
A deep learning model for lymph node metastasis prediction based on digital histopathological images of primary endometrial cancer
Introduction
Endometrial cancer (EC) is a common gynecological malignant tumor. In the past 20 years, the incidence of EC has shown an upward trend worldwide, becoming the most prevalent malignant tumor of the female reproductive system in some developed countries (1,2). Lymph node metastasis (LNM) is a common complication of EC and an independent risk factor that affects postoperative recurrence and prognosis. Clinical studies have shown that EC patients with LNM often have a poor prognosis (3,4). Indeed, patients without LNM have a 5-year overall survival (OS) rate of 96% which drops to 57% in the presence of systemic pelvic LNM and 49.4% with para-aortic LNM (3), and recurrence rates are as high as 60% (4). Therefore, accurate prediction of the presence of LNM improves prognosis evaluation and a risk prediction model has great clinical significance.
According to the National Comprehensive Cancer Network (NCCN) guidelines, “Mayo standards” and previous LNM prediction studies, CA125 preoperative serum level, tumor pathological type, size, deep muscle invasion, cervical invasion, adnexa invasion, vascular cancer thrombus and ascites are considered to be the main risk factors for LNM in EC (5,6). However, different models identify different risk factors, making unified clinical practice standards for LNM risk stratification difficult. In evaluator errors and inter-observer disagreements among pathologists lead to false negative or positive results. Both problems are fundamental and may affect individualized clinical decisions. Preoperative medical images, such as computed tomography (CT) and magnetic resonance imaging (MRI), are the most common current approaches for LNM assessment, especially enhanced pelvic MRI which is the preferred method from 2018 NCCN guidelines (5). However, a meta-analysis showed a sensitivity of 0.68, specificity of 0.96, AUC of 0.82 and accuracy of 0.75 for 18F-fluorodeoxyglucose-positron emission tomography (FDG-PET) or PET/CT in LNM preoperative prediction (7). Economic factors also restrict patients’ access to 18F-FDG-PET or PET/CT.
The hematoxylin and eosin (HE)-stained histopathological images which show tumor tissue structure and cellular characteristics are considered the “gold standard” for tumor diagnosis, generating richer information with multi-dimensional features than MRI/CT radiological or ultrasound images. However, since the derivation of information from pathological images has traditionally relied on subjective evaluation by pathologists, exploitation of the complexity and richness of the images has been difficult. The traditional method of artificial description has limited generation of a unified standard for risk prediction, diagnosis and treatment plans. Artificial intelligence (AI) technologies may offer solutions to this and other issues (8,9). AI models trained to interpret digital pathology imaging have been shown to predict specific clinical events, such as treatment response, prognosis assessment, classification, grading or scoring of different cancers (10-15) and detection of lymph node metastases (16). Takamatsu used Image J, a deep learning (DL) algorithm, to predict risk of preoperative LNM from pathological images of T1 colorectal cancer by extracting morphologic parameters from whole-slide images (WSI). The resulting areas under the curve (AUCs) were 0.938 for DL model and 0.826 for the conventional method (17), illustrating the successful combination of DL and pathological tissue morphology images for LNM prediction. However, to the best of our knowledge, in-depth analysis of histopathological images giving the essential characteristics of EC for DL has not been conducted.
We argue that AI has utility for predicting LNM in EC from analysis of histopathological images. The current study employed a DL neural network algorithm to fulfil a binary classification task and predict the presence or absence of LNM in EC. The model was verified using an external cohort. EC represents a group of heterogeneous tumors with morphological and histological differences and curettage specimens cannot comprehensively reflect tumor characteristics. Thus, pathological images of EC obtained from paraffin-embedded tissues after surgical resection was sued during the current study, in order to best observe the morphological characteristics of cancer tissue and obtain the most suggestive morphology information for LNM. DL features that proved to be most salient to LNM prediction are highlighted to inform pathologists an intuitive interpretation, and give transparency to the development of the multiple instance-learning (MIL) model. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-220/rc).
Methods
Study population and cases
Data of EC patients were retrospectively collated from West China Second University Hospital, Sichuan University with the following inclusion criteria: (I) patients underwent total hysterectomy (or extensive total hysterectomy), bilateral adnexectomy and pelvic lymphadenectomy; (II) no history of other malignant tumors; (III) no hormone therapy, radiotherapy or chemotherapy before surgery; (IV) diagnosed for the first time. The HE-stained sections from all cases were reviewed by double-blind microscopy observation by two senior attending physicians with experience in pathological clinical diagnosis. The status of lymph vascular space invasion (LVSI) and LNM was confirmed. The diagnosis of histological type was based on the 2020 World Health Organization (WHO) Classification of Tumors of the Breast and Female Genital Organs. Tumor staging and surgical pathological staging were undertaken according to the International Federation of Gynecology and Obstetrics (FIGO) Surgical-pathological staging standard. A total of 564 patients diagnosed with EC were enrolled of whom 230 had LNM (LNM+) and 334 did not (LNM−). External validation data were obtained by applying the same inclusion criteria to 261 patients, 114 LNM+ and 147 LNM−, from two independent medical institutions, Qingdao University, Affiliated Yantai Yu Huang Ding Hospital, Beijing Maternal and Child Health Care Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by our institutional review board. Informed consent was waived.
