In vivo diffusion spectrum imaging of non-human primate brain: initial experience in transcallosal fiber examination
Introduction
Non-human primates (NHPs) show high similarity of anatomical structures, functional connections and organizations to humans and are widely used in various studies in neuroscience and pharmaceutical development (1-4). Previous MRI studies have demonstrated that diffusion tensor imaging (DTI) is a robust approach to detect the microstructural abnormality of brain white matter in various NHP models such as stroke, HIV, development, and aging (5-10).
Fiber tractography, which can be non-invasively derived with DTI, illustrates the white matter fiber bundles in brain or spinal cord, showing good consistency with those observed with injected neural tract tracers (11). Currently, DTI and fiber tractography are popularly used on human and animal researches. However, DTI-based fiber tracking is hindered to characterize crossing or touching fibers within a single image voxel (12). In contrast, diffusion spectrum imaging (DSI) allows sufficient angular separation and anisotropy by measuring its diffusion density spectra estimator or the orientation density function (ODF) based on a densely acquired q-space. Hence, it can be used to examine crossing fiber tracts within a single voxel (13,14). DSI has been explored and applied in human studies in recent years (15-19). Prior results of fixed NHP brain tissues have demonstrated DSI could differentiate the cross fibers effectively (20) and showed good agreement between the DSI-identified long association pathways and those with autoradiographic tract tracing in formalin-fixed fascicularis monkey brains (14), suggesting DSI has substantial advantage over conventional DTI approaches and can be a useful tool for examining complex neuronal fiber pathways (14). Postmortem study usually requires sacrifice of live animals for sample collection and does not provide dynamic information. In contrast, in vivo examination allows for monitoring the longitudinal evolution of interested pathways and is much more clinically relevant. To the best of our knowledge, the application of DSI technique in in vivo NHP brain studies has not been reported.
Corpus callosum is the largest white matter structure in the central nerve system. It acts as a bridge between cerebral hemispheres and provides a communicative connection between homologous cortical areas (21). As the malformation of corpus callosum (such as callosal dysgenesis) is usually associated with functional and behavioral abnormality (22,23), it is of great importance to assess the transcallosal connections between the specific cortical regions in the two hemispheres. Based upon Witelson’s classification, corpus callosum can be segmented into seven sections, in which each segment is associated with specific cortical areas (24).DTI-based tractography has shown the capacity to evaluate the abnormal fiber connections to the dorsal and medial cortex, but not the lateral callosal projections in human and macaque monkeys (3), most likely due to the lateral projections crossing through multiple fiber bundles, such as longitude fasciculus.
Current high field clinical scanners provide convenient access for NHP imaging. However, NHP brain is much smaller than human’s (1,200-1,300 cc in human vs. ~100 cc in macaque), the DSI protocols used in human studies must be optimized in order to obtain optimal images of small subjects which require much higher spatial resolution. High resolution DSI of small subjects is readily achieved in dedicated research scanners but not in general whole body clinical scanners due to the limitation of their gradient system. Parallel imaging technique has demonstrated its effectiveness to improve DTI image quality in human and macaque brain studies with clinical scanners (25-28). In the present study, we aimed to establish a DSI protocol for examining NHP brains using parallel imaging technique on a clinical 3T scanner. The feasibility was assessed and demonstrated by tracking transcallosal fibers of adult rhesus monkey brains.
Methods and materials
Animals
Three adult rhesus monkeys (female, 8-12 years old, 9-12 kg) were used. All procedures were approved and in compliance with the Institutional Animal Care and Use Committees (IACUC) of Emory University and the NIH guide for the care and use of laboratory animals. During MRI scanning, animals were anesthetized with 1.0-1.5% isoflurane mixed with 100% O2 and immobilized with a custom-made head holder in the MRI scanner. Physiological parameters such as End-tidal CO2, inhaled CO2, O2 saturation, blood pressure, heart rate, respiration rate, and body temperature were monitored continuously and maintained in the normal range (29).
MRI experiments
MRI experiments were performed on a Siemens 3T TIM Trio scanner (Siemens Medical Solutions, Inc., PA, USA). To optimize the DSI sequence, DSI images were acquired with an 8-channel volume coil (INVIVO Inc, FL, USA) and 2D stimulated single-shot EPI sequence with the following acquisition parameters: TR =2,500 ms, FOV =96 mm × 96 mm, data matrix =80×80, band width =753 Hz, 1 average, slice thickness =1.2 mm, 30 slices, b =0, 4,000 s/mm2. The generalized autocalibrating partially parallel acquisition (GRAPPA) technique was used for image reconstruction (30). Different GRAPPA acceleration factors (R =2, 3, 4) were evaluated for optimization purpose with corresponding minimal TEs =107, 92, 83 ms, respectively. In addition, DSI images without parallel imaging was acquired with the minimal TE =160 ms for comparison purpose.
Optimized DSI protocol was employed to examine the macaque brains with the following imaging parameters: TE =107 ms, TR =7,600 ms, FOV =96 mm × 96 mm, band-width =753 Hz, data matrix =80×80, slice thickness =1.2 mm, 256 diffusion directions covering full q-space 3D grid with 4 radical grid size and b value =4,000 s/mm2 and b0 images acquired with b value =0 s/mm2, 6 averages, total acquisition time =3 hours. DTI images were acquired with the same setting and a dual spin-echo 2D EPI sequence with GRAPPA (R =3) and the following imaging parameters: TE =109 ms, TR =5,500 ms, FOV =96 mm × 96 mm, band-width =1,350 Hz, data matrix =80×80, 6 averages, slice thickness 1.2 mm, 60 gradient directions with b values of 0 and 1,000 s/mm2, total acquisition time =36 minutes. Whole brain field maps were acquired using a gradient echo sequence with TE =6.24 and 8.7 ms, TR =500 ms, FOV =96 mm × 96 mm, data matrix =74×74, voxel size =1.3 mm × 1.3 mm, and slice thickness =1.3 mm. T1-weighted images were acquired using 3D MPRage sequence with GRAPPA (R =2) and the following parameters: inversion time =950 ms, TE/TR =3.5 ms/3,000 ms, voxel size =1.0 mm × 1.0 mm × 1.0 mm.
Data processing
EPI image distortion correction was performed with the acquired field maps (31), and processed off-line with the FSL package (FMRIB, Oxford, UK) (32).
In order to compare the signal to noise ratio (SNR) of DSI images, the image noise level was defined as the standard derivation (SD) of the background signal outside the brain (33). One representative slice of DSI b0 images was selected for SNR comparison. DSI image SNR was calculated with the mean value of the brain tissue signal divided by the image noise level. The DSI b0 images with different GRAPPA acceleration factors (R =2, 3, 4) and without parallel acquisition were evaluated.
For both DTI and DSI data sets, the white matter tracts were reconstructed using deterministic fiber tracking with 0.5 mm step size (20,34) and the DSI-Studio software (http://dsi-studio.labsolver.org).
Corpus callosum was segmented into seven areas (Figure 1), including rostrum, genu, rostral body, anterior midbody, posterior midbody, isthmus and splenium (24), were registered (with 12 DOF linear affine transformation) to the DSI and DTI b0 images. A macaque monkey template with isotropic 0.5 mm spatial resolution built from averaged T1-weighed images of adult macaque monkeys in the Yerkes Research Center was utilized. With the seven segments as seeds, transcallosal fiber tracking of DTI and DSI data was performed separately with the DSI-Studio software. The acquired T1-weighed images were registered (with 12 DOF linear affine transformation) to the DSI and DTI b0 images and used to overlap the DSI and DTI reconstructed fibers.

