
Heritability of mammographic breast density
A recent paper in the journal Cancer Research (1) has revisited the issue of the heritability of mammographic density (MD). This topic achieved a fair deal of attention between 2002–2010, with a number of twin studies showing heritability of MD of between 53–70% (Table 1), by comparing correlation in monozygous versus dizygous twins (2-5). Twin study correlations allow a distinction between shared environment and shared genetics with identical twins sharing all genetic information with dizygous twins only sharing 50% on average. As such they are the perfect cohorts to assess the ‘hereditary component’. Dense areas that show white on X-ray mammography have been associated with an increased risk of breast cancer as well as masking of underlying cancers. There is an estimated 4–5 fold greater risk of breast cancer in women within the highest breast density quintile compared with the lowest quintile (after adjustment for age, menopausal status and BMI) (6,7). Because of the major impact of MD on risk even for BRCA1/2 mutation carriers (8), MD has been successfully included in several models to assess breast cancer risk (9-13).
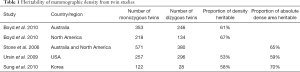
Full table
Despite the much higher heritability of MD than breast cancer itself, with only 27–30% of breast cancer being attributable to heritable factors (14,15), far less is known about the genes or genetic factors that cause this. Around 4–5% of breast cancer was thought to be due to a putative single high risk gene (16) and most of this is now accounted for by at least 13 genes that confer at least a 2-fold relative risk (17). Although the BRIDGES study (17) of 60,466 female breast cancer cases and 53,461 controls confirmed BRCA1, BRCA2 and PALB2 in the high-risk category and ATM, BARD1, CHEK2, RAD51C and RAD51D in the moderate risk category. The risk estimates for MSH6, NF1, PTEN, STK11 and TP53 were not stable mainly due to their rarity (excepting MSH6), but these syndromic genes may have been excluded and there is strong cohort data to show they confer moderate (NF1) or high risk (PTEN, STK11 and TP53) (18). Genome wide association studies have shown that 210 confirmed single nucleotide polymorphisms (SNPs) and at least an additional 103 imputed SNPs (19) now explain 37.1% of the heritable component of breast cancer. These SNPs work in a multiplicative fashion that can be used in breast cancer risk estimation as a polygenic risk score (PRS). With the Mendelian moderate and high risk genes this means that including SNPs at least 60% of the heritable component of breast cancer is explained.
In contrast, despite a genetic analysis of MD showing that the most parsimonious model was a Mendelian single major gene model in which an allele with population frequency 0.39 (95% CI, 0.33–0.46) influenced MD in an additive fashion (5). No such gene has yet been identified despite this ‘gene’ potentially explaining 66% of the residual variance. In addition, none of the known breast cancer risk genes have been linked to any direct association with MD and any increase in MD has been pretty much refuted for BRCA1 and BRCA2 (7). Indeed to date only around 30 SNPs have been associated with MD in 20 loci in the last decade (ESR1, LSP1, ZNF365, RAD51L, TBX5, 8p11.23, AREG, IGF1, PRDM6, TMEM184B, MKL1, NTN4, TAB2, 2p24.1, EBF1, MIR1972-2:FTO, 1q12.21, HABP2, INHBB and LINC01483) (19-28). These SNPs barely account for more than 5% of the heritability of MD of which 22 are independent—the others are in high linkage disequilibrium with another and probably reflect part of the same signal.
The first paper to report on heritability of MD in 2002 showed that after adjustment for age the correlation coefficient for percentage MD was 0.61 for monozygotic twin pairs in Australia, 0.67 for monozygotic pairs in North America, 0.25 for dizygotic pairs in Australia, and 0.27 for dizygotic pairs in North America (2). Heritability accounted for 60% of the variation in MD in Australian twins, 67% in North American twins. A follow up study on the same twin cohorts estimated heritability to be 65% for dense area and 66% for non-dense area (3). A Korean study showed heritability accounted for 70% for dense area, 52% for non-dense area, and 58% for percent dense area (4). Since the original work on heritability of MD digital mammography has replaced film-screen imaging, paving the way for a variety of automated approaches to density measurement in addition to subjective visual estimation. This should enable the reproducible detection of more subtle changes and quantification of image features, although methodologies may be influenced by imaging parameters. In this paper, the selection of dense area rather than percent density is likely to have reduced the impact of errors introduced by differences in patient positioning.