Slides and image acquisition
All slides were prepared by staining a 4-µm formalin-fixed paraffin-embedded section with HE; 1,017 LNM+ and 1,087 LNM− slides composed the internal dataset. In total, 2,104 slides were digitized through the following three types of digital slice scanning: The Hamamatsu Optics NanoZoomer 2.0 HT digital slice scanner, the Una Technology PRECICE 600 automatic digital slice scanner and the 3D HISTECH Panoramic SCAN 150 digital slice scanner; 388 LNM+ and 145 LNM− slides composed the external dataset and all 533 slides were digitized through 3D HISTECH Panoramic SCAN 150 digital slice scanner and ScanScope Aperio CS2 digital slice scanner (Leica Biosystems). Each WSI was given an LNM+ or LNM− label based on the results of final pathological diagnosis.
Data preparation
Tumor cells and glands were manually delineated using the ASAP software (version 1.9, https://computationalpathologygroup.github.io/ASAP/) at ×20 magnification (0.5 µm/pixel) by two expert pathologists. In this work, the pathologists first manually annotated the cancer regions on each WSI and used them as regions of interest (ROIs) for the following processing step. The ROIs were divided into a set of patches with a size of 512×512 pixels. Patches with less than 20% overlap with the ROIs were excluded before further analysis.
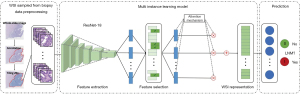
DL model
The framework of the proposed MIL model is illustrated in Figure 1, which consists of three components, i.e., instance-level (tile-level) feature extraction, instance-level (tile-level) feature selection, and bag-level (WSI-level) representation generation (18). The details of the components are illustrated in the remainder of this section. In the formalization of the MIL, each WSI is regarded as a bag and the patches tiled from the WSI are regarded as instances inside the bag. During the training phase, we chose to use the categorical cross-entropy loss, which is defined as:
where N denotes the number of samples and C represents the number of categories. The term δ (yi =c) is the indicator function of the ith observation belonging to the cth category. P (yi =c) is the predicted probability by the model.
Instance-level feature extractor
In our AI system, we employed the ResNet-18 model (19) as the instance-level feature extractor, which aimed at automatically learning useful features from patches. From another perspective, this component played the role of information compression, and each inputted patch was transformed into a low-dimensional feature space, which facilitated the following classification stage. In this work, we used a pretrained ResNet-18 model (trained on ImageNet) after removing the final fully connected layer to extract distinguishable features.
Feature selection strategy
The feature selection procedure chose the most discriminative instance-level features for generating the bag representation. Removing redundant or irrelevant features can also simplify the following learning task. Unlike most feature selection problems where there are feature-label pairs, in our task, the instance-level feature has no associated label and is assigned a bag-level label. We needed to build a bridge between the extracted instance feature and the bag label. We utilized the histogram as the bridge and the maximum mean discrepancy (20) as the criterion to evaluate the feature importance. For instance, when evaluating the k-th feature, feature k was chosen as the representation of the instance. A histogram of this feature was calculated in each bag, and then these histograms were utilized as the representations of a bag. After that, the maximum mean discrepancy (21) distance was calculated between positive bags and negative bags using these bag representations. The feature was regarded as discriminative if the maximum mean discrepancy distance was larger than a certain threshold (22).
WSI-level representation generator
The WSI-level representation generator in our pipeline generates the bag representation by integrating the extracted and selected most discriminative instance-level features. The attention-based deep MIL method (18) was applied, which made the WSI-level representation generator able to adaptively adjust the contribution of each tiled patch for the final decision. After iteratively and incrementally adjusting the attention weights on the feature of each patch during the training phase, the attention-based operator increased the contribution of the instances that were more related to the corresponding bag label, and vice versa. After obtaining the WSI-level representation of each pathological slide, a multilayer perceptron was utilized to map the representation to LNM prediction possibility.
Network training setting
We implemented the MIL system including both ResNet-18 and the attention-based MIL network with Python and PyTorch (22) framework. The network was trained with the Adam optimizer (9) with the learning rate of 10−4 under the supervision of the categorical cross-entropy loss. The early-stop strategy was leveraged with patience of 20 epoch to prevent over-fitting. To address the class imbalance problem during the bag-level classification stage, we employed the ‘weighted random sampler’ strategy (22) to prepare each training batch. The dataset is randomly split in the ratio of 6:2:2, that is, 1,264 WSIs in the training set, 420 WSIs in the validation set, and 420 WSIs in the testing set.
Model interpretability and feature visualization
Tiles were assigned an LNM probability score reflecting usefulness for LNM prediction. Heatmaps were generated from LNM probability scores to reflect the determination of each subregion of the WSI.
Baseline model based on “Mayo criteria”
Patients were divided into low-risk and high-risk groups of LNM, according to the widely accepted “Mayo criteria” (6). Mayo criteria recommend lymph node dissection for the following patients: (I) grade 1 or grade 2 endometrioid adenocarcinoma ≥2 cm and >50% myometrial invasion; (II) any grade 3 endometrioid adenocarcinoma and (III) all non-endometrioid adenocarcinomas (serous, clear cell, mixed, carcinosarcoma). Patients satisfying one or more of these conditions were considered to be with the high-risk factors of LNM. Mayo criteria were also used for the baseline feature model to predict LNM risk.
