
Machine learning for predicting accuracy of lung and liver tumor motion tracking using radiomic features
Introduction
Respiratory management has long been a crucial issue in radiotherapy (1-5). All tumor sites in the thorax and abdomen are affected by respiratory motion (6). Moreover, in the case of the lung and liver in particular, tumor motion is complex, uninterrupted, patient-dependent, and can occur in any direction (7). These factors pose significant challenges to image acquisition, treatment planning, and radiation delivery (8-10). To improve the accuracy of treatment planning and delivery, multiple approaches of respiratory management have been proposed and applied in clinical practice. For example, for tumors in the thoracic and abdominal regions, additional margins 5 mm or larger in each direction are added to the internal target volume (ITV) to generate a planning target volume (PTV) wherein the set-up error, respiratory motion, and its influence on optimized treatment planning are taken into account (11,12). However, although additional margins to the gross tumor volume (GTV) enable the delivery of sufficient doses of radiation to the tumor, they also lead to the delivery of high doses to adjacent organs at risk (OARs) (13-15). Thus, the risk of severe radiation-induced side effects on OARs, such as radiation pneumonia (16), esophagitis (17), and radiogenic liver damage (18), is increased. To reduce these risks caused by the extension of GTVs, real-time tumor tracking will have to be improved in a direct or indirect manner (19,20). Minimizing PTV margin is an important task in radiotherapy (19).
As a method for accommodating respiratory motion, real-time tumor tracking, which focuses on dynamically adjusting the radiation beam to accurately adapt to tumor site changes, is considered to be the most efficient motion compensation approach (21). In most cases, the direct observation of tumor movement requires the use of continuous fluoroscopy and/or radioactive markers. However, percutaneous implantation is invasive and uncomfortable for the patient, who, as a result of using fiducial markers, may suffer from complications such as bleeding, pneumothorax, and infection (22). The migration and stability of fiducial markers are also a concern (23). In addition, this method requires the orthogonal arrangement of two X-ray fluoroscopy systems and delivers additional imaging dose to the patient (24,25). It is thus necessary to infer the location of the tumor from external respiratory signals, especially if these tools are unavailable for the patient or in the hospital. Methods for obtaining respiratory signals are generally noninvasive, radiation dose-free, and widely applicable in respiratory gating, and include but are not limited to the use of infrared reflective markers, spirometry, real-time position monitoring, and surface-guided radiation therapy (SGRT) (2,12,26).
The correlation between external surrogates and tumor motion is patient-specific and varies widely among individuals, ranging from approximately 0.4 to nearly 1.0 (27-30). For patients with strong correlation coefficients (>0.8) between skin and tumor motion (31,32), the feasibility of using external surrogate motion to predict tumor locations in the lung or diaphragm has been proven (32,33). However, because the correlation is influenced by the observation direction (2), tumor site (30), marker location, and breathing pattern (24), tumor motion cannot be accurately predicted for some patients. Furthermore, tumor motion should be assessed individually because there is no correlation between tumor motion, size, location, or pulmonary function (34). The motivation for this study was to develop a method for accurately identifying the subset of patients with large errors in the internal–external correlation model using radiomics to inform the selection of an appropriate respiratory management approach before clinical practice.
Radiomics focuses on the improvement of image analysis using automated data characterization algorithms (35). Through the use of radiomics, quantitative radiographic phenotype features (tumor intensity, shape, texture, and transformed features) can be extracted from multiple medical image modalities (36). There have been numerous studies involving the use of radiomic features extracted from medical images in typifying tumor types and grades and producing prognoses (37). Several studies have discovered that some radiomic features in medical images of tumors are affected by respiratory motion (38,39). Prior research has suggested the potential feasibility of using radiomic features extracted via computed tomography (CT) to predict the relationship between internal tumor and external surrogate motions. In this study, we input radiomic features obtained from CT images into diverse classification algorithms based on the use of machine learning to evaluate the feasibility of classifying tumor tracking errors based on radiomics. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-621/rc).
Methods
Coordinate and radiomics dataset
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the ethics committee of the West China Hospital and individual consent for this retrospective analysis was waived. It was registered with the Chinese Clinical Trial Registration Center (registration number: “ChiCTR2100042714”). Image data obtained from a total of 108 lung cancer and 71 liver cancer patients treated at our radiation physics center were included in the study. A schematic of the workflow is shown in Figure 1.
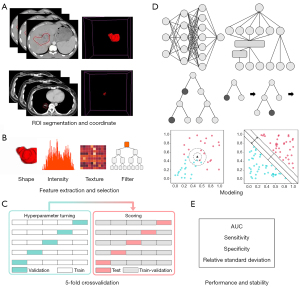
A four-dimensional computed tomography (4DCT) scan was performed in a free-breathing pattern in the treatment simulation room using a Revolution CT instrument (GE Healthcare, Waukesha, WI, USA). Each X-ray scan was synchronized with breathing signals obtained using a Varian Real-time Position Management (RPM) System (Varian Medical Systems, Palo Alto, CA) at the epigastrium below the xiphoid. The 4DCT image and RPM file were loaded into an Advantage V4.7 workstation (General Electric Co., Waukesha, WI), and the RPM motion signals were assigned and averaged according to 10 respiratory phases and recorded as external motion coordinate data; subsequently, 10 respiratory-phase CT (0–90%) images and an average intensity projection (AIP) CT (3 mm) image were produced.
Our radiation oncologist contoured the GTV range over 10 respiratory phases on 4DCT based on the Raystation treatment planning system V4.7.6 (RaySearch Laboratory, Stockholm, Sweden) and generated an ITV on the AIP CT that contained all motion ranges. The ITV was solely segmented from the AIP CT using an image computing platform (3D Slicer) (40), and then a set of radiomic features was extracted from the AIP-ITV using pyradiomics v3.0.1. The extracted features included 14 shape features describing the 3D physical appearance of the tumor, 18 first-order features describing the distribution of the region of interest (ROI) intensities excluding spatial relations, 68 texture features describing the inter-relationships among voxel intensities, and 1,118 transform-based features. In all, the radiomic features of 161 lung and 91 liver tumors were collected. The workflow was reviewed and supervised by radiation oncologists.
