
Improved second harmonic generation and two-photon excitation fluorescence microscopy-based quantitative assessments of liver fibrosis through auto-correction and optimal sampling
Introduction
Liver fibrosis associated with chronic liver disease, such as hepatitis B and hepatitis C, involves the excessive accumulation of extracellular matrix proteins (i.e., collagen), when hepatocytes are injured. Second harmonic generation (SHG) microscopy/two-photon excited fluorescence (TPEF) microscopy is widely used for the quantitative assessment of liver fibrosis (1-6). Researchers have observed a correlation between histological staging and the morphological features of collagen (6,7). The images obtained using SHG microscopy are largely free from variations introduced by the staining process. Computer-aided image analysis can be used to exclude inter-observer discrepancies among pathologists (8-10), and thereby achieve a more consistent evaluation of histopathological information.
The Metavir system, proposed by French METAVIR Cooperative Study Group, is one of most widely accepted scoring systems used to assess the extent of fibrosis by histopathological evaluation in a liver biopsy of patients with chronic liver disease. The system provides four stages of progression of fibrosis on a 5-point scale from 0 to 4: F0, no fibrosis; F1, portal fibrosis without septa; F2, portal fibrosis with few septa; F3, numerous septa without cirrhosis, and F4: cirrhosis. However, Metavir stage is limited by inter-observer variation in interpretation of liver biopsies because of heterogenicity of inflammation and fibrosis in different regions of the liver. Unfortunately, there are at present no well-established protocols for image acquisition or image analysis. Sampling discrepancy is a major limitation to the quantitative assessment of samples. Our ability to identify the collagen contributing to fibrosis can be influenced by the properties of the tissue sample as well as the area being scanned. Samples may include extraneous tissue, such as the capsule and longitudinal sections of vessels. Vessels in the tissue may be stretched during processing. Disturbing collagen (DC) should not be included in the quantitative assessment of fibrosis.
Despite improvements in efficiency due to advancements in scanning, there remains considerable room for improvement through the analysis of a smaller region of interest (ROI) rather than obtaining a full scan. If small ROI scans could be shown to provide the same correlative results as full scans with respect to Metavir stages, liver tissue obtained via a needle biopsy could undergo quantitative assessment using SHG/TPEF microscopy.
In this study, we sought to improve the accuracy of quantitative assessments by removing the signals from disturbing collagen in liver specimens. We also sought to optimize the sampling protocol with the aim of improving assessment efficiency.
Methods
Patients and image acquisition system
This study was approved by the Ethics Committee of National Taiwan University Hospital and conducted in accordance with approved guidelines. We included in this study 111 patients with chronic hepatitis B who underwent hepatic surgery at National Taiwan University Hospital between Jan. 2000 and Dec. 2010. Liver samples were obtained from specimens harvested during operations. Written informed consent was obtained from all patients before obtaining the liver samples. Full tissue scanning using the Genesis system (HistoIndex Pte. Ltd., Singapore) was performed on all of the liver samples. They were laser-excited at 780 nm, SHG signals were recorded at 390 nm to visualize collagen, and TPEF signals were recorded at 550 nm to identify tissue structures. Images were acquired at 20× magnification with 512×512 pixel resolution, and each image had a dimension of 200 µm × 200 µm. Multiple adjacent images were captured to encompass large areas. Chart records of all patients were retrospectively reviewed to obtain clinical data including sex, age, and Metavir fibrosis stage.
Removing signals from disturbing collagen (DC)
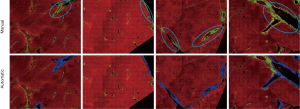
We adopted the labeling results of pathologist as a guide in the removal of three types of SHG signal during the quantified assessment of liver fibrosis (Figure 1). Specifically, we removed the signals associated with the following structures:
❖ Large patches of collagen near the boundary of capsules;
❖ Collagen around tubular structures, such as longitudinal sections of vessels;
❖ Collagen associated with distorted vessel walls (irregular shaped).