Statistical analysis
The statistical analyses were performed using SPSS 22.0 software. Age and tumor size were considered as numeric variables. Histological type, histological grade, depth of myometrial invasion and presence of LVSI were considered as categorical variables. Clinicopathological characteristics between cohorts were compared using Pearson’s χ2 or Fisher’s exact test. The AUC of the receiver operating characteristic (ROC) curves from cross-validation were calculated and plotted using “scikit-learn” in python to assess model performance. The optimal cut-off point of the ROC curves was determined by referring to the Youden index. AUC ranges from 0 to 1 and a model is considered to have a poor performance with a value of 0.5–0.6, fair 0.6–0.7 or good >0. Other standard metrics, sensitivity [true positive rate (TPR)], false positive rate (FPR), specificity, accuracy, recall-score, F1-score and positive predictive value (PPV)/negative predictive value (NPV) were also employed. The confidence intervals (CI) were calculated using the bootstrap method. All statistical tests were two sided and P values of less than 0.05 were considered to indicate statistical significance.
Results
Clinicopathological characteristics of EC patients
According to the inclusion and exclusion criteria, a total of 564 eligible patients were enrolled in this study; 230 patients were diagnosed with LNM+ EC of whom 114 (49.56%) cases were type I, 52 (22.61%) type II and 64 (27.83%) mixed type. A total of 143 (62.17%) cases were poorly differentiated while 87 (37.83%) cases were moderately-well differentiated. A total of 62 (26.96%), 79 (34.35%), 68 (29.56%), and 21 (9.13%) patients were diagnosed with FIGO stages I–IV, respectively; 334 patients were diagnosed with LNM− EC of whom 274 (82.03%) cases were type I, 19 (5.69%) type II and 41 (12.28%) mixed type; 112 (33.53%) cases were poorly differentiated and 222 (66.47%) cases were moderately-well differentiated. A total of 247 (73.95%), 61 (18.26%), 25 (7.49%) and 1 (0.30%) patients were diagnosed with FIGO stages I–IV, respectively. There were 336 patients in the training cohort and 228 patients in the independent test cohort after all WSIs were randomly divided into 8 (6+2):2. No significant differences in detailed characteristics between the training and independent test cohorts were present (all P>0.05; Table 1). Clinicopathological characteristics of EC patients in the external validation cohort are shown in Table S1.
Table 1
| Characteristics | All patients (n=564) | Training (n=336), n (%) | Test (n=228), n (%) | P |
|---|---|---|---|---|
| Age (years), mean ± SD | 53.90±8.89 | 54.46±9.29 | 53.08±8.23 | 0.070 |
| Tumor size (cm), mean ± SD | 3.55±1.78 | 3.39±1.58 | 3.79±2.01 | 0.091 |
| Tumor histological subtype | 0.158 | |||
| I | 388 | 237 (70.54) | 151 (66.23) | |
| II | 71 | 45 (13.39) | 26 (11.40) | |
| Mixed (I + II) | 105 | 54 (16.07) | 51 (22.37) | |
| Tumor grade (differentiation) | 0.062 | |||
| 1–2 (well/moderate differentiation) | 309 | 197 (58.63) | 112 (49.12) | |
| 3 (poor differentiation) | 255 | 139 (41.37) | 116 (50.88) | |
| Depth of tumor invasion | 0.340 | |||
| <1/2 | 258 | 166 (49.40) | 92 (40.35) | |
| ≥1/2 | 306 | 170 (50.60) | 136 (59.65) | |
| Lymph vascular space invasion | 0.739 | |||
| Yes | 297 | 175 (52.08) | 122 (53.51) | |
| No | 267 | 161 (47.92) | 106 (46.49) | |
| Lymph node metastasis | 0.381 | |||
| Yes | 230 | 132 (39.29) | 98 (42.98) | |
| No | 334 | 204 (60.71) | 130 (57.02) | |
| FIGO staging | ||||
| I | 309 | 186 (55.36) | 123 (53.95) | 0.297 |
| II | 140 | 86 (25.60) | 54 (23.68) | |
| III | 93 | 53 (15.77) | 40 (17.54) | |
| IV | 22 | 11 (3.27) | 11 (4.82) |
SD, standard deviation; FIGO, International Federation of Gynecology and Obstetrics.
The predictive performance of the “Mayo criteria”
Of 564 EC patients who underwent pelvic lymphadenectomy, 369 (65.43%) met the Mayo criteria for lymph node resection while the remaining 195 (34.57%) patients had comprehensive evaluations of clinical manifestations, pathological characteristics and other aspects; 195 (84.78%) of the 230 patients with LNM met the Mayo criteria, indicating that 35 (15.22%) belonged to the low-risk group for LNM prediction based on Mayo criteria. However, these 35 patients were found to have LNM after pathological diagnosis and 16 patients were FIGO I stage with 8 being grade 1 or grade 2 endometrioid adenocarcinoma <2 cm and <50% myometrial invasion; 174 (52.10%) of the 334 patients without LNM had high-risk LNM factors but no metastases were confirmed following a final lymph node examination. Therefore, 15.22% patients in the low-risk group according to existing Mayo criteria for LNM risk stratification will progress to develop LNM. Some patients were also exposed to unnecessary lymphadenectomy, corresponding to 52.10% in our comparative study. Thus, two rows and two columns were used to represent predicted and real values. The AUC was 0.666, TPR 0.878, FPR 0.545, accuracy score 0.635, PPV 0.544, NPV 0.833, F1_score 0.672 for LNM prediction in the test dataset. The finally clinical practices have shown that 37.97% of EC patients were exposed to unnecessary lymphadenectomy and approximatively 17.14% with a low-risk classification progressed to develop LNM in the test cohort. The statistical results are summarized in Table 2, and the ROC curve is shown in Figure 2A.
