A review of computer-aided diagnosis in thoracic and colonic imaging
Introduction
Medical imaging has been indispensable in medicine since the discovery of X-rays by Wilhelm C. Röntgen in 1895. Medical imaging offers useful information on patients’ medical conditions and provides clues to causes of their symptoms and diseases. As imaging technologies advance, a large number of medical images are produced which physicians/radiologists must interpret. Thus, computer aids are demanded and become indispensable in physicians’ decision making based on medical images. Consequently, computer-aided detection and diagnosis (CAD) (1-4) has been an active research area in medical imaging. CAD is defined as detection and/or diagnosis made by a radiologist/physician who takes into account the computer output as a “second opinion” (2). CAD is often categorized into two major groups: computer-aided detection (CADe) and computer-aided diagnosis (CADx). CADe focuses on a detection task, i.e., detection (or localization) of lesions in medical images. CADx focuses on a diagnosis (characterization) task, e.g., distinction between benign and malignant lesions, and classification among different lesion types.
A branch of the history of CAD started in 1955. A radiologist, Lee Lusted, mentioned the potential use of digital computers for large-scale data problems in medicine in (5) in 1955. It was only nine years after the first general-purpose computer, ENIAC, was introduced in 1946. In 1963, Lodwick et al. digitized chest radiographs for computer analysis (6). In 1964, Becker et al. developed automated measurement of the cardiothoracic ratio in chest radiographs (7,8). The first study on CADe of abnormalities in mammograms was published by Winsberg et al. (9) in 1967. In 1973, Toriwaki et al. (10) reported the first study on CADe of a focal abnormality in chest radiographs, and Roellinger et al. (11) reported the first study on CADe of a heart abnormality in chest radiographs. In the mid-1980’s, investigators in the Kurt Rossmann Laboratories in the Department of Radiology at the University of Chicago began studies on the development and evaluation of CAD. Chan et al. (12), Fujita et al. (13), Giger et al. (14), and Katsuragawa et al. (15) published a series of papers on CADe of microcalcifications in mammography, CAD for vessel size measurement in angiography, CADe of lung nodules in chest radiography, and CADe of interstitial lung disease in chest radiography, respectively. In 1988, a venture company, R2 Technology (now Hologic), which obtained licenses for CAD technologies from the University of Chicago, received approval for the first commercial CAD system for mammography from the U.S. Food and Drug Administration (FDA).
Evidence suggests that CAD can help improve the diagnostic performance of radiologists/physicians in their image interpretations (16-22). Consequently, many investigators have participated and developed CAD schemes such as those for detection of lung nodules in chest radiographs (14,23-25) and in thoracic CT (26-29), those for detection of microcalcifications/masses in mammography (12), breast MRI (30), and breast ultrasound (US) (31,32), and those for detection of polyps in CT colonography (CTC) (33-36).
In this paper, the progress and advancement of the development of CAD schemes of the thorax and colon are reviewed, including CAD schemes for detection and diagnosis of lung nodules in thoracic CT, and those for detection of polyps in CTC.
Generic architectures of CADe and CADx schemes
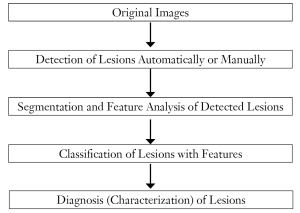
A flowchart for a generic CADe scheme of lesions in medical images is shown in Figure 1. A CADe scheme generally consists of four major steps and two optional steps: (I) segmentation of the organ of interest, (III) detection of lesion candidates from the segmented organ, (IV) segmentation and feature analysis of the detected lesion candidates, (V) classification of the lesion candidates by use of a classifier with features; optionally (II) enhancement of lesions between steps I and III, and (VI) reduction of false-positive (FP) detections. Segmentation of the organ of interest is the first necessary step that aims at making the rest of the steps focus on the organ. The development of the detection of lesion candidates generally aims at obtaining a high sensitivity level, because the sensitivity lost in this step cannot be recovered in the later steps. In the next step, the detected (or localized) lesion candidates are segmented, and connected-component labeling (37,38) is performed for identification of each segmented candidate as an individual isolated object. Pattern features such as gray-level-based features, texture features, and morphologic features are extracted from the segmented candidates. Finally, the detected lesion candidates are classified into lesions or non-lesions by use of a classifier. This final step is very important, because it determines the final performance of a CADe scheme when the additional step of FP reduction is not employed. The development of the classification step aims at removing as many non-lesions (i.e., FPs) as possible while minimizing the removal of lesions (i.e., true-positive detections).
A machine-learning technique is generally used in the step of classification of lesion candidates. The machine-learning technique is trained with sets of input features and correct class labels. This class of machine learning is referred to as feature-based machine learning, or simply as a classifier. The task of machine learning here is to determine “optimal” boundaries for separating classes in the multi-dimensional feature space which is formed by the input features (39). Feature-based machine-learning algorithms include linear discriminant analysis (LDA) (40), quadratic discriminant analysis (QDA) (40), multilayer perceptron [one of the most popular artificial neural network (ANN) models] (41), and support vector machines (SVMs) (42). The structure of an ANN may be designed by use of an automated design method such as sensitivity analysis (43,44). Recently, as available computational power has increased dramatically, pixel/voxel-based machine learning (45) emerged in medical image processing/analysis which uses pixel/voxel values in images directly, instead of features calculated from segmented regions, as input information; thus, feature calculation or segmentation is not required. Pixel-based machine learning has been used in the classification of the detected lesion candidates in CADe and CADx schemes.
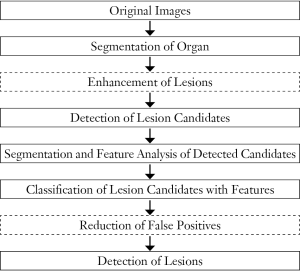
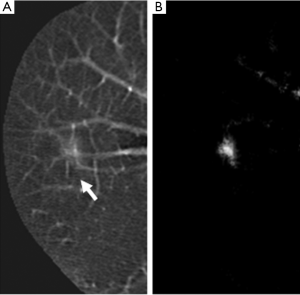
To improve the performance of CADe schemes, investigators sometimes employ an additional step that is enhancement of lesions after the step of the segmentation of the organ of interest. This additional step aims at improving the sensitivity for detection of lesion candidates in the subsequent step. It often helps improve the specificity as well. Figure 2 shows such an example. Suzuki (46) developed a supervised “lesion enhancement” filter based on a massive-training ANN (MTANN) for enhancing lesions and suppressing non-lesions in medical images. Figure 3 illustrates the enhancement of a lung nodule in a CT image by means of a trained MTANN lesion enhancement filter. In the output image, the lung nodule in the original CT image is enhanced, while normal structures such as lung vessels are suppressed substantially. By use of the MTANN lesion enhancement filter, the performance of the initial nodule candidate detection step in a CADe scheme was substantially improved from a 96% sensitivity with 19.3 FPs per section to a 97% sensitivity with 6.7 FPs per section.