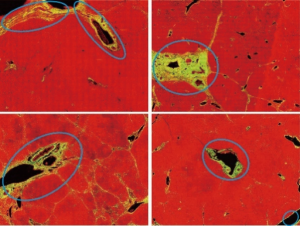
These forms of physiological collagen exist in normal liver tissue; however, they are regarded by pathologists as noise. In seeking to remove the signals from DC, we first selected 20 of the 111 full tissue SHG images for the extraction of image features from areas roughly labeled by pathologists. These feature sets were then used to identify the signals of DC in a training set to optimize the automation system (20 patients) and a validation set (91 patients). After training, the image analysis model was able to automatically detect signals from DC and exclude them from subsequent assessments of liver fibrosis. This auto-correction algorithm can be divided into three steps:
- Candidate segmentation: while the boundary and holes (tubular structures, distorted vessel) of tissues were detected from the TPEF channel, the DC candidates (the large patches of collagen) near these areas were detected from the SHG channel, both by the Otsu’s automatic threshold method (11);
- Feature extraction: the features of the candidates were quantified, including the length, width, area and density, the number of holes inside each DC candidate and the roundness of each hole;
- Model construction: based on the features of each DC candidate in the training set, a decision tree model was constructed using classification and regression tree (CART) method (12). This model was used to identify and remove DCs in the validation set.
We then compared quantitative assessments obtained before and after the removal of DC signals (i.e., with DC vs. without DC). Finally, we calculated the degree of correlation between image features and Metavir staging and compared the results obtained before and after DC removal.
Optimal sampling (full tissue scan vs. small ROI scans)
Normally, to image a tissue by SHG/TPEF microscope, many small tiles (1 tile = 200 µm × 200 µm) have to be imaged one by one. In our experiments, on average, 735 tiles were required to make a full scan. It is natural to think of using small ROI scans instead. To determine whether the small ROI scans were comparable with the full tissue scans in terms of accuracy, we compared units of 3×3 tiles and 5×5 tiles for the small ROI scans. We selected these sizes because specimens of this size could be obtained from the very thin liver samples obtained using needle biopsies. A 5×5 tile scan would be on the scale of a normal liver lobule (approximately 1 mm in diameter), whereas a 3×3 tile scan would cover the area containing two portal tracts and the central vein. Figure 2 presents example images of various sizes. We selected small ROIs at random (DC area was avoided) for 100 times. The correlation between image features and Metavir staging was evaluated using 3×3 tiles, 5×5 tiles, and full tissue scans.
Statistical analysis
The mean error of image features was calculated by Matlab (version 2015a; The MathWorks, Inc., Natick, MA). The correlation between image features and Metavir staging was calculated and compared using the Spearman's rank correlation coefficient. Statistical analysis was performed using SPSS 18 for Windows v. 18.0 (SPSS Inc, Chicago, IL, USA).
Results
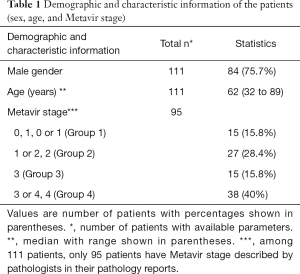
To validate our algorithm with clinical data, the demographics and fibrosis staging of 111 patients were collected as shown in Table 1. It should be noted that pathologists differ in their descriptions of Metavir stages; therefore, we divided the patients’ data into four groups (i.e., Group 1, 2, 3, and 4) in accordance with the specifics of those descriptions. Then the individual groups have strong correlation with Metavir staging.
Full table
Removing disturbing collagen improved the correlation between collagen quantification and stages of Metavir fibrosis
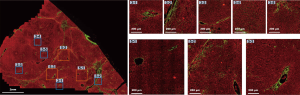
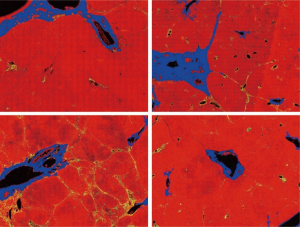
The DC signals were removed by the proposed auto-correction algorithm. An example of automatic labelling results in the training set (Figures 3, blue colors) were roughly the same as those recognized as disturbing collagen by a pathologist (Figure 1, blue circles). Images of the automatic labelling results from the validation sets further showed that automatic identification of DC performed well and were compatible with the areas of disturbing collagen recognized by the pathologists (Figure 4). As the algorithm was well-tuned to detect all the DC in the training set, DC detection accuracy was evaluated on the images from the validation set. The algorithm detected 88 of 97 (90.7%) DCs which were labeled by the pathologists.