Table 2
| Standard metrics | Internal test dataset | External validation dataset | ||
|---|---|---|---|---|
| Mayo criteria | MIL-model | MIL-model | ||
| AUC | 0.666 | 0.938 | 0.770 | |
| Accuracy | 0.635 | 0.847 | 0.630 | |
| Recall | 0.878 | 0.830 | 0.814 | |
| Precision (PPV) | 0.544 | 0.881 | 0.706 | |
| F1_score | 0.672 | 0.807 | 0.756 | |
| Sensitivity (TPR) | 0.878 | 0.830 | 0.814 | |
| Specificity | 0.455 | 0.911 | 0.520 | |
| FPR | 0.545 | 0.143 | 0.479 | |
| NPV | 0.833 | 0.872 | 0.665 | |
MIL, multiple instance-learning; LNM, lymph node metastasis; AUC, area under the curve; PPV, positive predictive value; TPR, true positive rate; FPR, false positive rate; NPV, negative predictive value.
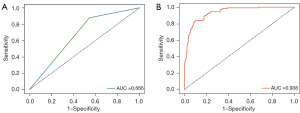
Predictive performance of the MIL model based on HE-stained images for the internal dataset
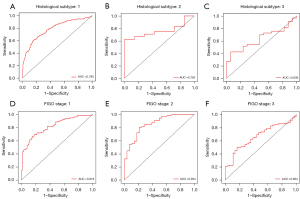
The training process was stopped after 30 epochs with 33,540 iterations and the best model, evaluated after each epoch, saved. After training, 420 test sets of WSIs were used to give an unbiased evaluation of the model. The mean AUC was 0.938 (95% CI: 91.3–96.2) on the WSI level, mean sensitivity 0.830 (95% CI: 75.9–88.9), mean specificity 0.911 (95% CI: 85.9–94.8), mean PPV 0.881 (95% CI: 81.5–92.3), mean NPV 0.872 (95% CI: 81.3–92.4) for the internal test data (Figure 2B, Figure 3 and Table 2). Clinical analysis showed that 21.51% of EC patients were exposed to unnecessary lymphadenectomy and approximatively 11.11% with low-risk classification progressed to develop LNM. Predictive performances for different histological subtypes [type I, type II and mixed (I and II) type] and different FIGO stages on the WSI level are presented in Table 3, Table 4 and Figure 4A-4F. For type I EC (endometrioid adenocarcinoma as the main histological type and mucinous adenocarcinoma), the mean AUC was 0.927 on the WSI level and 0.979 for type II and 0.929 for mixed type. Mean AUCs were 0.921, 0.938 and 0.955 for FIGO stages I, II and III on the WSI level. The PPV reached 0.999 for FIGO stage IV, perhaps because 95.45% (21/22) of stage IV patients are positive for LNM. The encouraging AUC value of the image model of 0.938, higher than the Mayo model at 0.666, indicated better LNM predictive performance for the AI model.
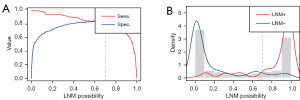
Table 3
| Standard metrics | Histological subtype: I (%) (95% CI) | Histological subtype: II (%) (95% CI) | Histological subtype: mixed (%) (95% CI) |
|---|---|---|---|
| Internal test dataset (420 WSIs from 228 patients) | |||
| AUC | 92.7 (89.8–95.7) | 97.9 (94.6–100.0) | 92.9 (88.1–97.7) |
| Sensitivity | 80.2 (70.6–87.8) | 97.0 (84.2–99.9) | 95.3 (86.9–99.0) |
| Specificity | 92.2 (88.4–95.0) | 95.1 (83.5–99.4) | 80.4 (66.1–90.6) |
| PPV | 76.8 (68.2–85.5) | 94.1 (80.6–99.8) | 87.1 (76.3–97.1) |
| NPV | 93.5 (89.5–95.9) | 97.5 (86.7–99.7) | 92.5 (80.1–96.7) |
| External validation dataset (533 WSIs from 261 patients) | |||
| AUC | 78.3 (74.1–82.5) | 76.2 (59.3–93.1) | 63.6 (48.3–79.0) |
| Sensitivity | 60.8 (54.5–66.8) | 62.5 (40.6–81.2) | 42.4 (25.5–60.8) |
| Specificity | 84.3 (78.4–89.1) | 100.0 (59.0–NaN) | 94.1 (71.3–99.9) |
| PPV | 83.3 (77.2–86.7) | 100.0 (75.5–100.0) | 93.3 (68.5–98.7) |
| NPV | 62.4 (56.2–71.6) | 43.8 (24.2–NaN) | 45.7 (28.1–97.2) |
Histological subtype I: endometrioid adenocarcinoma as the main histological type and mucinous adenocarcinoma; Histological subtype II: non endometrioid adenocarcinoma; Mixed subtype: composing of subtype I + II, where at least one component is either serous or clear cell. MIL, multiple instance-learning; WSI, whole slide image; AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value; NaN, not a number.