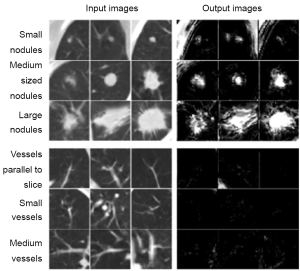
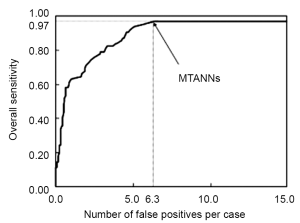
Investigators also often employ an additional step of reduction of FPs at the end in a CADe scheme. The FP reduction step aims at improving the specificity of the CADe scheme. Reduction of FPs is very important, because a large number of FPs could adversely affect the clinical application of CADe. A large number of FPs is likely to confound the radiologist’s task of image interpretation and thus lower his/her efficiency. In addition, radiologists may lose their confidence in CADe as a useful tool. Suzuki et al. developed an FP reduction technique based on MTANNs (26) for reduction of FPs in a CADe scheme for lung nodules in CT. The MTANNs were trained to enhance lung nodules and suppress various types of FPs (i.e., non-nodules) such as lung vessels. Figure 3 shows the results of the enhancement of various-sized lung nodules (a) and those of the suppression of various-sized lung vessels (b). Figure 4 shows a free-response receiver operating characteristic (FROC) curve (47) indicating the performance of the trained MTANNs in the CADe scheme. With the MTANNs, the specificity of the CADe scheme was improved from 15.0 to 6.3 FPs per case without sacrificing the original sensitivity of 97%.
After the development of a CADe scheme, the evaluation of the stand-alone performance of the developed scheme is the last step in CADe development. The evaluation of radiologists’ performance with the use of the developed CAD scheme is the important last step in CAD research.
The flowchart for a generic CADx scheme for diagnosis (or characterization) of detected lesions in medical images is shown in Figure 5. CADx schemes start from detected (localized) lesions either automatically or manually. In other words, the location of a lesion of interest is known. CADx schemes include distinction between benign and malignant status of detected lesions, and classification among different types of detected lesions. First, the detected lesions are segmented, and pattern features are extracted from the segmented lesions. With the extracted features, the lesions are classified into benign or malignant, or different lesion types. CADx schemes may provide the likelihood of malignancy or the likelihood of being a certain type of lesion, as opposed to categorical classification results (or classes).
CADe and CADx in thoracic imaging
Thoracic imaging for lung cancer detection and diagnosis
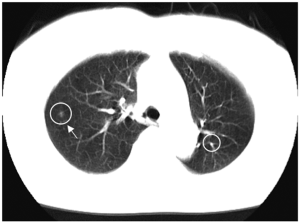
Lung cancer continues to rank as the leading cause of cancer deaths in the United States and in other countries such as Japan. Because CT is more sensitive than chest radiography in the detection of small nodules and of lung carcinoma at an early stage (48-51), lung cancer screening programs are being investigated in the United States (52,53), Japan (48,49), and other countries with low-dose (LD) helical CT as the screening modality. Evidence suggests that early detection of lung cancer may allow more timely therapeutic intervention and thus a more favorable prognosis for the patient (49,54). Helical CT, however, generates a large number of images that must be read by radiologists/physicians. This may lead to ‘‘information overload’’ for the radiologists/physicians. Furthermore, they may miss some cancers during their interpretation of CT images (55,56). Therefore, a CADe scheme for detection of lung nodules in CT images has been investigated as a tool for lung cancer screening. Figure 6 shows an example of CADe outputs on a CT image of the lungs. A CADe scheme detected a lung nodule correctly with an FP which was a branch of the lung vessels.
CADe of lung nodules in thoracic CT
In 1994, Giger et al. (57) developed a CADe scheme for detection of lung nodules in CT based on comparison of geometric features. They applied their CADe scheme to a database of thick-slice diagnostic CT scans of 8 patients with 47 nodules. They achieved a sensitivity of 94% with 1.25 FPs per case. In 1999, Armato et al. (28,58) extended the method to include 3D feature analysis, a rule-based scheme, and LDA for classification. They tested their CADe scheme with a database of thick-slice (10 mm) diagnostic CT scans of 43 patients with 171 nodules. They achieved a sensitivity of 70% with 42.2 FPs per case in a leave-one-out cross-validation test. Gurcan et al. (59) employed a similar approach, i.e., a rule-based scheme based on 2D and 3D features, followed by LDA for classification. They achieved a sensitivity of 84% with 74.4 FPs per case for a database of thick-slice (2.5-5 mm, mostly 5 mm) diagnostic CT scans of 34 patients with 63 nodules in a leave-one-out test. Lee et al. (60) employed a simpler approach which is a rule-based scheme based on 13 features for classification. They achieved a sensitivity of 72% with 30.6 FPs per case for a database of thick-slice (10 mm) diagnostic CT scans of 20 patients with 98 nodules.
Suzuki et al. (26) developed a pixel-based machine-learning technique called an MTANN for reduction of a single source of FPs and a multiple MTANN scheme for reduction of multiple sources of FPs that had not been removed by LDA. They achieved a sensitivity of 80.3% with 4.8 FPs per case for a database of thick-slice (10 mm) screening LDCT scans of 63 patients with 71 nodules with solid, part-solid, and non-solid patterns, including 66 cancers in a validation test. This MTANN approach did not require a large number of training cases: the MTANN was able to be trained with 10 positive and 10 negative cases (61-63), whereas feature-based classifiers generally require 400-800 training cases (61-63). Arimura et al. (27) employed a rule-based scheme followed by LDA or by the MTANN (26) for classification. They tested their scheme with a database of 106 thick-slice (10 mm) screening LDCT scans of 73 patients with 109 cancers that had solid, part-solid and non-solid patterns, and they achieved a sensitivity of 83% with 5.8 FPs per case in a validation test (or a leave-one-patient-out test for LDA). Farag et al. (64) developed a template-modeling approach that uses level sets for classification. They achieved a sensitivity of 93.3% with an FP rate of 3.4% for a database of thin-slice (2.5 mm) screening LDCT scans of 16 patients with 119 nodules and 34 normal patients. Ge et al. (65) incorporated 3D gradient field descriptors and ellipsoid features in LDA for classification. They employed Wilks’ lambda stepwise feature selection for selecting features before the LDA classification. They achieved a sensitivity of 80% with 14.7 FPs per case for a database of 82 thin-slice (1.0-2.5 mm) CT scans of 56 patients with 116 solid nodules in a leave-one-patient-out test. Matsumoto et al. (66) employed LDA with 8 features for classification. They achieved a sensitivity of 90% with 64.1 FPs per case for a database of thick-slice (5 or 7 mm) diagnostic CT scans of 5 patients (4 of which used contrast media) with 50 nodules in a leave-one-out test.