Tumor tracking accuracy
Measures of the relationship between external surrogate and internal tumor positions include but are not limited to correlation (41), phase discrepancy (27), external/internal correlation model, and tumor tracking error (42). However, correlation is insufficient for fully describing the relationship between external surrogate and internal tumor positions (27). To assess the accuracy of tumor tracking in clinical practice, direct indicators are necessary. Hence, we used tracking error as this indicator and decided not to calculate for correlation.
Currently, marker-based imaging using metal markers for target localization in real-time tumor-tracking radiotherapy has an accuracy of approximately 2 mm for single-projection imaging and 1 mm for several-view imaging (43). In line with the threshold generally recommended by research studies, by which the accelerator delivers a dose only when the marker is within 1–2 mm of its planned coordinates relative to the isocenter in treatment (44), we defined 2 mm as the accuracy threshold for tumor tracking, using RPM in all respiratory phases; patients who had an error greater than 2 mm between tumor motion and the predicted value in any phase were considered positive samples. The ratios of positive to negative samples were approximately 1:4 and 1:1 for lung and liver tumors, respectively.
Amplitude fitting
Amplitude fitting was used to predict internal tumor motion based on the geometric center location of each tumor determined for each phase. RPM motions were fitted with superior–inferior tumor motions over 10 phases using the least-squares method, as shown in Figure 2. The amplitude of the skin motion was magnified (or reduced) and translated to match the tumor motion using the following relation:

where xi and yi are the tumor and RPM coordinates in phase I, respectively, and k and b are selected to best minimize the fitting error.
Feature engineering
Feature engineering involves two steps: pre-processing and selection. The first step is performed to remove unit limits from the data and transform them into dimensionless pure values to enable the weighting of indices with different units or magnitudes. In our approach, the mean is subtracted from each feature and divided by the standard deviation to complete standardization. In the second step, 1,218 features are extracted from each image. Given that radiomics, like any high-throughput data mining approach, suffers from the curse of dimensionality, appropriate feature selection can be used to decrease the risk of data overfitting to improve performance on new samples (45-47). In our approach, the 5-fold cross-validation least absolute shrinkage and selection operator (LassoCV) is used to identify the principal features relevant to each binary-valued label (“0” for “negative” or “1” for “positive”).
Modeling
Supervised learning has been widely applied to classification tasks in machine learning (48-51). To find accurate and reliable classifiers using radiomics-derived models, we developed several models based on the following 13 algorithms: multilayer perceptron (MLP), wide and deep (W&D), categorical boosting (Cat), light gradient boosting machine (Light), extreme gradient boosting (XGB), adaptive boosting (Ada), random forest (RF), decision tree (DT), logistic regression via stochastic gradient descent (SGD), gaussian naive bayes (GNB), support vector classifiers (SVC), linear support vector classifiers (linearSVC), and K-nearest neighbor (KNN).
Training was performed using stratified random sampling and five-fold cross validation (Figure 1C), a reasonable and accurate evaluation approach in which data imbalances are addressed. Each of the 13 types of machine learning and deep learning algorithms developed in this study for use on the lung and liver utilized Bayesian optimization for hyperparameter tuning on a four-fold dataset via five-fold cross validation to obtain the best classification performance. Specifically, a total of 13,000 models were trained and tested [i.e., 13 algorithm types × 2 (for lung and liver) × 100 sample distributions per algorithm × 5 (five-fold cross validation)]. Furthermore, each model had different hyperparameters, and an additional five-fold cross validation was applied to each model to tune its hyperparameters. To train on each sample distribution, each model was subjected to hyperparameter tuning and testing on 4/5 and 1/5 of its dataset, respectively.
Prediction performance and stability
The classification performance of each algorithm was assessed using the area under the receiver operating characteristic curve (AUC), sensitivity, and specificity metrics. The averages of each of these metrics in five-fold cross validation were considered to be each classifier’s performance for a single sampling. We used the average of performance for 100 sampling times to determine the representative AUC, sensitivity, and specificity of each algorithm. Relative standard deviation (RSD) was used to assess the stabilities of the respective algorithms:
where σ and µ are the standard deviation and mean of 100 performance values, respectively.
To identify accurate algorithms, rank tables of AUC and scatterplots were created for the lung and liver. Algorithms above the median of performance and stability were identified as highly accurate and reliable. To determine whether there were statistically significant differences in performance between algorithms, we used the Wilcoxon rank-sum test, a non-parametric statistical significance test (52), to conduct a paired sample test between the algorithms above and below the AUC median.
Results
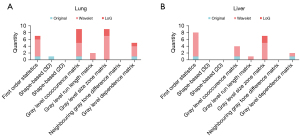
LassoCV selected 33 and 22 features for lung and liver tumors, respectively (Figure 3). The lung and liver classifiers were based on the same radiomic feature types, which included first-order statistics, gray-level co-occurrence matrix (GLCM), gray-level run length matrix (GLRLM), gray-level size zone matrix (GLSZM), and gray-level dependence matrix (GLDM), and the same image types, including original, wavelet, and Laplacian of Gaussian (LoG). In addition, one shape3D feature was presented among the lungs.
The AUC, sensitivity, specificity, and RSD were used to depict the predictive performance and stabilities of the different algorithms for the lung and liver (Table 1). For the lung, SVC and SGD performed best in terms of prediction (AUC: 0.941±0.010 and 0.941±0.011, respectively) (mean ± std), whereas DT (AUC: 0.848±0.026) performed worst. The most sensitive classifier was linearSVC (0.848±0.025); the least sensitive was XGB (0.723±0.036). The most specific classifiers were MLP (0.936±0.014) and W&D (0.936±0.014); the least specific was GNB (0.834±0.018).