Table 4
| Standard metrics | FIGO stage: I (%) (95% CI) | FIGO stage: II (%) (95% CI) | FIGO stage: III (%) (95% CI) | FIGO stage: IV (%) |
|---|---|---|---|---|
| Internal test dataset (420 WSIs from 228 patients) | ||||
| AUC | 92.1 (86.8–97.5) | 93.8 (90.2–97.4) | 95.5 (90.1–100.0) | – |
| Sensitivity | 90.6 (79.3–96.9) | 80.3 (68.7–89.1) | 94.0 (83.5–98.7) | 78.9 |
| Specificity | 87.6 (82.8–91.4) | 94.7 (87.1–98.5) | 95.3 (84.2–99.4) | – |
| PPV | 60.8 (51.4–83.3) | 93.0 (83.2–96.4) | 95.9 (85.9–99.2) | 99.9 |
| NPV | 97.8 (94.6–98.5) | 84.7 (74.9–95.4) | 93.2 (81.5–99.1) | – |
| External validation dataset (533 WSIs from 261 patients) | ||||
| AUC | 81.8 (75.8–85.8) | 85.4 (77.6–93.2) | 66.4 (56.9–76.1) | – |
| Sensitivity | 69.2 (57.8–79.2) | 80.6 (71.8–87.5) | 50.0 (39.7–60.3) | – |
| Specificity | 82.2 (75.2–88.0) | 78.6 (59.0–91.7) | 83.3 (67.2–93.6) | – |
| PPV | 66.7 (56.7–77.2) | 93.5 (85.1–96.1) | 89.1 (77.0–92.5) | – |
| NPV | 83.9 (76.0–89.1) | 51.2 (39.2–76.0) | 38.0 (28.8–64.3) | – |
FIGO stage I: tumour confined to the corpus uterus; FIGO stage II: tumour invades cervical stroma, but does not extend beyond the uterus; FIGO stage III: local and/or regional spread as specified here: the serosa of the corpus uterus or adnexae, vaginal or parametrial involvement, pelvic or para-aortic lymph nodes; FIGO stage IV: tumour invades bladder/bowel mucosa, and/or metastasizes remotely. MIL, multiple instance-learning; FIGO, International Federation of Gynecology and Obstetrics; CI, confidence interval; WSI, whole slide image; AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value.
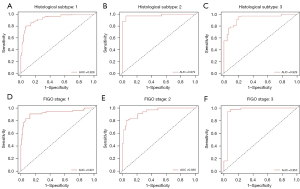
Predictive performance of the MIL model based on HE-stained images in external validation dataset
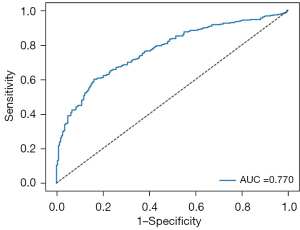
The proposed MIL model achieved an AUC of 0.770, a sensitivity of 0.814, a specificity of 0.520, a PPV of 0.706 and an NPV of 0.665 in the external validation dataset (Figure 5 and Table 2). Mean AUCs were 0.783 for type I, 0.762 for type II and 0.636 for mixed type on the WSI level (Figure 6A-6C and Table 3). Mean AUCs were 0.818, 0.854, 0.664 for FIGO stages I, II, and III on the WSI level (Figure 6D-6F). All stage IV cases included in the external validation dataset were positive for LNM, so that no analysis of predicted results was possible. In summary, the predictive performance of the proposed MIL model was not as good for the external as for the internal cohort.
Interpretation of predictive performance of the MIL model
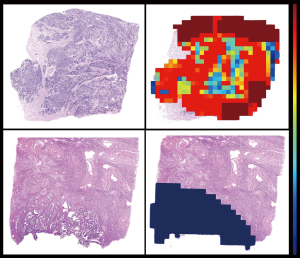
The LNM prediction probability distribution in each WSI was evaluated, with LNM probability of each subregion (tile) calculated and visualized, to illustrate the decision-mechanism of the model (Figure 7). Tiles with high (near 1) and low (near 0) LNM probability scores had distinguishable features to support LNM prediction. Enhanced texture, voluminous extracellular mucin, cleft-like structures and elongated angular glandular lumen were “histomics” features with significant alterations between LNM+ and LNM− WSIs, indicating an increased rate of lymph node involvement. These features were consistent with previous studies (23-25) and provide an interpretative reliability to the predictive results of the AI-model.
Discussion
At present, the use of cutting-edge DL technology to predict the preoperative LNM status of tumors, including gastric cancer (26,27), breast cancer (28), colorectal cancer (29), lung cancer (30), urothelial cancer (31) and EC (7,32), usually relies on the readily-available MRI/CT and ultrasound imaging. However, WSI technology and advances in machine learning algorithms have enabled digital transformation of pathological images for cancer research and sparked a global trend. The current study applied a DL neural network algorithm to the binary classification task of LNM+/LNM− in EC, based on a comprehensive analysis of postoperative pathological specimens on the WSI-level. Prediction efficiency was validated in external samples. The MIL DL model of LNM prediction achieved AUCs of 0.938 and 0.770 in the internal and external cohorts, regardless of the specific histological EC subtype. In addition, a heat map was generated to visualize the contributions made by regions on the WSI to the LNM diagnosis and to explain the end-to-end process of the MIL model. The DL model has predictive utility for specimens of endometrioid and non-endometrioid cancer with relatively intact structure obtained before or during surgery. The pathological images also have the potential to be used as an AI-based biomarker for LNM prediction.