The current Swedish study (1) using mammographic screening history and detailed questionnaire data from 56,820 women from the KARMA prospective cohort study also estimated heritability of mammographic features. The main aim of this study was to estimate the heritability of mammographically dense area (MDA), but also previously unstudied mammographic features including microcalcifications (mean number of clusters in both breasts), masses (not defined), and MD change (cm2/year) and was estimated using 1,940 sister pairs. Heritability was estimated to be 58% (95% CI: 48–67%) for MD, 23% (95% CI: 2–45%) for microcalcifications, and 13% (95% CI: 1–25%) for masses. There was no association with MD change over time. The authors also found a positive association between breast cancer family history and PRS with MD (P<0.0001) as well as microcalcifications. The findings of a heritability component to two other mammographically measurable features (microcalcifications and masses) is novel and has important implications for assessment of future breast cancer risk. However, the heritable components of these additional risk factors are far smaller than MD. The absence of an association with another known factors associated with breast cancer risk, that of MD change association is perhaps not surprising. This is likely to be due to stochastic effects as well as non-genetic risk factors such as exposure to exogenous hormones (increase) and tamoxifen (decrease), which are known to change MD over time. There are also issues associated with the reproducibility of imaging.
The KARMA study paper is very well executed and although it contains large numbers of women does still produce data with wide confidence intervals that are close to showing no effect at the lower 95% CI for masses and calcifications. The study emphasises the complexity of using MD and other mammographic features in breast cancer risk estimation. Some of these factors may be associated with a short-term risk, whereas MD itself confers increased risk for as long as the MD is higher than for a woman of the same age. The lack of overlap between MD SNPs in a PRS and family history have implications in ensuring that these interlinked factors are not double or triple counted in studies using multiple factors for risk estimation (9,12). However, the PRS also predicts MD in the KARMA study. There is still a very long way to go to determine markers of the hereditary component of MD. At present the discovered SNPs only scratch the surface of the strong inherited component of MD confirmed in this study.
Acknowledgments
Funding:DGE, SA and AH are supported by the National Institute for Health Research (NIHR) BRC Manchester (Grant Reference Number 1215-200074). Work referred to was also supported by Prevent Breast Cancer.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-2020-20). DGE reports grants from Prevent Breast Cancer, personal fees from Astrazeneca, during the conduct of the study. EVV reports grants from Prevent Breast Cancer, during the conduct of the study. SA reports grants from National Institute for Health Research (NIHR) BRC Manchester, during the conduct of the study. The other authors have no conflicts of interest to declare.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Holowko N, Eriksson M, Kuja-Halkola R, Azam S, He W, Hall P, Czene K. Heritability of Mammographic Breast Density, Density Change, Microcalcifications, and Masses. Cancer Res 2020;80:1590-600. [Crossref] [PubMed]
- Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, Giles GG, Tritchler D, Chiarelli A, Yaffe MJ, Hopper JL. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med 2002;347:886-94. [Crossref] [PubMed]
- Stone J, Dite GS, Gunasekara A, English DR, McCredie MR, Giles GG, Cawson JN, Hegele RA, Chiarelli AM, Yaffe MJ, Boyd NF, Hopper JL. The heritability of mammographically dense and nondense breast tissue. Cancer Epidemiol Biomarkers Prev 2006;15:612-7. [Crossref] [PubMed]
- Sung J, Song YM, Stone J, Lee K, Jeong JI, Kim SS. Genetic influences on mammographic density in Korean twin and family: the Healthy Twin study. Breast Cancer Res Treat 2010;124:467-74. [Crossref] [PubMed]