Yuan et al. (67) tested a commercially available CADe system (ImageChecker CT, LN-1000, by R2 Technology, Sunnyvale, CA; Hologic now). They achieved a sensitivity of 73% with 3.2 FPs per case for a database of thin-slice (1.25 mm) CT scans of 150 patients with 628 nodules in an independent test. Pu et al. (68) developed a scoring method based on the similarity distance of medial axis-like shapes for classification. They achieved a sensitivity of 81.5% with 6.5 FPs per case for a database of thin-slice (2.5 mm) screening CT scans of 52 patients with 184 nodules, including 16 non-solid nodules. Retico et al. (69) used a voxel-based neural approach (i.e., a class of the MTANN approach) with pixel values in a subvolume as input for classification. They obtained sensitivities of 80-85% with 10-13 FPs per case for a database of thin-slice (1 mm) screening CT scans of 39 patients with 102 nodules. Ye et al. (70) used a rule-based scheme followed by a weighted SVM for classification. They achieved a sensitivity of 90.2% with 8.2 FPs per case for a database of thin-slice (1 mm) screening CT scans of 54 patients with 118 nodules including 17 non-solid nodules in an independent test. Golosio et al. (71) used a fixed-topology ANN for classification, and they evaluated their CADe scheme with a publicly available database from the Lung Image Database Consortium (LIDC) (72). They achieved a sensitivity of 79% with 4 FPs per case for a database of thin-slice (1.5-3.0 mm) CT scans of 83 patients with 148 nodules that one radiologist detected from an LIDC database in an independent test.
Murphy et al. (73) used a k-nearest-neighbor classifier with features selected from 135 features for classification. They achieved a sensitivity of 80 with 4.2 FPs per case for a large database of thin-slice screening CT scans of 813 patients with 1,525 nodules in an independent test. Tan et al. (74) developed a feature-selective classifier based on a genetic algorithm and ANNs for classification. They achieved a sensitivity of 87.5% with 4 FPs per case for a database of thin-slice CT scans of 125 patients with 80 nodules that 4 radiologists agreed from the LIDC database in an independent test. Messay et al. (75) developed a sequential forward selection process for selecting the optimum features for LDA and quadratic discriminant analysis (QDA). They obtained a sensitivity of 83% with 3 FPs per case for a database of thin-slice CT scans of 84 patients with 143 nodules from the LIDC database in a 7-fold cross-validation test. Riccard et al. (76) used a heuristic approach based on geometric features, followed by an SVM for classification. They achieved a sensitivity of 71% with 6.5 FPs per case for a database of thin-slice CT scans of 154 patients with 117 nodules that 4 radiologists agreed on from the LIDC database in a 2-fold cross-validation test.
Thus, various approaches have been proposed for CADe schemes for lung nodules in CT. Sensitivities for detection of lung nodules in CT range from 70% to 95%, with from a few to 70 FPs per case. Major sources of FPs are various-sized lung vessels. Major sources of false negatives are ground glass nodules, nodules attached to vessels, and nodules attached to the lung wall (i.e., juxtapleural nodules). Ground glass nodules are difficult to detect, because they are subtle, of low-contrast, and have ill-defined boundaries. The MTANN approach was able to enhance and thus detect ground-glass nodules (26). The cause of false negatives due to vessel-attached nodules and juxtapleural nodules is mis-segmentation and thus inaccurate feature calculation. Because the MTANN approach does not require segmentation or feature calculation, it was able to detect such nodules (26).
CADx of lung nodules in thoracic CT
Although CT has been shown to be sensitive to the detection lung nodules, it may be difficult for radiologists to distinguish between benign and malignant nodules on LDCT images. In a screening program with LDCT in New York, 88% (206/233) of suspicious lesions were found to be benign on follow-up examinations (50). In a screening program in Japan, only 83 (10%) among 819 scans with suspicious lesions were diagnosed to be cancer cases (56). According to recent findings at the Mayo Clinic, 2,792 (98.6%) of 2,832 nodules detected by a multidetetor CT were benign, and 40 (1.4%) nodules were malignant (52). Thus, a large number of benign nodules were found with CT; follow-up examinations such as high-resolution CT (HRCT) and/or biopsy were performed on these patients. Therefore, CADx schemes for distinction between benign and malignant nodules in LDCT would be useful for reducing the number of “unnecessary” follow-up examinations.
A number of researchers developed CADx schemes distinguishing malignant nodules from benign nodules automatically and/or determining the likelihood of malignancy for the detected nodules. The performance of the schemes was generally evaluated by means of receiver-operating-characteristic (ROC) analysis (77), because this task is a two-class classification. The area under the ROC curve (AUC) (78) was often used as a performance index.
In 1999, McNitt-Gray et al. (79) developed a classification scheme based on LDA for distinction between malignant and benign nodules in HRCT. They achieved a correct classification rate of 90.3% for a database of 17 malignant and 14 benign nodules. Matsuki et al. (80) used an ANN with subjective features determined by radiologists for classification between 99 malignant and 56 benign nodules in HRCT and achieved an AUC value of 0.951. Aoyama et al. (81) used LDA for distinction between malignant and benign nodules in thick-slice (10 mm) screening LDCT. They achieved an AUC value of 0.846 for a database of 73 patients with 76 primary cancers and 342 patients with 413 benign nodules. Mori et al. (82) developed a classification scheme for distinction between malignant and benign nodules in contrast-enhanced (CE) CT by using LDA with 3 features (i.e., attenuation, shape index, and curvedness value). They used a database of thin-slice (2 mm) CE-CT scans of 35 malignant and 27 benign nodules for testing their CADx scheme. They achieved AUC values of 0.91 and 1.0 with non-CE CT and CE-CT, respectively, in a leave-one-out test.
Shah et al. (83) employed different classifiers such as logistic regression and QDA with features selected from a group of 31 by using stepwise feature selection based on the Akaike information criterion. Their scheme with logistic regression achieved an AUC value of 0.92 in the distinction between 19 malignant and 16 benign nodules in thin-slice CE-CT. Suzuki et al. (84) developed a pixel-based machine-learning technique called a multiple MTANN scheme for the classification task. They achieved an AUC value of 0.88 for thick-slice screening LDCT scans of 73 patients with 76 primary cancers and 342 patients with 413 benign nodules. Iwano et al. (85) achieved a sensitivity of 76.9% and a specificity of 80% with their scheme based on LDA with 2 features in their evaluation of HRCT images of 52 malignant and 55 benign nodules. Way et al. (86) incorporated nodule surface features into their classification based on LDA or an SVM, and they achieved an AUC value of 0.857 in the classification of 124 malignant and 132 benign nodules in 152 patients. Chen et al. (87) employed an ANN ensemble to classify 19 malignant and 13 benign nodules, and they achieved an AUC value of 0.915. Lee et al. (88) developed a two-step supervised learning scheme combining a genetic algorithm with a random subspace method, and they achieved an AUC value of 0.889 in the classification between 62 malignant and 63 benign nodules.