Table 1
| Location | Algorithm | AUC | Sensitivity | Specificity | |||||
|---|---|---|---|---|---|---|---|---|---|
| Performance | Stability (%) | Performance | Stability (%) | Performance | Stability (%) | ||||
| Lung | SVC | 0.941±0.010 | 1.07 | 0.799±0.033 | 4.10 | 0.930±0.016 | 1.77 | ||
| SGD | 0.941±0.011 | 1.22 | 0.828±0.027 | 3.25 | 0.919±0.016 | 1.72 | |||
| linearSVC | 0.939±0.013 | 1.33 | 0.848±0.025 | 2.92 | 0.909±0.018 | 2.02 | |||
| MLP | 0.934±0.011 | 1.22 | 0.802±0.019 | 2.33 | 0.936±0.014 | 1.47 | |||
| W&D | 0.934±0.010 | 1.02 | 0.801±0.023 | 2.82 | 0.936±0.014 | 1.54 | |||
| KNN | 0.912±0.010 | 1.15 | 0.774±0.023 | 2.99 | 0.913±0.013 | 1.41 | |||
| Light | 0.911±0.011 | 1.23 | 0.802±0.030 | 3.74 | 0.886±0.014 | 1.55 | |||
| RF | 0.908±0.010 | 1.09 | 0.754±0.020 | 2.69 | 0.912±0.015 | 1.64 | |||
| Cat | 0.904±0.010 | 1.13 | 0.757±0.023 | 3.08 | 0.910±0.015 | 1.60 | |||
| Ada | 0.890±0.023 | 2.62 | 0.783±0.035 | 4.43 | 0.900±0.019 | 2.06 | |||
| XGB | 0.889±0.013 | 1.50 | 0.723±0.036 | 5.04 | 0.899±0.021 | 2.31 | |||
| GNB | 0.881±0.010 | 1.18 | 0.838±0.017 | 2.09 | 0.834±0.018 | 2.15 | |||
| DT | 0.848±0.026 | 3.08 | 0.738±0.046 | 6.26 | 0.843±0.027 | 3.24 | |||
| Liver | SGD | 0.892±0.020 | 2.28 | 0.829±0.042 | 5.07 | 0.796±0.039 | 4.90 | ||
| MLP | 0.887±0.019 | 2.19 | 0.862±0.035 | 4.05 | 0.744±0.035 | 4.68 | |||
| SVC | 0.887±0.019 | 2.20 | 0.807±0.044 | 5.49 | 0.805±0.041 | 5.05 | |||
| linearSVC | 0.882±0.025 | 2.79 | 0.781±0.047 | 5.95 | 0.829±0.039 | 4.66 | |||
| W&D | 0.833±0.021 | 2.48 | 0.717±0.042 | 5.88 | 0.782±0.042 | 5.43 | |||
| KNN | 0.809±0.021 | 2.63 | 0.859±0.039 | 4.52 | 0.618±0.035 | 5.61 | |||
| GNB | 0.807±0.026 | 3.19 | 0.794±0.044 | 5.54 | 0.677±0.045 | 6.69 | |||
| RF | 0.801±0.031 | 3.81 | 0.818±0.049 | 5.96 | 0.627±0.042 | 6.66 | |||
| Ada | 0.772±0.032 | 4.15 | 0.755±0.056 | 7.38 | 0.661±0.050 | 7.61 | |||
| Cat | 0.761±0.027 | 3.48 | 0.766±0.046 | 5.97 | 0.618±0.028 | 4.47 | |||
| DT | 0.739±0.034 | 4.60 | 0.789±0.058 | 7.39 | 0.610±0.053 | 8.66 | |||
| XGB | 0.735±0.034 | 4.59 | 0.736±0.047 | 6.40 | 0.612±0.038 | 6.13 | |||
| Light | 0.677±0.031 | 4.62 | 0.769±0.053 | 6.84 | 0.566±0.021 | 3.78 | |||
Results are ranked in descending order of AUC value. The value is presented as means ± standard deviation. AUC, area under the receiver operating characteristic curve; RSD, relative standard deviation; SVC, support vector classifier; SGD, logistic regression via stochastic gradient descent; linearSVC, linear support vector classifier; MLP, multilayer perceptron; W&D, wide and deep; KNN, K-nearest neighbor; Light, light gradient boosting machine; RF, random forest; Cat, categorical boosting; Ada, adaptive boosting; XGB, extreme gradient boosting; GNB, gaussian naive bayes; DT, decision tree.
To comprehensively evaluate the models, both classification performance and stability were evaluated using scatterplots (Figure 4). The results suggest MLP and W&D to be the preferable algorithms because these were the only models, among those tested, whose performance and stability were both greater than or equal to the median [AUC: (0.911, 1.219); sensitivity: (0.799, 3.077); specificity: (0.901, 1.719) (performance and stability)]. The results of Wilcoxon rank-sum tests between the above- and below-median AUC groups are listed in Table 2. Against a null hypothesis that the population median of the difference between a pair of samples was zero, all P values were less than 0.05, except for KNN compared with Light (P value: 0.4422).