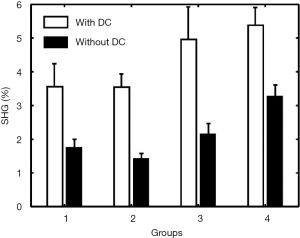
The comparison of the quantitative assessments (mean and standard deviation of SHG percentage) obtained before and after removal of DC signals in the four groups was shown in Figure 5. The standard deviation reduced in all the four groups (indicating that more homogeneous within each group), while the mean difference between groups 3 and group 4 increased (indicating enhancing ability to distinguish between different groups).
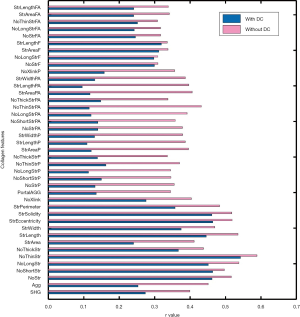
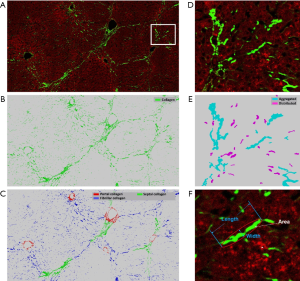
Furthermore, the image quantification (3 to 4) was applied to as-obtained images as well as images that underwent DC removal. Removal of DC signals significantly improved the correlation between Metavir stage and quantification of collagen features (Figure 6). These collagen features were developed in our previous studies and had already been validated in quantitative analysis of liver fibrosis (7,13). The abbreviation and full description of collagen features in Figure 6 were listed in Table 2. An example of illustrations of collagen features were shown in Figure 7. These collagen features were parameters derived from physical characteristics of an image (e.g., length, width, area) which can be automatically calculated during quantitative assessment. Each collagen feature reflects different aspect of a scanning image according to its definition, and could be regarded as a unique parameter during processing image analysis by algorithm.
Optimal sampling of small ROI scans
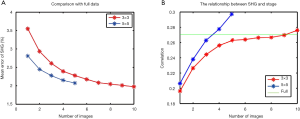
We compared the total percentage of collagen in full scans against the normalized average in small ROIs from the same sample. It was found that increasing the number of scans greatly reduced the mean error rates. Five scans of 5×5 tiles or ten scans of 3×3 tiles were sufficient to minimize the mean errors (Figure 8A). We obtained similar results in the correlation test, where the correlation coefficients between the total percentage of collagen and Metavir stage in the small ROI scans exceeded the same evaluation in the full scan which required 735 tiles on average in our experiments (Figure 8B).
Discussion
Our experiment results demonstrate that the automated removal of DC signals can greatly improve the accuracy of SHG assessment for liver fibrosis and that optimizing the sampling process using small ROIs can improve efficiency. We identified three types of DC that are likely to interfere with the quantification of samples performed by doctors (or computer algorithms):
These forms of physiological collagen exist in normal liver tissue and must be disregarded during the examination of images for signs of fibrosis. Pathologists learn through experience to disregard DC in order to correctly identify correlations between image features and Metavir stages. Our findings demonstrate that the same results can be achieved by an algorithm in an automated image analysis program. The proposed algorithm could be used to pre-process full scans in order to improve quantification result. It could also be applied to low-resolution full scans in order guide the scanning machine or its operator to avoid areas containing DC.
Full tissue scans are currently the standard for this type of assessment; however, this requires large specimen obtained via surgical biopsy and the scanning process is time consuming (up to several hours depending on resolution). The very thin liver samples obtained via needle biopsies (approximately 1 mm in thickness) are inapplicable to full tissue scanning analysis; however, we found that multiple small ROI scans (five scans of 5×5 tiles or ten scans of 3×3 tiles) could achieve correlation results on par with those of full tissue scans. Furthermore, the small area of these samples would greatly reduce the time required for scanning.