Thus, various approaches to CADx schemes have been proposed. The database size varied in different studies; CT scans in the databases included screening LDCT, standard diagnostic CT, and HRCT.
CADe in colonic imaging
Colonic imaging for colorectal cancer detection
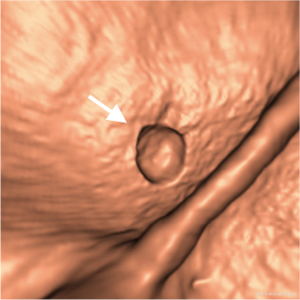
Colorectal cancer is the second leading cause of cancer deaths in the United States (89). Evidence suggests that early detection and removal of polyps (which are precursors of colorectal cancer) can reduce the incidence of colorectal cancer (90,91). Consequently, the American Cancer Society recommends that a person who is at average risk for developing colorectal cancer, beginning at age 50, should have colorectal cancer screening with examinations including optical colonoscopy and CTC. CTC, also known as virtual colonoscopy, is a technique for detecting colorectal neoplasms by use of CT scans of the colon (92). The diagnostic performance of CTC in detecting polyps, however, remains uncertain due to a propensity for perceptual errors in the detection of polyps (93). CADe of polyps has been investigated in an effort to address this issue with CTC (94-96). CADe has the potential to (I) increase radiologists’ diagnostic accuracy in the detection of polyps, (II) decrease reader variability, and (III) reduce radiologists’ interpretation time when CAD is used during the primary read (94,95). A number of investigators have developed automated or semi-automated CADe schemes for the detection of polyps in CTC (33,97-102). Figure 7 shows an example of a CADe output for detection of polyps in an endoluminal view in CTC. A CADe scheme detected a polyp correctly.
CADe of polyps in CTC
In 2000, Summers et al. (36) developed a CADe scheme for detection of polyps in CTC based on curvature analysis. They tested the feasibility of it with simulated polyps. In 2001, they tested their CADe scheme on 50 actual polyps. In 2001, Yoshida et al. (33) developed a CADe scheme based on curvature analysis called a shape index. They tested their CADe scheme on a database of 43 patients with 12 polyps. They achieved a by-patient sensitivity of 100% with 2.0 FPs per case. In 2001, Gokturk et al. (103) employed an SVM with histogram input that is used as a shape signature for classification. They achieved a by-patient (by-polyp) sensitivity of 100% (95%) with a specificity of 0.69 (0.74) [14.3 FPs/patient (12.0 FPs/patient)] for CTC data (2.5-3.0 mm collimation) of 48 patients in either supine or prone position, containing 40 polyps (2-15 mm). Näppi et al. (104) developed a CADe scheme based on LDA or QDA with 54 volumetric features (9 statistics of 6 features). They achieved a by-patient (by-polyp) sensitivity of 100% (95%) with 2.4 FPs/patient for CTC data (5 mm collimation) of 40 patients in both supine and prone positions, including 12 polyps in 11 patients.
Acar et al. (105) used edge-displacement fields to model the changes in consecutive cross-sectional views of CTC data and QDA for classification. They achieved a sensitivity of 100% (95%) with a specificity of 0.47 (0.56) for CTC data (2.5-3.0 mm collimation) of 48 patients in either supine or prone position, containing 40 polyps (2-15 mm). Jerebko et al. (106) used a multilayer perceptron to classify polyp candidates in their CADe scheme and improved the performance by incorporating a committee of multilayer perceptrons (107) and a committee of SVMs (108). They used CTC data (5 mm collimation) of 40 patients in both supine and prone positions, including 21 polyps (5-25 mm) for evaluating their CADe scheme. They finally achieved a sensitivity of 86.7% for larger polyps (≥10 mm) and 75% for other polyps with 3 FPs/patient in a independent test. Wang et al. (109) developed a classification method based on LDA with internal features (geometric, morphologic, and textural) of polyps. They achieved a sensitivity of 100% and 100% for larger polyps (≥10 mm) and other polyps with 4 and 6.9 FPs/patient, respectively, for CTC data (5 mm collimation) of 153 patients in both supine and prone positions, including 61 polyps (4-30 mm) in 45 patients. Näppi et al. (110) developed a method for false-positive reduction (i.e., classification between lesions and non-lesions) based on supine-prone correspondence of CTC scans where patients are scanned in the supine and prone positions. Näppi et al. (111) also developed feature-guided analysis in colon segmentation and feature analysis of polyp candidates and compared them with their previously developed fuzzy clustering. They were able to reduce 70-75% of FPs with their new method, which combines both a non-machine-learning-based method and a feature-based classifier.
Suzuki et al. (112) developed a pixel-based machine-learning technique called a 3D MTANN by extending the structure of a 2D MTANN (26) to process 3D volume data in CTC. Their CADe scheme was based on a Bayesian ANN with texture and geometric features, followed by t3D MTANNs. They removed FPs due to rectal tubes by using a single 3D MTANN (112) and multiple sources of FPs by developing and using a mixture of expert 3D MTANNs (35). They achieved a by-polyp (by-patient) sensitivity of 96.4% (100%) with 2.1 FPs/patient in a leave-one-lesion-out cross-validation test of the classification part for CTC data (1.25-5 mm collimation) of 73 patients in both supine and prone positions, including 28 polyps (5-25 mm) in 15 patients. Li et al. (113) developed a classification method based on an SVM classifier with wavelet-based features. They obtained a sensitivity of 71% with 5.4 FPs/patient in a 4-fold cross-validation test of the classification part for CTC data of 44 patients containing 45 polyps (6-9 mm). Wang et al. (114) improved the SVM performance by using nonlinear dimensionality reduction (i.e., a diffusion map and locally linear embedding). They used CTC data (1.25-2.5 mm collimation) of 792 patients in both supine and prone positions, including 226 polyps (>6 mm) for evaluating their CADe scheme. They achieved a sensitivity of 83% for polyps (6-9 mm) with 9 FPs/patient.