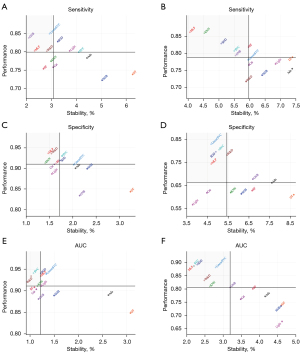
Table 2
| Location | > Median | ≤ Median | ||||||
|---|---|---|---|---|---|---|---|---|
| Light | RF | Cat | Ada | XGB | GNB | DT | ||
| Lung | SVC | 5.510×10−18 | 3.955×10−18 | 3.954×10−18 | 5.189×10−18 | 3.955×10−18 | 3.955×10−18 | 3.956×10−18 |
| SGD | 9.313×10−18 | 4.330×10−18 | 3.955×10−18 | 4.077×10−18 | 3.952×10−18 | 3.953×10−18 | 3.955×10−18 | |
| linearSVC | 2.410×10−17 | 7.010×10−18 | 5.034×10−18 | 4.077×10−18 | 4.076×10−18 | 3.953×10−18 | 3.956×10−18 | |
| MLP | 3.333×10−17 | 9.741×10−18 | 6.029×10−18 | 5.190×10−18 | 4.076×10−18 | 3.955×10−18 | 3.955×10−18 | |
| W&D | 5.805×10−18 | 4.075×10−18 | 3.951×10−18 | 4.330×10−18 | 3.955×10−18 | 3.955×10−18 | 3.956×10−18 | |
| KNN | 0.4422 | 1.502×10−4 | 2.440×10−11 | 1.539×10−13 | 7.223×10−18 | 4.462×10−18 | 3.955×10−18 | |
| Liver | SVC | 3.955×10−18 | 3.954×10−18 | 3.950×10−18 | 3.954×10−18 | 3.954×10−18 | 3.955×10−18 | 3.955×10−18 |
| SGD | 3.954×10−18 | 3.953×10−18 | 3.953×10−18 | 3.953×10−18 | 3.954×10−18 | 4.075×10−18 | 3.956×10−18 | |
| linearSVC | 3.954×10−18 | 3.953×10−18 | 3.953×10−18 | 3.955×10−18 | 3.954×10−18 | 4.076×10−18 | 3.955×10−18 | |
| MLP | 3.955×10−18 | 3.954×10−18 | 3.952×10−18 | 3.955×10−18 | 3.952×10−18 | 3.955×10−18 | 3.955×10−18 | |
| W&D | 3.954×10−18 | 2.243×10−15 | 3.953×10−18 | 5.512×10−18 | 3.954×10−18 | 5.654×10−12 | 3.955×10−18 | |
| KNN | 3.955×10−18 | 5.435×10−3 | 8.641×10−18 | 5.065×10−15 | 8.646×10−18 | 0.5047 | 1.066×10−17 | |
Median values of AUC are considered reference values. AUC, area under the receiver operating characteristic curve; Light, light gradient boosting machine; RF, random forest; Cat, categorical boosting; Ada, adaptive boosting; XGB, extreme gradient boosting; GNB, gaussian naive bayes; DT, decision tree; SVC, support vector classifier; SGD, logistic regression via stochastic gradient descent; linearSVC, linear support vector classifier; MLP, multilayer perceptron; W&D, wide and deep; KNN, K-nearest neighbor.
For the liver, SGD had the best predictive performance (AUC: 0.892±0.020), whereas Light performed worst (AUC: 0.677±0.031) (Table 1). The most sensitive classifier was MLP (0.862±0.035); the least sensitive was W&D (0.717±0.042). The most specific classifier was linearSVC (0.829±0.039); the least specific was Light (0.566±0.021). A comprehensive evaluation revealed three classifiers—SGD, MLP, and SVC—to be the preferable algorithms for the liver based on classification performance and stabilities greater than or equal to the median [AUC (0.807, 3.192), sensitivity (0.789, 5.954), and specificity (0.661, 5.430)]. The only P value greater than 0.05 was for the comparison between KNN and GNB (P value: 0.5047) (Table 2).
The performance and stabilities of the classifiers were significantly better on the lung than on the liver, except in terms of sensitivity, where there was no significant difference (Wilcoxon rank-sum test, P>0.05). In each scatterplot shown in Figure 4, MLP is the only algorithm located in the upper-left quadrant (indicating superior results in terms of both performance and sensitivity).
Discussion
In this study, a novel approach using artificial intelligence to accurately predict internal tumor motion using external respiratory signals based on a radiomics-based system for predicting lung and liver tumor motion-tracking errors was proposed. The proposed method applies the least-squares method to fit internal tumor motion with RPM motion, based on CT images used as inputs, to identify the fitting error. Thirteen models from different classifier families were developed and validated through comprehensive and unbiased analysis involving 100-time five-fold cross-validation on each model to highlight the repeatability of the model training.
The MLP model demonstrated excellent classification performance and stability for both lung and liver tumors, which proved that the ITV segmented from AIP CT images and produced via the proposed method contains sufficient information on the relationship between RPM and tumor motion. The many parallel processing units and multiple embedded hidden layers of MLP provide it with processing abilities and informational advantages well matched to the challenges of processing radiomic data (53-56). The performance of MLP can then be further improved through the use of sufficient and balanced data, given that the weight parameters used in most neural networks are designed to be optimized using sufficient and balanced data (57,58).
Although the selected features used by the lung and liver models developed in this study came from nearly identical image and feature types, the models generally performed better in terms of classification performance and stability on the lung, except in terms of the sensitivity metric. This difference can potentially be attributed to differences between the lungs and liver in terms of the tissue density change caused by tumor motion. For the same respiratory pattern, tissue density changes in the beam’s eye view for a centrally located liver tumor will be small, given the smaller difference between the tumor and surrounding liver tissue; by contrast, tumor motion caused by respiration can cause huge tissue density changes due to the air/tissue interfaces in the lung and liver dome regions. Another possible reason is the difference in respiratory patterns between the lung and liver. Liver tumors tend to rotate more in all three directions than lung tumors do (4,59-61); the effectiveness of radiomic features as predictors of tumor motion has been shown to be affected by tumor rotation (39). Compared with that in lung tissue, respiratory-induced deformation is larger in the liver, and localization error increases with tumor deformation (62-64). Therefore, the geometric center location is not sufficient to describe tumor motion in translation. In addition, lung samples tend to be more unevenly distributed than liver samples; given that, for imbalanced data, the performance of the classifier is skewed toward the majority class, the accuracy tends to be poor for the minority class (65).
