion
In our study, we proposed a deep learning framework based on the EfficientNet-V2 model to predict EGFR mutations by noninvasively fusing PET and CT images. The results show that our model can feasibly predict EGFR mutation. The main advantage of our model is that it can acquire more features by fusing PET and CT images, and it can automatically learn the features of PET/CT images without extracting features manually.
Our method is a noninvasive assay designed to avoid invasive injury when surgery or biopsy are inconvenient. In addition, CT and PET images are readily available throughout the treatment period to detect EGFR mutations. The acquisition of CT and PET images spends relatively few resources of cost and time. Our method does not need the doctor’s domain knowledge; it can predict EGFR mutations more economically and conveniently.
Previous studies have shown that EGFR mutations can be reflected based on quantitative radiomics characteristics (42,43) and CT characteristics (15,44). However, these can only reflect simple visual traits. Some EGFR abrupt states are difficult to be represented by artificial feature engineering. In this case, the introduction of a PET/CT fusion image can better reflect some abstract features and pathological features. Deep learning also demonstrates its benefits by being able to extract more abstract features that are challenging to articulate but necessary for detecting EGFR mutations.
The deep learning approach has the following benefits compared to artificial feature extraction: (I) the deep learning model can extract the abstract multilevel features associated with the EGFR mutation information from the visual data using the hierarchical neural network structure; (II) the deep learning model does not need to label the tumor boundary artificially, which is a great advantage compared with the artificial feature extraction; and (III) the deep learning algorithm is quick and simple to use and only requires the original CT and PET images as input in order to predict the EGFR mutation without the need for additional artificial input.
Our study used a deep learning model to predict EGFR mutations. In validated dataset predictions, EGFR mutations were detected in 83.64% of AUC, 81.92% of ACC, and 78.37% of F1. The AUC of the radiomics model was 82.41%, the ACC was 80.62%, and the F1 was 76.35%. Our model was superior to the radiomics model. In addition, radiomics models require radiologists to identify lesion areas in advance. These results indicate that our model method can predict EGFR mutation effectively and feasibly. However, using noninvasive image analysis to predict gene mutations cannot always replace biopsies. The main advantage of image analysis over pathological biopsy is that image analysis can be supplementary to biopsy if the patient’s body is in an unsuitable condition. The model method used can be tracked repeatedly during the exploration of tumor therapy.
Although we obtained satisfactory prediction results in predicting EGFR mutations, our model still has limitations. First, the datasets were small, and some were incomplete. Furthermore, very few public datasets contain PET and CT images; therefore, the model’s generalizability needs to be further improved. In future studies, we will collect additional datasets and use image data augmentation to build our model to avoid situations such as overfitting.
Finally, we also need to optimize the model itself. The model we used can adopt the spatial attention mechanism in terms of the attention mechanism, and the preprocessing part of the image still needs to be optimized. In future research, we will collect more relevant methods for model optimization, such as using particle swarm optimization, genetic algorithm (45), and Bayesian optimization.
Conclusions
In our work, we developed a deep learning approach to detect EGFR mutations using a noninvasive fusion of PET and CT images. The model was built using a dataset of 150 patients obtained from a partner institution, and its accuracy in identifying EGFR mutations was 81.92%. The outcome suggests that our model approach is a workable method for predicting EGFR mutations. The integration of PET and CT images in our model maximizes the extraction of feature information. It provides a noninvasive supplementary diagnostic tool appropriate for preventing intrusive harm in situations where both surgery and biopsy are unfeasible. Additionally, because our technique does not require medical field knowledge, it is a more affordable and practical way to identify EGFR mutations.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Guangdong Province in China (No. 2020A1515010733), the Guangdong Innovation Platform of Translational Research for Cerebrovascular Diseases of China, and National High-Level Hospital Clinical Research Funding (No. 2022-PUMCH-B-070).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-760/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-760/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics board of Yunnan Cancer Hospital, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Motono N, Funasaki A, Sekimura A, Usuda K, Uramoto H. Prognostic value of epidermal growth factor receptor mutations and histologic subtypes with lung adenocarcinoma. Med Oncol 2018;35:22. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, Massey D, Zazulina V, Shahidi M, Schuler M. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Ding Y, Wang Y, Hu Q. Recent advances in overcoming barriers to cell‐based delivery systems for cancer immunotherapy. Exploration 2022;2:20210106. [Crossref]
- Guo S, Li K, Hu B, Li C, Zhang M, Hussain A, Wang X, Cheng Q, Yang F, Ge K, Zhang J, Chang J, Liang XJ, Weng Y, Huang Y. Membrane‐destabilizing ionizable lipid empowered imaging‐guided siRNA delivery and cancer treatment. Exploration 2021;1:35-49. [Crossref]
- Huang H, Dong C, Chang M, Ding L, Chen L, Feng W, Chen Y. Mitochondria-specific nanocatalysts for chemotherapy-augmented sequential chemoreactive tumor therapy. Exploration 2021;1:50-60. [Crossref]
- Jiang J, Mei J, Ma Y, Jiang S, Zhang J, Yi S, Feng C, Liu Y, Liu Y. Tumor hijacks macrophages and microbiota through extracellular vesicles. Exploration 2022;2:20210144. [Crossref]
- Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039-49. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Itakura H, Achrol AS, Mitchell LA, Loya JJ, Liu T, Westbroek EM, Feroze AH, Rodriguez S, Echegaray S, Azad TD, Yeom KW, Napel S, Rubin DL, Chang SD, Harsh GR 4th, Gevaert O. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015;7:303ra138. [Crossref] [PubMed]
- Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association Between Younger Age and Targetable Genomic Alterations and Prognosis in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:313-20. [Crossref] [PubMed]
- Loughran CF, Keeling CR. Seeding of tumour cells following breast biopsy: a literature review. Br J Radiol 2011;84:869-74. [Crossref] [PubMed]
- Girard N, Sima CS, Jackman DM, Sequist LV, Chen H, Yang JC, Ji H, Waltman B, Rosell R, Taron M, Zakowski MF, Ladanyi M, Riely G, Pao W. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J 2012;39:366-72. [Crossref] [PubMed]
- Rios Velazquez E, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH, Gillies R, Quackenbush J, Aerts HJWL. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. [Crossref] [PubMed]
- Digumarthy SR, Padole AM, Gullo RL, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore) 2019;98:e13963. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Rizzo S, Raimondi S, de Jong EEC, van Elmpt W, De Piano F, Petrella F, Bagnardi V, Jochems A, Bellomi M, Dingemans AM, Lambin P. Genomics of non-small cell lung cancer (NSCLC): Association between CT-based imaging features and EGFR and K-RAS mutations in 122 patients-An external validation. Eur J Radiol 2019;110:148-55. [Crossref] [PubMed]
- Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, Ye Z, Gillies RJ. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-448.e6. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Liu C, Hu SC, Wang C, Lafata K, Yin FF. Automatic detection of pulmonary nodules on CT images with YOLOv3: development and evaluation using simulated and patient data. Quant Imaging Med Surg 2020;10:1917-29. [Crossref] [PubMed]
- Wang Y, Zhou L, Wang M, Shao C, Shi L, Yang S, Zhang Z, Feng M, Shan F, Liu L. Combination of generative adversarial network and convolutional neural network for automatic subcentimeter pulmonary adenocarcinoma classification. Quant Imaging Med Surg 2020;10:1249-64. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, Liu Y, Gevaert O, Wang K, Zhu Y, Zhou H, Liu Z, Tian J. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019;53:1800986. [Crossref] [PubMed]
- Xiong JF, Jia TY, Li XY, Yu W, Xu ZY, Cai XW, Fu L, Zhang J, Qin BJ, Fu XL, Zhao J. Identifying epidermal growth factor receptor mutation status in patients with lung adenocarcinoma by three-dimensional convolutional neural networks. Br J Radiol 2018;91:20180334. [Crossref] [PubMed]
- Li XY, Xiong JF, Jia TY, Shen TL, Hou RP, Zhao J, Fu XL. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J Thorac Dis 2018;10:6624-35. [Crossref] [PubMed]
- Xiong J, Li X, Lu L, Lawrence SH, Fu X, Zhao J, Zhao B. Implementation strategy of a CNN model affects the performance of CT assessment of EGFR mutation status in lung cancer patients. IEEE Access 2019;7:64583-91.
- Dong Y, Hou L, Yang W, Han J, Wang J, Qiang Y, Zhao J, Hou J, Song K, Ma Y, Kazihise NGF, Cui Y, Yang X. Multi-channel multi-task deep learning for predicting EGFR and KRAS mutations of non-small cell lung cancer on CT images. Quant Imaging Med Surg 2021;11:2354-75. [Crossref] [PubMed]
- Oquab M, Bottou L, Laptev I, Sivic J. editors. Learning and transferring mid-level image representations using convolutional neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2014.
- Gondara L. editor. Medical image denoising using convolutional denoising autoencoders. 2016 IEEE 16th international conference on data mining workshops (ICDMW); 2016: IEEE.
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;546:686. [Crossref] [PubMed]
- Shen W, Zhou M, Yang F, Dong D, Yang C, Zang Y, Tian J. editors. Learning from experts: Developing transferable deep features for patient-level lung cancer prediction. International conference on medical image computing and computer-assisted intervention; Springer; 2016.
- Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. [Crossref] [PubMed]
- Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognition 2017;61:663-73. [Crossref]
- Setio AA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MM, Naqibullah M, Sanchez CI, van Ginneken B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. [Crossref] [PubMed]
- Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 2017;12:e0188290. [Crossref] [PubMed]
- Guan Q, Huang Y. Multi-label chest X-ray image classification via category-wise residual attention learning. Pattern Recognition Letters 2020;130:259-66. [Crossref]
- Liu M, Zhang J, Adeli E, Shen D. Deep Multi-Task Multi-Channel Learning for Joint Classification and Regression of Brain Status. Med Image Comput Comput Assist Interv 2017;10435:3-11.
- Pellegrino E, Jacques C, Beaufils N, Nanni I, Carlioz A, Metellus P, Ouafik L. Machine learning random forest for predicting oncosomatic variant NGS analysis. Sci Rep 2021;11:21820. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. editors. Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision; 2017.
- Yano M, Sasaki H, Kobayashi Y, Yukiue H, Haneda H, Suzuki E, Endo K, Kawano O, Hara M, Fujii Y. Epidermal growth factor receptor gene mutation and computed tomographic findings in peripheral pulmonary adenocarcinoma. J Thorac Oncol 2006;1:413-6. [Crossref] [PubMed]
- Zhou JY, Zheng J, Yu ZF, Xiao WB, Zhao J, Sun K, Wang B, Chen X, Jiang LN, Ding W, Zhou JY. Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with ALK rearrangements or EGFR mutations. Eur Radiol 2015;25:1257-66. [Crossref] [PubMed]
- Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y, Ye Z, Gillies RJ. CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology 2016;280:271-80. [Crossref] [PubMed]
- Pellegrino E, Brunet T, Pissier C, Camilla C, Abbou N, Beaufils N, Nanni I, Metellus P, Ouafik L. Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1 / BRCA2 Pathogenicity NGS Analysis. BioMedInformatics 2022;2:244-67. [Crossref]