Yao at al. (115) employed a topographic height map for calculating features for an SVM classifier. They used a large database containing CTC data (1.25-2.5 mm collimation) of 792 patients in both supine and prone positions, including 226 polyps (>6 mm) for evaluating their CADe scheme. They obtained a sensitivity of 93% and 76% for larger polyps (≥10 mm) and other polyps with 1.2 and 3.1 FPs/patient, respectively, in a 10-fold cross-validation test of the classification part. Suzuki et al. (116) tested a CADe scheme based on a Bayesian ANN with texture and geometric features together with MTANNs (i.e., a pixel-based machine-learning technique). They used CTC data (1.25-5 mm collimation) of 24 patients in both supine and prone positions, including 23 polyps (6-25 mm) and a mass (35 mm), that had been “missed” by radiologists (117) in a multicenter clinical trial (118). They achieved a by-polyp (by-patient) sensitivity of 96.4% (100%) with 1.1 FPs/patient in a leave-one-lesion-out cross-validation test of the classification part.
Suzuki et al. (119,120) also improved the efficiency of the MTANN approach by incorporating principal-component analysis-based and Laplacian eigenmap-based dimension reduction techniques. Xu and Suzuki (121) showed that other nonlinear regression models such as support vector and nonlinear Gaussian process regression models instead of the ANN regression model could be used as the core model in the MTANN framework. Zhou et al. (122) developed projection features for an SVM classifier. They achieved by-polyp sensitivities of 93.1% and 80.6% for larger polyps (≥10 mm) and other polyps with 1.9 and 5.2 FPs/patient, respectively, for CTC data (1.25-5.0 mm collimation) of 325 patients in the supine and/or prone positions, including 347 polyps and masses (5-60 mm). Wang et al. (123) improved the performance of a CAD scheme by adding statistical curvature features in multiple-kernel learning. They obtained a sensitivity of 83% with 5 FPs/patient in a leave-one-out cross-validation test of the classification part for a database containing CTC data (1.25-2.5 mm collimation) of 66 patients in supine and/or prone positions, including 96 polyps.
Thus, various approaches have been proposed for CADe schemes for polyps in CTC. Most of the methods for the initial detection of polyp candidates employed morphologic analysis, including curvature analysis such as the shape index. Curvatures are often calculated by using the Hessian matrix. Classifiers used in CADe schemes include LDA, QDA, an SVM, and a Bayesian ANN. Sensitivities for detection of polyps in CTC range from 70% to 100%, with 1 to 30 FPs per case. Major sources of FPs include haustral folds, residual stool, rectal tubes, the ileocecal valve, and extra-colonic structures such as the small bowel and stomach. Existing CADe schemes tend to miss superficially elevated neoplasms (often called flat lesions) (124,125). Recently, Suzuki et al. developed a CADe scheme for detection of superficially elevated neoplasms (126). Detection of superficially elevated neoplasms is very important, because they are histologically aggressive, and because they are often missed by radiologists in CTC as well as by gastroenterologists in optical colonoscopy.
Summary
In this paper, CADe and CADx schemes for detection and diagnosis of lung nodules in thoracic CT and those for detection of polyps in CTC have been reviewed. A CADe scheme (i.e., detection of lesions) generally consists of four major steps and two optional steps: segmentation of the organ of interest, detection of lesion candidates from the segmented organ, segmentation and feature analysis of the detected lesion candidates, and classification of the lesion candidates by use of a classifier with features; and optionally, enhancement of lesions and reduction of FPs. Feature-based machine learning (or classifier), including LDA, QDA, an ANN, a Bayesian ANN, and an SVM, was used in the step of the classification of the lesion candidates. To improve the performance of CADe schemes further, investigators sometimes employ additional steps of enhancement of lesions and FP reduction. CADe schemes that employ those addition steps tend to yield a high performance. Sensitivities for detection of lung nodules in CT range from 70% to 95%, with a few to 70 FPs per case. On the other hand, sensitivities for detection of polyps in CTC range from 70% to 100%, with 1 to 30 FPs per case. Thus, the sensitivities of CADe schemes are relatively high, but the number of FPs is high-compared to radiologists’ performance. Therefore, further improvement in specificity is necessary in future research.
A CADx scheme (i.e., diagnosis or characterization of detected lesions) generally consists of two major steps, which are segmentation and feature analysis of the detected lesions and classification of the lesions. CADx schemes start from detected (localized) lesions either automatically or manually. The major component of CADx schemes is classification of lesions by use of a classifier with features. The performance of CADx schemes for distinction between benign and malignant lung nodules in CT range from AUC values of 0.85 to 0.95. Thus, the computer performance is relatively high compared to radiologists’ performance.
I hope that this review will be useful for researchers to advance the development of CAD schemes in thoracic and colonic imaging as well as for those who want to start the development of CAD schemes in other imaging areas.
Acknowledgements
The author is grateful to Ms. E. F. Lanzl for improving the manuscript. This work was partly supported by Grant Number R01CA120549 from the National Cancer Institute/National Institutes of Health and by the NIH S10 RR021039 and P30 CA14599.
Disclosure: CAD and machine-learning technologies developed at the University of Chicago have been licensed to companies including R2 Technology (Hologic), Riverain Medical (Riverain Technologies), Deus Technology, Median Technologies, Mitsubishi Space Software, General Electric, and Toshiba. It is the policy of the University of Chicago that investigators disclose publicly actual or potential significant financial interests that may appear to be affected by research activities.
References
- Giger ML, Suzuki K. Computer-Aided Diagnosis (CAD). In: Feng DD. eds. Biomedical Information Technology. Academic Press, 2007:359-74.
- Doi K. Current status and future potential of computer-aided diagnosis in medical imaging. Br J Radiol 2005;78:S3-S19. [PubMed]
- Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph 2007;31:198-211. [PubMed]
- Giger ML, Chan HP, Boone J. Anniversary paper: History and status of CAD and quantitative image analysis: the role of Medical Physics and AAPM. Med Phys 2008;35:5799-820. [PubMed]
- Lusted LB. Medical electronics. N Engl J Med 1955;252:580-5. [PubMed]
- Lodwick GS, Keats TE, Dorst JP. The Coding of Roentgen Images for Computer Analysis as Applied to Lung Cancer. Radiology 1963;81:185-200. [PubMed]
- Becker HC, Nettleton WJ Jr, Meyers PH, et al. Digital Computer Determination of a Medical Diagnostic Index Directly from Chest X-Ray Images. IEEE Trans Biomed Eng 1964;11:67-72. [PubMed]
- Meyers PH, Nice CM Jr, Becker HC, et al. Automated Computer Analysis of Radiographic Images. Radiology 1964;83:1029-34. [PubMed]
- Winsberg F, Elkin M, Macy J Jr, et al. Detection of Radiographic Abnormalities in Mammograms by Means of Optical Scanning and Computer Analysis. Radiology 1967;89:211-5.
- Toriwaki J, Suenaga Y, Negoro T, et al. Pattern recognition of chest x-ray images. Comput Graph Image Process 1973;2:252-71.