Conclusions
In conclusion, the automated removal of DC signals using small ROI scans as an alternative to full tissue scans can greatly improve the accuracy and efficiency of image acquisition and image analysis for the quantitative assessment of liver fibrosis based on SHG/TPEF microscopy.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-394). The authors have no conflicts of interest to declare.
Ethical Statement: The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional committee and written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sun W, Chang S, Tai DC, Tan N, Xiao G, Tang H, Yu H. Nonlinear optical microscopy: use of second harmonic generation and two-photon microscopy for automated quantitative liver fibrosis studies. J Biomed Opt 2008;13:064010. [Crossref] [PubMed]
- Tai DC, Tan N, Xu S, Kang CH, Chia SM, Cheng CL, Wee A, Wei CL, Raja AM, Xiao G, Chang S, Rajapakse JC, So PT, Tang HH, Chen CS, Yu H. Fibro-C-Index: comprehensive, morphology-based quantification of liver fibrosis using second harmonic generation and two-photon microscopy. J Biomed Opt 2009;14:044013. [Crossref] [PubMed]
- Kottmann RM, Sharp J, Owens K, Salzman P, Xiao GQ, Phipps RP, Sime PJ, Brown EB, Perry SW. Second harmonic generation microscopy reveals altered collagen microstructure in usual interstitial pneumonia versus healthy lung. Respir Res 2015;16:61. [Crossref] [PubMed]
- Liu F, Chen L, Rao HY, Teng X, Ren YY, Lu YQ, Zhang W, Wu N, Liu FF, Wei L. Automated evaluation of liver fibrosis in thioacetamide, carbon tetrachloride, and bile duct ligation rodent models using second-harmonic generation/two-photon excited fluorescence microscopy. Lab Invest 2017;97:84-92. [Crossref] [PubMed]
- Liu F, Zhao JM, Rao HY, Yu WM, Zhang W, Theise ND, Wee A, Wei L. Second Harmonic Generation Reveals Subtle Fibrosis Differences in Adult and Pediatric Nonalcoholic Fatty Liver Disease. Am J Clin Pathol 2017;148:502-12. [Crossref] [PubMed]
- Wang TH, Chen TC, Teng X, Liang KH, Yeh CT. Automated biphasic morphological assessment of hepatitis B-related liver fibrosis using second harmonic generation microscopy. Sci Rep 2015;5:12962. [Crossref] [PubMed]
- Xu S, Wang Y, Tai DCS, Wang S, Cheng CL, Peng Q, Yan J, Chen Y, Sun J, Liang X, Zhu Y, Rajapakse JC, Welsch RE, So PTC, Wee A, Hou J, Yu H. qFibrosis: a fully-quantitative innovative method incorporating histological features to facilitate accurate fibrosis scoring in animal model and chronic hepatitis B patients. J Hepatol 2014;61:260-9. [Crossref] [PubMed]
- The French METAVIR Cooperative Study Group. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology 1994;20:15-20. [Crossref] [PubMed]
- Westin J, Lagging LM, Wejstål R, Norkrans G, Dhillon AP. Interobserver study of liver histopathology using the Ishak score in patients with chronic hepatitis C virus infection. Liver 1999;19:183-7. [Crossref] [PubMed]
- Grønbaek K, Christensen PB, Hamilton-Dutoit S, Federspiel BH, Hage E, Jensen OJ, Vyberg M. Interobserver variation in interpretation of serial liver biopsies from patients with chronic hepatitis C. J Viral Hepat 2002;9:443-9. [Crossref] [PubMed]
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern Syst 1979;9:62-6. [Crossref]
- Lemon SC, Roy J, Clark MA, Friedmann PD, Rakowski W. Classification and regression tree analysis in public health: methodological review and comparison with logistic regression. Ann Behav Med 2003;26:172-81. [Crossref] [PubMed]
- Wang Y, Vincent R, Yang J, Asgharpour A, Liang X, Idowu MO, Contos MJ, Daitya K, Siddiqui MS, Mirshahi F, Sanyal AJ. Dual-photon microscopy-based quantitation of fibrosis-related parameters (q-FP) to model disease progression in steatohepatitis. Hepatology 2017;65:1891-903. [Crossref] [PubMed]