The classification performance results observed in this study likely originated from different patterns and/or degrees of tumor motion rather than lesions representing different phenotypes. Radiomics aims to quantify image patterns, allowing the differentiation of tumor phenotypes; these patterns can be influenced by tumor motion. For example, first-order, GLCM, and GLRLM have smaller mean concordance correlation coefficients than those of Shape for 3D/4D images produced by either CT or positron emission tomography (39). Du et al. reported that first-order, GLCM, GLRLM, GLSZM, and GLDM have larger percentages of instability than those of other radiomic feature types extracted using 4DCT (66). Larue et al. determined that features based on unfiltered images are more robust than wavelet-filtered features (38). The radiomic features that were inferred to be sensitive to respiratory motion are highly consistent with those on which our model is based. It is necessary to inspect the tracking error of tumor motion before each session because of the variability of respiratory motion during inter-fraction treatment and stability during intra-fraction treatment when the respiratory pattern is regular (67,68). Radiomic features can be extracted from cone-beam CT (CBCT) or 4D-CBCT images, which also contain tumor motion information from the duration of the treatment.
Artificial intelligence algorithms based on the radiomic features of CT can accurately classify whether a patient is suitable for indirect tumor tracking using RPM; this approach is also applicable to tumor tracking using SGRT. Most methods for estimating tumor motion and for indirect tumor tracking achieve accuracy in tracking performance through the use of complicated systems that ignore patient specificity (69). For some patients, it is possible to use simple and common techniques to achieve accurate tumor tracking from external respiratory signals because of the strong correlation coefficients between their external signals and internal tumor motion (31). For patients with acceptable tumor tracking accuracies, the tumor tracking error may be considered as the margin to be added to the ITV to generate the PTV, reducing the risk of side effects on OARs. However, for 4DCT, the correlation between tumor motion and external surrogates cannot be directly observed in clinical settings; the accuracy of tumor tracking is generally obtained via manual extraction, preprocessing, and fitting of tumor motion and surrogate signal information. The assessment process can cause additional clinical workload, especially when performed on every patient. Our study focused on distinguishing this type of patient from a general pool of patients based on CT imaging and then applying a simpler and more rapid process than the commonly used approach of manually extracting tumor and body surface motion information. The generation principle used in AIP also makes it suitable as an input for determining the appropriate respiratory management approach for scanning 4DCT images using other respiratory signals (70,71).
Study limitations
Indirect tumor tracking using external markers relies on the assumption that the correlation between the external surface and internal tumor position remains constant both inter- and intra-fractionally (72). This presented a limitation to our approach, in which we inspected the respiratory circle before treatment to classify the accuracy of tracking. In response to radiation therapy and other potential concomitant therapies, tumor and normal tissues can shrink and grow; furthermore, respiratory motion patterns vary daily (6,73). The 4DCT scans we took presented only snapshots of the times before treatment, forcing us to ignore correlation variations in the intra- and inter-fractions in this study. To help overcome these problems, we should ensure that the accuracy of the correlation model used in indirect tumor tracking is guaranteed over the entire course of radiotherapy treatment. A feasible solution is the use of CBCT and audiovisual biofeedback for respiratory guidance, which can be applied prior to each treatment to adjust the respiratory baseline, track tumor motion, and assess variations in the tumor and organ volumes to guarantee the reproducibility of the respiratory cycle in 4DCT, update the correlation model for each treatment session, or identify whether an additional 4DCT scan should be conducted (26,74).
Conclusions
Respiratory-sensitive radiomic features extracted from CT images of lung and liver tumors were proved to contain sufficient information for establishing an external/internal motion relationship. We developed a rapid and accurate method based on radiomics for classifying the accuracy of monitoring a patient’s external surface for lung and liver tumor tracking. Several machine learning algorithms—in particular, MLP—demonstrated excellent classification performance and stability.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant number 81972848), and the Science and Technology Department of Sichuan Province (grant number 2021YFS0143).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-621/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-621/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics committee of West China Hospital and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhao YT, Liu ZK, Wu QW, Dai JR, Zhang T, Jia AY, Jin J, Wang SL, Li YX, Wang WH. Observation of different tumor motion magnitude within liver and estimate of internal motion margins in postoperative patients with hepatocellular carcinoma. Cancer Manag Res 2017;9:839-48. [Crossref] [PubMed]
- Zhang J, Wang L, Li X, Huang M, Xu B. Quantification of Intrafraction and Interfraction Tumor Motion Amplitude and Prediction Error for Different Liver Tumor Trajectories in Cyberknife Synchrony Tracking. Int J Radiat Oncol Biol Phys 2021;109:1588-605. [Crossref] [PubMed]
- Yu ZH, Lin SH, Balter P, Zhang L, Dong L. A comparison of tumor motion characteristics between early stage and locally advanced stage lung cancers. Radiother Oncol 2012;104:33-8. [Crossref] [PubMed]