- Roellinger FX Jr, Kahveci AE, Chang JK, et al. Computer analysis of chest radiographs. Comput Graph Image Process 1973;2:232-51.
- Chan HP, Doi K, Galhotra S, et al. Image feature analysis and computer-aided diagnosis in digital radiography. I. Automated detection of microcalcifications in mammography. Med Phys 1987;14:538-48. [PubMed]
- Fujita H, Doi K, Fencil LE, et al. Image feature analysis and computer-aided diagnosis in digital radiography. 2. Computerized determination of vessel sizes in digital subtraction angiography. Med Phys 1987;14:549-56. [PubMed]
- Giger ML, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields. Med Phys 1988;15:158-66. [PubMed]
- Katsuragawa S, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography: detection and characterization of interstitial lung disease in digital chest radiographs. Med Phys 1988;15:311-9. [PubMed]
- Chan HP, Sahiner B, Helvie MA, et al. Improvement of radiologists’ characterization of mammographic masses by using computer-aided diagnosis: an ROC study. Radiology 1999;212:817-27. [PubMed]
- Li F, Aoyama M, Shiraishi J, et al. Radiologists’ performance for differentiating benign from malignant lung nodules on high-resolution CT using computer-estimated likelihood of malignancy. AJR Am J Roentgenol 2004;183:1209-15. [PubMed]
- Li F, Arimura H, Suzuki K, et al. Computer-aided detection of peripheral lung cancers missed at CT: ROC analyses without and with localization. Radiology 2005;237:684-90. [PubMed]
- Dean JC, Ilvento CC. Improved cancer detection using computer-aided detection with diagnostic and screening mammography: prospective study of 104 cancers. AJR Am J Roentgenol 2006;187:20-8. [PubMed]
- Suzuki K, Hori M, McFarland E, et al. Can CAD help improve the performance of radiologists in detection of difficult polyps in CT colonography? Proceedings of RSNA Annual Meeting, Chicago, IL 2009.
- Dachman AH, Obuchowski NA, Hoffmeister JW, et al. Effect of computer-aided detection for CT colonography in a multireader, multicase trial. Radiology 2010;256:827-35. [PubMed]
- Petrick N, Haider M, Summers RM, et al. CT colonography with computer-aided detection as a second reader: observer performance study. Radiology 2008;246:148-56. [PubMed]
- Suzuki K, Shiraishi J, Abe H, et al. False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network. Acad Radiol 2005;12:191-201. [PubMed]
- van Ginneken B, ter Haar Romeny BM, Viergever MA. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans Med Imaging 2001;20:1228-41. [PubMed]
- Chen S, Suzuki K, MacMahon H. Development and evaluation of a computer-aided diagnostic scheme for lung nodule detection in chest radiographs by means of two-stage nodule enhancement with support vector classification. Med Phys 2011;38:1844-58. [PubMed]
- Suzuki K, Armato SG 3rd, Li F, et al. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose computed tomography. Med Phys 2003;30:1602-17. [PubMed]
- Arimura H, Katsuragawa S, Suzuki K, et al. Computerized scheme for automated detection of lung nodules in low-dose computed tomography images for lung cancer screening. Acad Radiol 2004;11:617-29. [PubMed]
- Armato SG 3rd, Giger ML, Moran CJ, et al. Computerized detection of pulmonary nodules on CT scans. Radiographics 1999;19:1303-11. [PubMed]
- Armato SG 3rd, Li F, Giger ML, et al. Lung cancer: performance of automated lung nodule detection applied to cancers missed in a CT screening program. Radiology 2002;225:685-92. [PubMed]
- Gilhuijs KG, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys 1998;25:1647-54. [PubMed]
- Horsch K, Giger ML, Vyborny CJ, et al. Performance of computer-aided diagnosis in the interpretation of lesions on breast sonography. Acad Radiol 2004;11:272-80. [PubMed]
- Drukker K, Giger ML, Metz CE. Robustness of computerized lesion detection and classification scheme across different breast US platforms. Radiology 2005;237:834-40. [PubMed]
- Yoshida H, Näppi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imaging 2001;20:1261-74. [PubMed]
- Suzuki K, Yoshida H, Näppi J, et al. Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes. Med Phys 2006;33:3814-24. [PubMed]
- Suzuki K, Yoshida H, Näppi J, et al. Mixture of expert 3D massive-training ANNs for reduction of multiple types of false positives in CAD for detection of polyps in CT colonography. Med Phys 2008;35:694-703. [PubMed]
- Summers RM, Beaulieu CF, Pusanik LM, et al. Automated polyp detector for CT colonography: feasibility study. Radiology 2000;216:284-90. [PubMed]
- He L, Chao Y, Suzuki K, et al. Fast connected-component labeling. Pattern Recognit 2009;42:1977-87.
- Suzuki K, Horiba I, Sugie N. Linear-time connected-component labeling based on sequential local operations. Comput Vis Image Underst 2003;89:1-23.
- Duda RO, Hart PE, Stork DG. In: Pattern Recognition, 2nd ed. Hoboken, NJ: Wiley Interscience, 2001.
- Fukunaga K. eds. Introduction to Statistical Pattern Recognition. San Diego: Academic Press, 1990.
- Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986;323:533-6.
- Vapnik VN. eds. The Nature of Statistical Learning Theory. Berlin: Springer-Verlag, 1995.
- Suzuki K. Determining the receptive field of a neural filter. J Neural Eng 2004;1:228-37. [PubMed]
- Suzuki K, Horiba I, Sugie N. A simple neural network pruning algorithm with application to filter synthesis. Neural Process Lett 2001;13:43-53.
- Suzuki K. Pixel-based machine learning in medical imaging. Int J Biomed Imaging 2012;2012:792079.
- Suzuki K. A supervised “lesion-enhancement” filter by use of a massive-training artificial neural network (MTANN) in computer-aided diagnosis (CAD). Phys Med Biol 2009;54:S31-45. [PubMed]
- Bunch PC, Hamilton JF, Sanderson KG, et al. A free-response approach to the measurement and characterization of radiographic-observer performance. J of Appl Photogr Eng 1978;4:166-71.