Clinical experience has illustrated that the existing LNM risk stratification criteria, including the Mayo criteria and NCCN guidelines, will expose 75% of EC patients to unnecessary lymphadenectomy (33) and about 10% of low-risk and 15% of early-stage ECs will progress to LNM (34). In our internal cohort, 52.10% of EC patients underwent unnecessary lymphadenectomy and approximatively 15.22% of those with low-risk developed LNM when assessed according to the accepted “Mayo criteria”. These observations are basically consistent with the results of a recent prospective multicenter study (35). The current analysis reveals two possible explanations: (I) racial differences and (II) existing NCCN guidelines and Mayo standards rely on the professional skills and subjective judgments of pathologists, making it difficult to achieve unified quality control. Such clinical phenomenon also prompts us to re-understand the existing criteria, and explores more accurate morphological characteristics of pathological images to predict the LNM risk. The current AI model relies on HE-stained histopathological images producing more accurate prediction with a mean AUC of 0.938, higher than that based on Mayo criteria. Thus, machine learning may be superior to human subjective experience and is more accurate, objective, stable and highly repeatable.
Most of the existing research to produce LNM prediction models by comprehensive analysis the clinicopathological features and laboratory examination data, such as nomogram and Bayesian model, has focused on type I EC or endometrioid adenocarcinoma (36-38). Such approaches are accurate but often ignore the heterogeneity of EC and do not apply to type II EC or other histological subtypes. The current study included a variety of histological subtypes of EC, including endometrioid adenocarcinoma, serous adenocarcinoma (adenoid serous adenocarcinoma and papillary serous adenocarcinoma), clear cell adenocarcinoma, mucinous adenocarcinoma, mixed endometrial adenocarcinoma, mesonephric adenocarcinoma and adeno-squamous carcinoma. HE-stained histopathological images on WSI are thus more complicated and extraction of image features for consensus among different observers is more difficult. Fortunately, MIL technology enables fast and objective analysis by integrating recurring patterns from complex images (39). The resulting AUC for prediction of LNM reached 0.938 without distinguishing specific histological types. LNM prediction rates were 0.927, 0.979 and 0.929 for type I, type II and mixed type based on pathomorphological diagnosis (Table 3). LNM prediction rates were 0.783, 0.762 and 0.636 for type I, II and mixed type from the external validation dataset. The current MIL model performed less well for the external cohort than the internal, but was still superior to predictions based on Mayo criteria, regardless of histological subtype. Thus, even when the specific histological subtype was unclear before or during surgical operation, a reliable prediction result could be reached. For low risk early FIGO (FIGO I), mean AUCs were 0.921 and 0.818 in internal and external datasets, potentially enabling more accurate individualized LNM stratification in early stage EC.
The lower predictive performance in the external dataset may be due to the following reasons: (I) pathology technicians from different medical institutions may prepare specimens with different slice thicknesses, staining intensities and uniformities. (II) Differences in automatic dyeing machine models do not completely overcome this problem. (III) Definition and resolution of the digitized WSI images are different due to variations in scanner models. However, expansion of the sample size and continued optimization may improve future performance.
Hierarchical clustering enabled heatmap visualization of each tumor region in the WSI and its contribution to LNM prediction with the heatmap mapped to the original image by the jet color space. Tiles were assigned an LNM probability score to measure the contribution to LNM prediction, red subregions represented a higher and blue a lower contribution to LNM. Figure 7 shows the “histomics” features with significant alterations between LNM+ and LNM− patients in top-value tiles, enhanced texture, voluminous extracellular mucin, cleft-like structures and elongated angular glandular lumen, which distinguish LNM+ from LNM− patients well. These suggested features from the red subregions appear to be highly consistent with pathologists’ experience and previous studies (23-25). Feature visualization demonstrated that output results of our model have realistic interpretability.
We acknowledge some limitations to the current study. Firstly, digital pathology images from paraffin tissues after surgical resection of EC patients were used retrospectively. Considering that predictions mainly depend on preoperative curettage specimens, preoperative data from multiple centers are indeed required to further confirm the utility of such DL models in clinical practice. Secondly, although a variety of histological EC subtypes, including endometrioid and non-endometrioid carcinoma were analyzed, less common subtypes, such as carcinosarcoma, was not collected in the current study. Thirdly, the inclusion of additional features, such as CA125 and CA19-9 expression, ascites and other risk factors that are related to LNM, may confirm to improve predictive accuracy. DL experience of unusual cases may improve predictive accuracy and reduce false-positives and false-negatives.
Conclusions
In summary, the main contribution of our study is the development of a DL model for LNM prediction in EC based on HE-stained histopathological images with different subtypes for the first time. The model showed particular accuracy for early staging patients and advances the approach of pathological slide analysis based on AI. The most significant features for DL of LNM predictions were highlighted to allow intuitive interpretation of the MIL model by pathologists. To confirm the utility of the proposed MIL model in clinical practice, preoperative curettage specimens and further multicenter prospective validation is indeed required.