- Xu Q, Hanna G, Grimm J, Kubicek G, Pahlajani N, Asbell S, Fan J, Chen Y, LaCouture T. Quantifying rigid and nonrigid motion of liver tumors during stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2014;90:94-101. [Crossref] [PubMed]
- Seppenwoolde Y, Shirato H, Kitamura K, Shimizu S, van Herk M, Lebesque JV, Miyasaka K. Precise and real-time measurement of 3D tumor motion in lung due to breathing and heartbeat, measured during radiotherapy. Int J Radiat Oncol Biol Phys 2002;53:822-34. [Crossref] [PubMed]
- Keall PJ, Mageras GS, Balter JM, Emery RS, Forster KM, Jiang SB, Kapatoes JM, Low DA, Murphy MJ, Murray BR, Ramsey CR, Van Herk MB, Vedam SS, Wong JW, Yorke E. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phys 2006;33:3874-900. [Crossref] [PubMed]
- George R, Suh Y, Murphy M, Williamson J, Weiss E, Keall P. On the accuracy of a moving average algorithm for target tracking during radiation therapy treatment delivery. Med Phys 2008;35:2356-65. [Crossref] [PubMed]
- Wong JW, Sharpe MB, Jaffray DA, Kini VR, Robertson JM, Stromberg JS, Martinez AA. The use of active breathing control (ABC) to reduce margin for breathing motion. Int J Radiat Oncol Biol Phys 1999;44:911-9. [Crossref] [PubMed]
- Shirato H, Shimizu S, Kunieda T, Kitamura K, van Herk M, Kagei K, Nishioka T, Hashimoto S, Fujita K, Aoyama H, Tsuchiya K, Kudo K, Miyasaka K. Physical aspects of a real-time tumor-tracking system for gated radiotherapy. Int J Radiat Oncol Biol Phys 2000;48:1187-95. [Crossref] [PubMed]
- Rietzel E, Chen GT, Choi NC, Willet CG. Four-dimensional image-based treatment planning: Target volume segmentation and dose calculation in the presence of respiratory motion. Int J Radiat Oncol Biol Phys 2005;61:1535-50. [Crossref] [PubMed]
- Purdie TG, Moseley DJ, Bissonnette JP, Sharpe MB, Franks K, Bezjak A, Jaffray DA. Respiration correlated cone-beam computed tomography and 4DCT for evaluating target motion in Stereotactic Lung Radiation Therapy. Acta Oncol 2006;45:915-22. [Crossref] [PubMed]
- Oh SA, Yea JW, Kim SK. Statistical Determination of the Gating Windows for Respiratory-Gated Radiotherapy Using a Visible Guiding System. PLoS One 2016;11:e0156357. [Crossref] [PubMed]
- Wolthaus JW, Sonke JJ, van Herk M, Belderbos JS, Rossi MM, Lebesque JV, Damen EM. Comparison of different strategies to use four-dimensional computed tomography in treatment planning for lung cancer patients. Int J Radiat Oncol Biol Phys 2008;70:1229-38. [Crossref] [PubMed]
- Depuydt T, Poels K, Verellen D, Engels B, Collen C, Buleteanu M, Van den Begin R, Boussaer M, Duchateau M, Gevaert T, Storme G, De Ridder M. Treating patients with real-time tumor tracking using the Vero gimbaled linac system: implementation and first review. Radiother Oncol 2014;112:343-51. [Crossref] [PubMed]
- Kamerling CP, Fast MF, Ziegenhein P, Menten MJ, Nill S, Oelfke U. Real-time 4D dose reconstruction for tracked dynamic MLC deliveries for lung SBRT. Med Phys 2016;43:6072. [Crossref] [PubMed]
- Livingston GC, Last AJ, Shakespeare TP, Dwyer PM, Westhuyzen J, McKay MJ, Connors L, Leader S, Greenham S. Toxicity and dosimetric analysis of non-small cell lung cancer patients undergoing radiotherapy with 4DCT and image-guided intensity modulated radiotherapy: a regional centre's experience. J Med Radiat Sci 2016;63:170-8. [Crossref] [PubMed]
- Hashimoto T, Shirato H, Kato M, Yamazaki K, Kurauchi N, Morikawa T, Shimizu S, Ahn YC, Akine Y, Miyasaka K. Real-time monitoring of a digestive tract marker to reduce adverse effects of moving organs at risk (OAR) in radiotherapy for thoracic and abdominal tumors. Int J Radiat Oncol Biol Phys 2005;61:1559-64. [Crossref] [PubMed]
- Chen G, Han Y, Zhang H, Tu W, Zhang S. Radiotherapy-Induced Digestive Injury: Diagnosis, Treatment and Mechanisms. Front Oncol 2021;11:757973. [Crossref] [PubMed]
- Kanoulas E, Aslam JA, Sharp GC, Berbeco RI, Nishioka S, Shirato H, Jiang SB. Derivation of the tumor position from external respiratory surrogates with periodical updating of the internal/external correlation. Phys Med Biol 2007;52:5443-56. [Crossref] [PubMed]
- Yan H, Zhu G, Yang J, Lu M, Ajlouni M, Kim JH, Yin FF. The investigation on the location effect of external markers in respiratory-gated radiotherapy. J Appl Clin Med Phys 2008;9:57-68. [Crossref] [PubMed]
- Schweikard A, Shiomi H, Adler J. Respiration tracking in radiosurgery without fiducials. Int J Med Robot 2005;1:19-27. [Crossref] [PubMed]
- Steinfort DP, Siva S, Kron T, Chee RR, Ruben JD, Ball DL, Irving LB. Multimodality guidance for accurate bronchoscopic insertion of fiducial markers. J Thorac Oncol 2015;10:324-30. [Crossref] [PubMed]
- Imura M, Yamazaki K, Shirato H, Onimaru R, Fujino M, Shimizu S, Harada T, Ogura S, Dosaka-Akita H, Miyasaka K, Nishimura M. Insertion and fixation of fiducial markers for setup and tracking of lung tumors in radiotherapy. Int J Radiat Oncol Biol Phys 2005;63:1442-7. [Crossref] [PubMed]
- Li G, Citrin D, Camphausen K, Mueller B, Burman C, Mychalczak B, Miller RW, Song Y. Advances in 4D medical imaging and 4D radiation therapy. Technol Cancer Res Treat 2008;7:67-81. [Crossref] [PubMed]
- Poulsen PR, Cho B, Sawant A, Ruan D, Keall PJ. Dynamic MLC tracking of moving targets with a single kV imager for 3D conformal and IMRT treatments. Acta Oncol 2010;49:1092-100. [Crossref] [PubMed]
- Fassi A, Schaerer J, Fernandes M, Riboldi M, Sarrut D, Baroni G. Tumor tracking method based on a deformable 4D CT breathing motion model driven by an external surface surrogate. Int J Radiat Oncol Biol Phys 2014;88:182-8. [Crossref] [PubMed]
- Hoisak JD, Sixel KE, Tirona R, Cheung PC, Pignol JP. Correlation of lung tumor motion with external surrogate indicators of respiration. Int J Radiat Oncol Biol Phys 2004;60:1298-306. [Crossref] [PubMed]
- Gallego-Ortiz N, de Xivry JO, Descampe A, et al. Respiratory motion variations from skin surface on lung cancer patients from 4D CT data. Medical Imaging 2014;9034:90342Y.