- Kaneko M, Eguchi K, Ohmatsu H, et al. Peripheral lung cancer: screening and detection with low-dose spiral CT versus radiography. Radiology 1996;201:798-802. [PubMed]
- Sone S, Takashima S, Li F, et al. Mass screening for lung cancer with mobile spiral computed tomography scanner. Lancet 1998;351:1242-5. [PubMed]
- Henschke CI, McCauley DI, Yankelevitz DF, et al. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet 1999;354:99-105. [PubMed]
- Miettinen OS, Henschke CI. CT screening for lung cancer: coping with nihilistic recommendations. Radiology 2001;221:592-6; discussion 597. [PubMed]
- Swensen SJ, Jett JR, Hartman TE, et al. Lung cancer screening with CT: Mayo Clinic experience. Radiology 2003;226:756-61. [PubMed]
- Henschke CI, Yankelevitz DF, Naidich DP, et al. CT screening for lung cancer: suspiciousness of nodules according to size on baseline scans. Radiology 2004;231:164-8. [PubMed]
- Heelan RT, Flehinger BJ, Melamed MR, et al. Non-small-cell lung cancer: results of the New York screening program. Radiology 1984;151:289-93. [PubMed]
- Gurney JW. Missed lung cancer at CT: imaging findings in nine patients. Radiology 1996;199:117-22. [PubMed]
- Li F, Sone S, Abe H, et al. Lung cancers missed at low-dose helical CT screening in a general population: comparison of clinical, histopathologic, and imaging findings. Radiology 2002;225:673-83. [PubMed]
- Giger ML, Bae KT, MacMahon H. Computerized detection of pulmonary nodules in computed tomography images. Invest Radiol 1994;29:459-65. [PubMed]
- Armato SG 3rd, Giger ML, MacMahon H. Automated detection of lung nodules in CT scans: preliminary results. Med Phys 2001;28:1552-61. [PubMed]
- Gurcan MN, Sahiner B, Petrick N, et al. Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer-aided diagnosis system. Med Phys 2002;29:2552-8. [PubMed]
- Lee Y, Hara T, Fujita H, et al. Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. IEEE Trans Med Imaging 2001;20:595-604. [PubMed]
- Suzuki K, Doi K. How can a massive training artificial neural network (MTANN) be trained with a small number of cases in the distinction between nodules and vessels in thoracic CT? Acad Radiol 2005;12:1333-41. [PubMed]
- Chan HP, Sahiner B, Wagner RF, et al. Classifier design for computer-aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys 1999;26:2654-68. [PubMed]
- Sahiner B, Chan HP, Hadjiiski L. Classifier performance prediction for computer-aided diagnosis using a limited dataset. Med Phys 2008;35:1559-70. [PubMed]
- Farag AA, El-Baz A, Gimelfarb G, et al. Quantitative nodule detection in low dose chest CT scans: new template modeling and evaluation for CAD system design. Med Image Comput Comput Assist Interv 2005;8:720-8. [PubMed]
- Ge Z, Sahiner B, Chan HP, et al. Computer-aided detection of lung nodules: false positive reduction using a 3D gradient field method and 3D ellipsoid fitting. Med Phys 2005;32:2443-54. [PubMed]
- Matsumoto S, Kundel HL, Gee JC, et al. Pulmonary nodule detection in CT images with quantized convergence index filter. Med Image Anal 2006;10:343-52. [PubMed]
- Yuan R, Vos PM, Cooperberg PL. Computer-aided detection in screening CT for pulmonary nodules. AJR Am J Roentgenol 2006;186:1280-7. [PubMed]
- Pu J, Zheng B, Leader JK, et al. An automated CT based lung nodule detection scheme using geometric analysis of signed distance field. Med Phys 2008;35:3453-61. [PubMed]
- Retico A, Delogu P, Fantacci ME, et al. Lung nodule detection in low-dose and thin-slice computed tomography. Comput Biol Med 2008;38:525-34. [PubMed]
- Ye X, Lin X, Dehmeshki J, et al. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Trans Biomed Eng 2009;56:1810-20. [PubMed]
- Golosio B, Masala GL, Piccioli A, et al. A novel multithreshold method for nodule detection in lung CT. Med Phys 2009;36:3607-18. [PubMed]
- Armato SG 3rd, McLennan G, McNitt-Gray MF, et al. Lung image database consortium: developing a resource for the medical imaging research community. Radiology 2004;232:739-48. [PubMed]
- Murphy K, van Ginneken B, Schilham AM, et al. A large-scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classification. Med Image Anal 2009;13:757-70. [PubMed]
- Tan M, Deklerck R, Jansen B, et al. A novel computer-aided lung nodule detection system for CT images. Med Phys 2011;38:5630-45. [PubMed]
- Messay T, Hardie RC, Rogers SK. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 2010;14:390-406. [PubMed]
- Riccardi A, Petkov TS, Ferri G, et al. Computer-aided detection of lung nodules via 3D fast radial transform, scale space representation, and Zernike MIP classification. Med Phys 2011;38:1962-71. [PubMed]
- Metz CE. ROC methodology in radiologic imaging. Invest Radiol 1986;21:720-33. [PubMed]
- Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983;148:839-43. [PubMed]
- McNitt-Gray MF, Hart EM, Wyckoff N, et al. A pattern classification approach to characterizing solitary pulmonary nodules imaged on high resolution CT: preliminary results. Med Phys 1999;26:880-8. [PubMed]
- Matsuki Y, Nakamura K, Watanabe H, et al. Usefulness of an artificial neural network for differentiating benign from malignant pulmonary nodules on high-resolution CT: evaluation with receiver operating characteristic analysis. AJR Am J Roentgenol 2002;178:657-63. [PubMed]
- Aoyama M, Li Q, Katsuragawa S, et al. Automated computerized scheme for distinction between benign and malignant solitary pulmonary nodules on chest images. Med Phys 2002;29:701-8. [PubMed]
- Mori K, Niki N, Kondo T, et al. Development of a novel computer-aided diagnosis system for automatic discrimination of malignant from benign solitary pulmonary nodules on thin-section dynamic computed tomography. J Comput Assist Tomogr 2005;29:215-22. [PubMed]
- Shah SK, McNitt-Gray MF, Rogers SR, et al. Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Acad Radiol 2005;12:1310-9. [PubMed]
- Suzuki K, Li F, Sone S, et al. Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose CT by use of massive training artificial neural network. IEEE Trans Med Imaging 2005;24:1138-50. [PubMed]
- Iwano S, Nakamura T, Kamioka Y, et al. Computer-aided differentiation of malignant from benign solitary pulmonary nodules imaged by high-resolution CT. Comput Med Imaging Graph 2008;32:416-22. [PubMed]
- Way TW, Sahiner B, Chan HP, et al. Computer-aided diagnosis of pulmonary nodules on CT scans: improvement of classification performance with nodule surface features. Med Phys 2009;36:3086-98. [PubMed]
- Chen H, Xu Y, Ma Y, et al. Neural network ensemble-based computer-aided diagnosis for differentiation of lung nodules on CT images: clinical evaluation. Acad Radiol 2010;17:595-602. [PubMed]
- Lee MC, Boroczky L, Sungur-Stasik K, et al. Computer-aided diagnosis of pulmonary nodules using a two-step approach for feature selection and classifier ensemble construction. Artif Intell Med 2010;50:43-53. [PubMed]
- Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin 2005;55:10-30. [PubMed]
- Winawer S, Fletcher R, Rex D, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 2003;124:544-60. [PubMed]
- Dachman AH. eds. Atlas of Virtual Colonoscopy. New York: Springer-Verlag, 2003.