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Funding: This work was supported by the National Key Research and Development Program (No. 2017YFC0113908); Technological Innovation Project of Chengdu New Industrial Technology Research Institute (No. 2017-CY02-00026-GX); 1.3.5 project for disciplines of excellence, West China Hospital (No. ZYGD18012).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-220/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-220/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by our institutional review board. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Arend RC, Jones BA, Martinez A, Goodfellow P. Endometrial cancer: Molecular markers and management of advanced stage disease. Gynecol Oncol 2018;150:569-80. [Crossref] [PubMed]
- Gonthier C, Douhnai D, Koskas M. Lymph node metastasis probability in young patients eligible for conservative management of endometrial cancer. Gynecol Oncol 2020;157:131-5. [Crossref] [PubMed]
- Vetter MH, Smith B, Benedict J, Hade EM, Bixel K, Copeland LJ, Cohn DE, Fowler JM, O'Malley D, Salani R, Backes FJ. Preoperative predictors of endometrial cancer at time of hysterectomy for endometrial intraepithelial neoplasia or complex atypical hyperplasia. Am J Obstet Gynecol 2020;222:60.e1-7. [Crossref] [PubMed]
- Koh WJ, Abu-Rustum NR, Bean S, Bradley K, Campos SM, Cho KR, et al. Uterine Neoplasms, Version 1.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:170-99. [Crossref] [PubMed]
- Suh-Burgmann E, Hung YY, Armstrong MA. Complex atypical endometrial hyperplasia: the risk of unrecognized adenocarcinoma and value of preoperative dilation and curettage. Obstet Gynecol 2009;114:523-9. [Crossref] [PubMed]
- Hu J, Zhang K, Yan Y, Zang Y, Wang Y, Xue F. Diagnostic accuracy of preoperative 18F-FDG PET or PET/CT in detecting pelvic and para-aortic lymph node metastasis in patients with endometrial cancer: a systematic review and meta-analysis. Arch Gynecol Obstet 2019;300:519-29. [Crossref] [PubMed]
- Higgins C. Applications and challenges of digital pathology and whole slide imaging. Biotech Histochem 2015;90:341-7. [Crossref] [PubMed]
- Yao J, Zhu X, Jonnagaddala J, Hawkins N, Huang J. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med Image Anal 2020;65:101789. [Crossref] [PubMed]
- Bhowal P, Sen S, Velasquez JD, Sarkar R. Fuzzy ensemble of deep learning models using choquet fuzzy integral, coalition game and information theory for breast cancer histology classification. Expert Syst Appl 2022;190:116167. [Crossref]
- Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, Wey N, Wild PJ, Rüschoff JH, Claassen M. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 2018;8:12054. [Crossref] [PubMed]
- Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat 2019;173:455-63. [Crossref] [PubMed]
- Balagopal A, Morgan H, Dohopolski M, Timmerman R, Shan J, Heitjan DF, Liu W, Nguyen D, Hannan R, Garant A, Desai N, Jiang S. PSA-Net: Deep learning-based physician style-aware segmentation network for postoperative prostate cancer clinical target volumes. Artif Intell Med 2021;121:102195. [Crossref] [PubMed]
- Lee H, Lee DE, Park S, Kim TS, Jung SY, Lee S, Kang HS, Lee ES, Sim SH, Park IH, Lee KS, Kwon YM, Kong SY, Joo J, Jeong HJ, Kim SK. Predicting Response to Neoadjuvant Chemotherapy in Patients With Breast Cancer: Combined Statistical Modeling Using Clinicopathological Factors and FDG PET/CT Texture Parameters. Clin Nucl Med 2019;44:21-9. [Crossref] [PubMed]
- Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 2020;395:350-60. [Crossref] [PubMed]
- Golden JA. Deep Learning Algorithms for Detection of Lymph Node Metastases From Breast Cancer: Helping Artificial Intelligence Be Seen. JAMA 2017;318:2184-6. [Crossref] [PubMed]
- Takamatsu M, Yamamoto N, Kawachi H, Chino A, Saito S, Ueno M, Ishikawa Y, Takazawa Y, Takeuchi K. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Programs Biomed 2019;178:155-61. [Crossref] [PubMed]
- Wang M, Deng W. Deep visual domain adaptation: A survey. Neurocomputing 2018;312:135-53. [Crossref]
- Yang Z, Shi J, Asyrofi MH, Lo D. Revisiting Neuron Coverage Metrics and Quality of Deep Neural Networks. In: 2022 IEEE International Conference on Software Analysis, Evolution and Reengineering (SANER). Honolulu, HI, USA: IEEE, 2022.