- Lee D, Greer PB, Paganelli C, Ludbrook JJ, Kim T, Keall P. Audiovisual biofeedback improves the correlation between internal/external surrogate motion and lung tumor motion. Med Phys 2018;45:1009-17. [Crossref] [PubMed]
- Fayad H, Pan T, Clement JF, Visvikis D. Technical note: Correlation of respiratory motion between external patient surface and internal anatomical landmarks. Med Phys 2011;38:3157-64. [Crossref] [PubMed]
- Ozhasoglu C, Murphy MJ. Issues in respiratory motion compensation during external-beam radiotherapy. Int J Radiat Oncol Biol Phys 2002;52:1389-99. [Crossref] [PubMed]
- Ahn S, Yi B, Suh Y, Kim J, Lee S, Shin S, Shin S, Choi E. A feasibility study on the prediction of tumour location in the lung from skin motion. Br J Radiol 2004;77:588-96. [Crossref] [PubMed]
- Seregni M, Cerveri P, Riboldi M, Pella A, Baroni G. Robustness of external/internal correlation models for real-time tumor tracking to breathing motion variations. Phys Med Biol 2012;57:7053-74. [Crossref] [PubMed]
- Stevens CW, Munden RF, Forster KM, Kelly JF, Liao Z, Starkschall G, Tucker S, Komaki R. Respiratory-driven lung tumor motion is independent of tumor size, tumor location, and pulmonary function. Int J Radiat Oncol Biol Phys 2001;51:62-8. [Crossref] [PubMed]
- Xue C, Yuan J, Lo GG, Chang ATY, Poon DMC, Wong OL, Zhou Y, Chu WCW. Radiomics feature reliability assessed by intraclass correlation coefficient: a systematic review. Quant Imaging Med Surg 2021;11:4431-60. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Alic L, Niessen WJ, Veenland JF. Quantification of heterogeneity as a biomarker in tumor imaging: a systematic review. PLoS One 2014;9:e110300. [Crossref] [PubMed]
- Larue RTHM, Van De Voorde L, van Timmeren JE, Leijenaar RTH, Berbée M, Sosef MN, Schreurs WMJ, van Elmpt W, Lambin P. 4DCT imaging to assess radiomics feature stability: An investigation for thoracic cancers. Radiother Oncol 2017;125:147-53. [Crossref] [PubMed]
- Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of Image Features Computed from Conventional and Respiratory-Gated PET/CT Images of Lung Cancer. Transl Oncol 2015;8:524-34. [Crossref] [PubMed]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Onodera Y, Nishioka N, Yasuda K, Fujima N, Torres M, Kamishima T, Ooyama N, Onimaru R, Terae S, Ooizumi S, Nishimura M, Shirato H. Relationship between diseased lung tissues on computed tomography and motion of fiducial marker near lung cancer. Int J Radiat Oncol Biol Phys 2011;79:1408-13. [Crossref] [PubMed]
- Malinowski K, McAvoy TJ, George R, Dietrich S, D'Souza WD. Incidence of changes in respiration-induced tumor motion and its relationship with respiratory surrogates during individual treatment fractions. Int J Radiat Oncol Biol Phys 2012;82:1665-73. [Crossref] [PubMed]
- Riboldi M, Orecchia R, Baroni G. Real-time tumour tracking in particle therapy: technological developments and future perspectives. Lancet Oncol 2012;13:e383-91. [Crossref] [PubMed]
- Shirato H, Onimaru R, Ishikawa M, Kaneko J, Takeshima T, Mochizuki K, Shimizu S, Umegaki K. Real-time 4-D radiotherapy for lung cancer. Cancer Sci 2012;103:1-6. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Avanzo M, Stancanello J, El Naqa I. Beyond imaging: The promise of radiomics. Phys Med 2017;38:122-39. [Crossref] [PubMed]
- Pekalska E, Duin RPW. The Dissimilarity Representation for Pattern Recognition: foundations and applications. Series in Machine Perception and Artificial Intelligence: Volume 64. World Scientific; 2005.
- Choudhary R, Gianey HK. Comprehensive review on supervised machine learning algorithms. 2017 International Conference on Machine Learning and Data Science, IEEE, 2017:37-43.