- Kataoka M, Antoniou A, Warren R, Leyland J, Brown J, Audley T, Easton D. Genetic models for the familial aggregation of mammographic breast density. Cancer Epidemiol Biomarkers Prev 2009;18:1277-84. [Crossref] [PubMed]
- Santen RJ, Boyd NF, Chlebowski RT, Cummings S, Cuzick J, Dowsett M, Easton D, Forbes JF, Key T, Hankinson SE, Howell A, Ingle J. Breast Cancer Prevention Collaborative Group. Critical assessment of new risk factors for breast cancer: considerations for development of an improved risk prediction model. Endocr Relat Cancer 2007;14:169-87. [Crossref] [PubMed]
- Astley SM, Harkness EF, Sergeant JC, Warwick J, Stavrinos P, Warren R, Wilson M, Beetles U, Gadde S, Lim Y, Jain A, Bundred S, Barr N, Reece V, Brentnall AR, Cuzick J, Howell T, Evans DG. A comparison of five methods of measuring mammographic density: a case-control study. Breast Cancer Res 2018;20:10. [Crossref] [PubMed]
- Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, Mattison J, Cook M, Warsi I, Evans DG, Eccles D, Douglas F, Paterson J, Hodgson S, Izatt L, Cole T, Burgess L, Eeles R, Easton DF. Mammographic Density and Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. Cancer Res 2006;66:1866-72. [Crossref] [PubMed]
- Brentnall AR, Harkness EF, Astley SM, Donnelly LS, Stavrinos P, Sampson S, Fox L, Sergeant JC, Harvie MN, Wilson M, Beetles U, Gadde S, Lim Y, Jain A, Bundred S, Barr N, Reece V, Howell A, Cuzick J, Evans DG. Mammographic density adds accuracy to both the Tyrer-Cuzick and Gail breast cancer risk models in a prospective UK screening cohort. Breast Cancer Res 2015;17:147. [Crossref] [PubMed]
- Warwick J, Birke H, Stone J, Warren RM, Pinney E, Brentnall AR, Duffy SW, Howell A, Cuzick J. Mammographic breast density refines Tyrer-Cuzick estimates of breast cancer risk in high-risk women: findings from the placebo arm of the International Breast Cancer Intervention Study I. Breast Cancer Res 2014;16:451. [Crossref] [PubMed]
- Tice JA, Bissell MCS, Miglioretti DL, Gard CC, Rauscher GH, Dabbous FM, Kerlikowske K. Validation of the breast cancer surveillance consortium model of breast cancer risk. Breast Cancer Res Treat 2019;175:519-23. [Crossref] [PubMed]
- Lee A, Mavaddat N, Wilcox AN, Cunningham AP, Carver T, Hartley S, Babb de Villiers C, Izquierdo A, Simard J, Schmidt MK, Walter FM, Chatterjee N, Garcia-Closas M, Tischkowitz M, Pharoah P, Easton DF, Antoniou AC. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Version 2. Genet Med 2019;21:1708-18. [Crossref] [PubMed]
- Vilmun BM, Vejborg I, Lynge E, Lillholm M, Nielsen M, Nielsen MB, Carlsen JF. Impact of adding breast density to breast cancer risk models: A systematic review. Eur J Radiol 2020;127:109019. [Crossref] [PubMed]
- Claus EB, Risch N, Thompson WD. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet 1991;48:232-42. [PubMed]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A. N. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland Engl J Med 2000;343:78-84.
- Möller S, Mucci LA, Harris JR, Scheike T, Holst K, Halekoh U, Adami HO, Czene K, Christensen K, Holm NV, Pukkala E, Skytthe A, Kaprio J, Hjelmborg JB. The Heritability of Breast Cancer among Women in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev 2016;25:145-50. [Crossref] [PubMed]
- Dorling L, Carvalho S, Allen J, et al. Breast cancer risk genes: association analysis of rare coding variants in 34 genes in 60,466 cases and 53,461 controls. N Engl J Med 2020. In Press.
- Evans DG, Howell SJ, Frayling IM, Peltonen J. Gene panel testing for breast cancer should not be used to confirm syndromic gene associations. NPJ Genom Med 2018;3:32. [Crossref] [PubMed]
- Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G. at al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet 2020;52:572-81. [Crossref] [PubMed]
- Lindström S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, Brown J, Leyland J, Audley T, Wareham NJ, Loos RJ, Paterson AD, Rommens J, Waggott D, Martin LJ, Scott CG, Pankratz VS, Hankinson SE, Hazra A, Hunter DJ, Hopper JL, Southey MC, Chanock SJ, Silva Idos S, Liu J, Eriksson L, Couch FJ, Stone J, Apicella C, Czene K, Kraft P, Hall P, Easton DF, Boyd NF, Tamimi RM. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet 2011;43:185-7. [Crossref] [PubMed]