- Zhou L, Zhao Y, Yang J, Yu Q, Xu X. Deep multiple instance learning for automatic detection of diabetic retinopathy in retinal images. IET Image Process 2018;12:563-71. [Crossref]
- Chen Z, Chen Z, Liu J, Zheng Q, Zhu Y, Zuo Y, Wang Z, Guan X, Wang Y, Li Y. Weakly Supervised Histopathology Image Segmentation With Sparse Point Annotations. IEEE J Biomed Health Inform 2021;25:1673-85. [Crossref] [PubMed]
- Bhattacharjee K, Pant M, Zhang YD, Satapathy SC. Multiple Instance Learning with Genetic Pooling for medical data analysis. Pattern Recognit Lett 2020;133:247-55. [Crossref]
- Musa F, Huang M, Adams B, Pirog E, Holcomb K. Mucinous histology is a risk factor for nodal metastases in endometrial cancer. Gynecol Oncol 2012;125:541-5. [Crossref] [PubMed]
- Duzguner S, Turkmen O, Kimyon G, Duzguner IN, Karalok A, Basaran D, Tasci T, Ureyen I, Turan T. Mucinous endometrial cancer: Clinical study of the eleven cases. North Clin Istanb 2019;7:60-4. [PubMed]
- Kihara A, Yoshida H, Watanabe R, Takahashi K, Kato T, Ino Y, Kitagawa M, Hiraoka N. Clinicopathologic Association and Prognostic Value of Microcystic, Elongated, and Fragmented (MELF) Pattern in Endometrial Endometrioid Carcinoma. Am J Surg Pathol 2017;41:896-905. [Crossref] [PubMed]
- Jin C, Jiang Y, Yu H, Wang W, Li B, Chen C, Yuan Q, Hu Y, Xu Y, Zhou Z, Li G, Li R. Deep learning analysis of the primary tumour and the prediction of lymph node metastases in gastric cancer. Br J Surg 2021;108:542-9. [Crossref] [PubMed]
- Dong D, Fang MJ, Tang L, Shan XH, Gao JB, Giganti F, Wang RP, Chen X, Wang XX, Palumbo D, Fu J, Li WC, Li J, Zhong LZ, De Cobelli F, Ji JF, Liu ZY, Tian J. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol 2020;31:912-20. [Crossref] [PubMed]
- Xu F, Zhu C, Tang W, Wang Y, Zhang Y, Li J, Jiang H, Shi Z, Liu J, Jin M. Predicting Axillary Lymph Node Metastasis in Early Breast Cancer Using Deep Learning on Primary Tumor Biopsy Slides. Front Oncol 2021;11:759007. [Crossref] [PubMed]
- Schurink NW, Lambregts DMJ, Beets-Tan RGH. Diffusion-weighted imaging in rectal cancer: current applications and future perspectives. Br J Radiol 2019;92:20180655. [Crossref] [PubMed]
- Wu Y, Liu J, Han C, Liu X, Chong Y, Wang Z, Gong L, Zhang J, Gao X, Guo C, Liang N, Li S. Preoperative Prediction of Lymph Node Metastasis in Patients With Early-T-Stage Non-small Cell Lung Cancer by Machine Learning Algorithms. Front Oncol 2020;10:743. [Crossref] [PubMed]
- Frączek M, Kamecki H, Kamecka A, Sosnowski R, Sklinda K, Czarniecki M, Królicki L, Walecki J. Evaluation of lymph node status in patients with urothelial carcinoma-still in search of the perfect imaging modality: a systematic review. Transl Androl Urol 2018;7:783-803. [Crossref] [PubMed]
- Nougaret S, Horta M, Sala E, Lakhman Y, Thomassin-Naggara I, Kido A, Masselli G, Bharwani N, Sadowski E, Ertmer A, Otero-Garcia M, Kubik-Huch RA, Cunha TM, Rockall A, Forstner R. Endometrial Cancer MRI staging: Updated Guidelines of the European Society of Urogenital Radiology. Eur Radiol 2019;29:792-805. [Crossref] [PubMed]
- Vargas R, Rauh-Hain JA, Clemmer J, Clark RM, Goodman A, Growdon WB, Schorge JO, Del Carmen MG, Horowitz NS, Boruta DM 2nd. Tumor size, depth of invasion, and histologic grade as prognostic factors of lymph node involvement in endometrial cancer: a SEER analysis. Gynecol Oncol 2014;133:216-20. [Crossref] [PubMed]
- Karalok A, Turan T, Basaran D, Turkmen O, Comert Kimyon G, Tulunay G, Tasci T. Lymph Node Metastasis in Patients With Endometrioid Endometrial Cancer: Overtreatment Is the Main Issue. Int J Gynecol Cancer 2017;27:748-53. [Crossref] [PubMed]
- Koskas M, Fournier M, Vanderstraeten A, Walker F, Timmerman D, Vergote I, Amant F. Evaluation of models to predict lymph node metastasis in endometrial cancer: A multicentre study. Eur J Cancer 2016;61:52-60. [Crossref] [PubMed]
- Bendifallah S, Genin AS, Naoura I, Chabbert Buffet N, Clavel Chapelon F, Haddad B, Luton D, Darai E, Rouzier R, Koskas M. A nomogram for predicting lymph node metastasis of presumed stage I and II endometrial cancer. Am J Obstet Gynecol 2012;207:197.e1-8. [Crossref] [PubMed]
- Taşkın S, Şükür YE, Varlı B, Koyuncu K, Seval MM, Ateş C, Yüksel S, Güngör M, Ortaç F. Nomogram with potential clinical use to predict lymph node metastasis in endometrial cancer patients diagnosed incidentally by postoperative pathological assessment. Arch Gynecol Obstet 2017;296:803-9. [Crossref] [PubMed]
- Reijnen C, Gogou E, Visser NCM, Engerud H, Ramjith J, van der Putten LJM, et al. Preoperative risk stratification in endometrial cancer (ENDORISK) by a Bayesian network model: A development and validation study. PLoS Med 2020;17:e1003111. [Crossref] [PubMed]
- Puttagunta M, Ravi S. Medical image analysis based on deep learning approach. Multimed Tools Appl 2021;80:24365-98. [Crossref] [PubMed]