- Meng Y, Zhang H, Li Q, Xing P, Liu F, Cao K, Fang X, Li J, Yu J, Feng X, Ma C, Wang L, Jiang H, Lu J, Bian Y, Shao C. Noncontrast Magnetic Resonance Radiomics and Multilayer Perceptron Network Classifier: An approach for Predicting Fibroblast Activation Protein Expression in Patients With Pancreatic Ductal Adenocarcinoma. J Magn Reson Imaging 2021;54:1432-43. [Crossref] [PubMed]
- Wang S, Dong D, Zhang W, Hu H, Li H, Zhu Y, Zhou J, Shan X, Tian J. Specific Borrmann classification in advanced gastric cancer by an ensemble multilayer perceptron network: a multicenter research. Med Phys 2021;48:5017-28. [Crossref] [PubMed]
- Yun J, Park JE, Lee H, Ham S, Kim N, Kim HS. Radiomic features and multilayer perceptron network classifier: a robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci Rep 2019;9:5746. [Crossref] [PubMed]
- Schober P, Vetter TR. Nonparametric Statistical Methods in Medical Research. Anesth Analg 2020;131:1862-3. [Crossref] [PubMed]
- Yu H, Samuels DC, Zhao YY, Guo Y. Architectures and accuracy of artificial neural network for disease classification from omics data. BMC Genomics 2019;20:167. [Crossref] [PubMed]
- Tkachev V, Sorokin M, Borisov C, Garazha A, Buzdin A, Borisov N. Flexible Data Trimming Improves Performance of Global Machine Learning Methods in Omics-Based Personalized Oncology. Int J Mol Sci 2020; [Crossref] [PubMed]
- Castiglioni I, Rundo L, Codari M, Di Leo G, Salvatore C, Interlenghi M, Gallivanone F, Cozzi A, D'Amico NC, Sardanelli F. AI applications to medical images: From machine learning to deep learning. Phys Med 2021;83:9-24. [Crossref] [PubMed]
- Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of Deep Learning in Biomedicine. Mol Pharm 2016;13:1445-54. [Crossref] [PubMed]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform 2017;18:851-69. [PubMed]
- Cui S, Tseng HH, Pakela J, Ten Haken RK, El Naqa I. Introduction to machine and deep learning for medical physicists. Med Phys 2020;47:e127-47. [Crossref] [PubMed]
- Dhont J, Harden SV, Chee LYS, Aitken K, Hanna GG, Bertholet J. Image-guided Radiotherapy to Manage Respiratory Motion: Lung and Liver. Clin Oncol (R Coll Radiol) 2020;32:792-804. [Crossref] [PubMed]
- Huang CY, Tehrani JN, Ng JA, Booth J, Keall P. Six degrees-of-freedom prostate and lung tumor motion measurements using kilovoltage intrafraction monitoring. Int J Radiat Oncol Biol Phys 2015;91:368-75. [Crossref] [PubMed]
- Bertholet J, Worm ES, Fledelius W, Høyer M, Poulsen PR. Time-Resolved Intrafraction Target Translations and Rotations During Stereotactic Liver Radiation Therapy: Implications for Marker-based Localization Accuracy. Int J Radiat Oncol Biol Phys 2016;95:802-9. [Crossref] [PubMed]
- Seppenwoolde Y, Wunderink W, Wunderink-van Veen SR, Storchi P, Méndez Romero A, Heijmen BJ. Treatment precision of image-guided liver SBRT using implanted fiducial markers depends on marker-tumour distance. Phys Med Biol 2011;56:5445-68. [Crossref] [PubMed]
- Lu XQ, Shanmugham LN, Mahadevan A, Nedea E, Stevenson MA, Kaplan I, Wong ET, La Rosa S, Wang F, Berman SM. Organ deformation and dose coverage in robotic respiratory-tracking radiotherapy. Int J Radiat Oncol Biol Phys 2008;71:281-9. [Crossref] [PubMed]
- von Siebenthal M, Székely G, Lomax AJ, Cattin PC. Systematic errors in respiratory gating due to intrafraction deformations of the liver. Med Phys 2007;34:3620-9. [Crossref] [PubMed]
- Kaur H, Pannu HS, Malhi AK. A systematic review on imbalanced data challenges in machine learning: applications and solutions. ACM Comput Surv 2020;52:1-36. [Crossref]
- Du Q, Baine M, Bavitz K, McAllister J, Liang X, Yu H, Ryckman J, Yu L, Jiang H, Zhou S, Zhang C, Zheng D. Radiomic feature stability across 4D respiratory phases and its impact on lung tumor prognosis prediction. PLoS One 2019;14:e0216480. [Crossref] [PubMed]
- Goossens S, Senny F, Lee JA, Janssens G, Geets X. Assessment of tumor motion reproducibility with audio-visual coaching through successive 4D CT sessions. J Appl Clin Med Phys 2014;15:4332. [Crossref] [PubMed]
- Chen H, Zhong Z, Yang Y, Chen J, Zhou L, Zhen X, Gu X. Internal Motion Estimation by Internal-external Motion Modeling for Lung Cancer Radiotherapy. Sci Rep 2018;8:3677. [Crossref] [PubMed]
- Zhao W, Shen L, Islam MT, Qin W, Zhang Z, Liang X, Zhang G, Xu S, Li X. Artificial intelligence in image-guided radiotherapy: a review of treatment target localization. Quant Imaging Med Surg 2021;11:4881-94. [Crossref] [PubMed]
- Li XA, Stepaniak C, Gore E. Technical and dosimetric aspects of respiratory gating using a pressure-sensor motion monitoring system. Med Phys 2006;33:145-54. [Crossref] [PubMed]
- Vedam SS, Keall PJ, Kini VR, Mostafavi H, Shukla HP, Mohan R. Acquiring a four-dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol 2003;48:45-62. [Crossref] [PubMed]
- Berbeco RI, Nishioka S, Shirato H, Chen GT, Jiang SB. Residual motion of lung tumours in gated radiotherapy with external respiratory surrogates. Phys Med Biol 2005;50:3655-67. [Crossref] [PubMed]
- Ge J, Santanam L, Noel C, Parikh PJ. Planning 4-dimensional computed tomography (4DCT) cannot adequately represent daily intrafractional motion of abdominal tumors. Int J Radiat Oncol Biol Phys 2013;85:999-1005. [Crossref] [PubMed]
- De Ruysscher D, Faivre-Finn C, Nestle U, Hurkmans CW, Le Péchoux C, Price A, Senan S. European Organisation for Research and Treatment of Cancer recommendations for planning and delivery of high-dose, high-precision radiotherapy for lung cancer. J Clin Oncol 2010;28:5301-10. [Crossref] [PubMed]