- Macari M, Bini EJ. CT colonography: where have we been and where are we going? Radiology 2005;237:819-33. [PubMed]
- Fletcher JG, Booya F, Johnson CD, et al. CT colonography: unraveling the twists and turns. Curr Opin Gastroenterol 2005;21:90-8. [PubMed]
- Yoshida H, Dachman AH. Computer-aided diagnosis for CT colonography. Semin Ultrasound CT MR 2004;25:419-31. [PubMed]
- Yoshida H, Dachman AH. CAD techniques, challenges, and controversies in computed tomographic colonography. Abdom Imaging 2005;30:26-41. [PubMed]
- Suzuki K, Dachman AH. Computer-aided diagnosis in CT colonography. In: Dachman AH, Laghi A. eds. Atlas of Virtual Colonoscopy. 2nd ed. New York: Springer, 2011:163-82.
- Yoshida H, Masutani Y, MacEneaney P, et al. Computerized detection of colonic polyps at CT colonography on the basis of volumetric features: pilot study. Radiology 2002;222:327-36. [PubMed]
- Yoshida H, Näppi J, MacEneaney P, et al. Computer-aided diagnosis scheme for detection of polyps at CT colonography. Radiographics 2002;22:963-79. [PubMed]
- Summers RM, Johnson CD, Pusanik LM, et al. Automated polyp detection at CT colonography: feasibility assessment in a human population. Radiology 2001;219:51-9. [PubMed]
- Paik DS, Beaulieu CF, Rubin GD, et al. Surface normal overlap: a computer-aided detection algorithm with application to colonic polyps and lung nodules in helical CT. IEEE Trans Med Imaging 2004;23:661-75. [PubMed]
- Kiss G, Van Cleynenbreugel J, Thomeer M, et al. Computer-aided diagnosis in virtual colonography via combination of surface normal and sphere fitting methods. Eur Radiol 2002;12:77-81. [PubMed]
- Summers RM, Yao J, Pickhardt PJ, et al. Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology 2005;129:1832-44. [PubMed]
- Göktürk SB, Tomasi C, Acar B, et al. A statistical 3-D pattern processing method for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging 2001;20:1251-60. [PubMed]
- Näppi J, Yoshida H. Automated detection of polyps with CT colonography: evaluation of volumetric features for reduction of false-positive findings. Acad Radiol 2002;9:386-97. [PubMed]
- Acar B, Beaulieu CF, Göktürk SB, et al. Edge displacement field-based classification for improved detection of polyps in CT colonography. IEEE Trans Med Imaging 2002;21:1461-7. [PubMed]
- Jerebko AK, Summers RM, Malley JD, et al. Computer-assisted detection of colonic polyps with CT colonography using neural networks and binary classification trees. Med Phys 2003;30:52-60. [PubMed]
- Jerebko AK, Malley JD, Franaszek M, et al. Multiple neural network classification scheme for detection of colonic polyps in CT colonography data sets. Acad Radiol 2003;10:154-60. [PubMed]
- Jerebko AK, Malley JD, Franaszek M, et al. Support vector machines committee classification method for computer-aided polyp detection in CT colonography. Acad Radiol 2005;12:479-86. [PubMed]
- Wang Z, Liang Z, Li L, et al. Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy. Med Phys 2005;32:3602-16. [PubMed]
- Näppi J, Okamura A, Frimmel H, et al. Region-based supine-prone correspondence for the reduction of false-positive CAD polyp candidates in CT colonography. Acad Radiol 2005;12:695-707. [PubMed]
- Näppi J, Yoshida H. Feature-guided analysis for reduction of false positives in CAD of polyps for computed tomographic colonography. Med Phys 2003;30:1592-601. [PubMed]
- Suzuki K, Yoshida H, Näppi J, et al. Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes. Med Phys 2006;33:3814-24. [PubMed]
- Li J, Van Uitert R, Yao J, et al. Wavelet method for CT colonography computer-aided polyp detection. Med Phys 2008;35:3527-38. [PubMed]
- Wang S, Yao J, Summers RM. Improved classifier for computer-aided polyp detection in CT colonography by nonlinear dimensionality reduction. Med Phys 2008;35:1377-86. [PubMed]
- Yao J, Li J, Summers RM. Employing topographical height map in colonic polyp measurement and false positive reduction. Pattern Recognit 2009;42:1029-40. [PubMed]
- Suzuki K, Rockey DC, Dachman AH. CT colonography: advanced computer-aided detection scheme utilizing MTANNs for detection of “missed” polyps in a multicenter clinical trial. Med Phys 2010;37:12-21. [PubMed]
- Doshi T, Rusinak D, Halvorsen RA, et al. CT colonography: false-negative interpretations. Radiology 2007;244:165-73. [PubMed]
- Rockey DC, Paulson E, Niedzwiecki D, et al. Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: prospective comparison. Lancet 2005;365:305-11. [PubMed]
- Suzuki K, Zhang J, Xu J. Massive-training artificial neural network coupled with Laplacian-eigenfunction-based dimensionality reduction for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging 2010;29:1907-17. [PubMed]
- Suzuki K, Xu J, Zhang J, et al. Principal-Component Massive-Training Machine-Learning Regression for False-Positive Reduction in Computer-Aided Detection of Polyps in CT Colonography. Machine Learning in Medical Imaging 2012;6357:182-9.
- Xu JW, Suzuki K. Massive-training support vector regression and Gaussian process for false-positive reduction in computer-aided detection of polyps in CT colonography. Med Phys 2011;38:1888-902. [PubMed]
- Zhu H, Liang Z, Pickhardt PJ, et al. Increasing computer-aided detection specificity by projection features for CT colonography. Med Phys 2010;37:1468-81. [PubMed]
- Wang S, Yao J, Petrick N, et al. Combining Statistical and Geometric Features for Colonic Polyp Detection in CTC Based on Multiple Kernel Learning. Int J Comput Intell Appl 2010;9:1-15. [PubMed]
- Lostumbo A, Suzuki K, Dachman AH. Flat lesions in CT colonography. Abdom Imaging 2010;35:578-83. [PubMed]
- Lostumbo A, Wanamaker C, Tsai J, et al. Comparison of 2D and 3D views for evaluation of flat lesions in CT colonography. Acad Radiol 2010;17:39-47. [PubMed]
- Suzuki K, Sheu I, Kawaler E, et al. Computer-aided detection (CADe) of flat lesions in CT colonography (CTC) by means of a spinning-tangent technique. Program of RSNA 2010.