- Stevens KN, Lindstrom S, Scott CG, Thompson D, Sellers TA, Wang X, Wang A, Atkinson E, Rider DN, Eckel-Passow JE, Varghese JS, Audley T, Brown J, Leyland J, Luben RN, Warren RM, Loos RJ, Wareham NJ, Li J, Hall P, Liu J, Eriksson L, Czene K, Olson JE, Pankratz VS, Fredericksen Z, Diasio RB, Lee AM, Heit JA, DeAndrade M, Goode EL, Vierkant RA, Cunningham JM, Armasu SM, Weinshilboum R, Fridley BL, Batzler A, Ingle JN, Boyd NF, Paterson AD, Rommens J, Martin LJ, Hopper JL, Southey MC, Stone J, Apicella C, Kraft P, Hankinson SE, Hazra A, Hunter DJ, Easton DF, Couch FJ, Tamimi RM, Vachon CM. Identification of a novel percent mammographic density locus at 12q24. Hum Mol Genet 2012;21:3299-305. [Crossref] [PubMed]
- Vachon CM, Scott CG, Fasching PA, Hall P, Tamimi RM, Li J, Stone J, Apicella C, Odefrey F, Gierach GL, Jud SM, Heusinger K, Beckmann MW, Pollan M, Fernández-Navarro P, Gonzalez-Neira A, Benitez J, van Gils CH, Lokate M, Onland-Moret NC, Peeters PH, Brown J, Leyland J, Varghese JS, Easton DF, Thompson DJ, Luben RN, Warren RM, Wareham NJ, Loos RJ, Khaw KT, Ursin G, Lee E, Gayther SA, Ramus SJ, Eeles RA, Leach MO, Kwan-Lim G, Couch FJ, Giles GG, Baglietto L, Krishnan K, Southey MC, Le Marchand L, Kolonel LN, Woolcott C, Maskarinec G, Haiman CA, Walker K, Johnson N, McCormack VA, Biong M, Alnaes GI, Gram IT, Kristensen VN, Børresen-Dale AL, Lindström S, Hankinson SE, Hunter DJ, Andrulis IL, Knight JA, Boyd NF, Figuero JD, Lissowska J, Wesolowska E, Peplonska B, Bukowska A, Reszka E, Liu J, Eriksson L, Czene K, Audley T, Wu AH, Pankratz VS, Hopper JL, dos-Santos-Silva I. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol Biomarkers Prev 2012;21:1156-66. [Crossref] [PubMed]
- Lindström S, Thompson DJ, Paterson AD, Li J, Gierach GL, Scott C, Stone J, Douglas JA, dos-Santos-Silva I, Fernandez-Navarro P, Verghase J, Smith P, Brown J, Luben R, Wareham NJ, Loos RJ, Heit JA, Pankratz VS, Norman A, Goode EL, Cunningham JM, deAndrade M, Vierkant RA, Czene K, Fasching PA, Baglietto L, Southey MC, Giles GG, Shah KP, Chan HP, Helvie MA, Beck AH, Knoblauch NW, Hazra A, Hunter DJ, Kraft P, Pollan M, Figueroa JD, Couch FJ, Hopper JL, Hall P, Easton DF, Boyd NF, Vachon CM, Tamimi RM. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nat Commun 2014;5:5303. [Crossref] [PubMed]
- Brand JS, Humphreys K, Thompson DJ, Li J, Eriksson M, Hall P, Czene K. Volumetric mammographic density: heritability and association with breast cancer susceptibility loci. J Natl Cancer Inst 2014;106:dju334. [Crossref] [PubMed]
- Fernandez-Navarro P, González-Neira A, Pita G, Díaz-Uriarte R, Tais Moreno L, Ederra M, Pedraz-Pingarrón C, Sánchez-Contador C, Vázquez-Carrete JA, Moreo P, Vidal C, Salas-Trejo D, Stone J, Southey MC, Hopper JL, Pérez-Gómez B, Benitez J, Pollan M. Genome wide association study identifies a novel putative mammographic density locus at 1q12-q21. Int J Cancer 2015;136:2427-36. [Crossref] [PubMed]
- Stone J, Thompson DJ, Dos Santos Silva I, Scott C, Tamimi RM, Lindstrom S, Kraft P, Hazra A, Li J, Eriksson L, Czene K, Hall P, Jensen M, Cunningham J, Olson JE, Purrington K, Couch FJ, Brown J, Leyland J, Warren RM, Luben RN, Khaw KT, Smith P, Wareham NJ, Jud SM, Heusinger K, Beckmann MW, Douglas JA, Shah KP, Chan HP, Helvie MA, Le Marchand L, Kolonel LN, Woolcott C, Maskarinec G, Haiman C, Giles GG, Baglietto L, Krishnan K, Southey MC, Apicella C, Andrulis IL, Knight JA, Ursin G, Alnaes GI, Kristensen VN, Borresen-Dale AL, Gram IT, Bolla MK, Wang Q, Michailidou K, Dennis J, Simard J, Pharoah P, Dunning AM, Easton DF, Fasching PA, Pankratz VS, Hopper JL, Vachon CM. Novel Associations between Common Breast Cancer Susceptibility Variants and Risk-Predicting Mammographic Density Measures. Cancer Res 2015;75:2457-67. [Crossref] [PubMed]
- Brand JS, Li J, Humphreys K, Karlsson R, Eriksson M, Ivansson E, Hall P, Czene K. Identification of two novel mammographic density loci at 6Q25.1. Breast Cancer Res 2015;17:75. [Crossref] [PubMed]
- Brand JS, Humphreys K, Li J, Karlsson R, Hall P, Czene K. Common genetic variation and novel loci associated with volumetric mammographic density. Breast Cancer Res 2018;20:30. [Crossref] [PubMed]