Results and discussion
In the present study, a parallel imaging-based DSI protocol was optimized for in vivo rhesus monkey brain study on a 3T clinic scanner equipped with an 8-channel phased-array volume coil and evaluated in transcallosal fiber examination.
Because DSI usually requires stronger diffusion gradient than DTI (currently b=4,000 s/mm2 in DSI vs. b=1,000 s/mm2 in DTI), the minimal TE can be increased dramatically in clinical scanners which have much lower gradient strength than dedicated animal scanners, resulting in severe susceptibility artifacts. As seen in Figure 2, the usage of paralleling imaging technique significantly reduced the minimal TE and improved the image quality. Also, the highest image SNR (in arbitrary units) was achieved with least parallel acquisition acceleration (SNR =9.3 at R =2), while the SNR was 8.5 at R =3, 7.5 at R =4, and 5.7 without parallel imaging acquisition. In comparison with the whole brain SNR, the SNR in corpus callosum was slightly lower (SNR =8.1, 7.2, 6.0, 4.5 with R =2, 3, 4 and without parallel acquisition, respectively).

As illustrated in Figure 3, DSI-derived ODF clearly shows multiple fiber orientations in the interested region of white matter bundles, including dorsal and lateral fibers in the conjunction of primary motor and somato-sensory cortices. In contrast, DTI-derived ODF only exhibits one principle fiber orientation, demonstrating that DSI has the unique capacity for imaging crossing fibers in brain tissue.

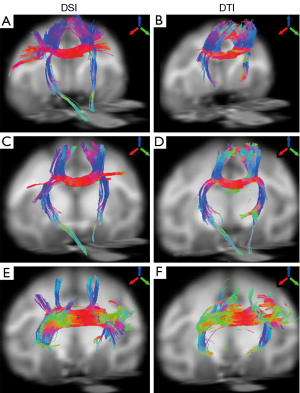
The fiber bundles passing through different segments of corpus callosum were examined with DSI and DTI. As demonstrated in Figure 4, the DTI and DSI-derived fiber bundles across rostrum, genu, rostral body, and anterior midbody of corpus callosum are revealed. Obviously, DSI-based tractography revealed more extensive fibers than those from DTI in these interested regions. In addition, long association pathways such as arcuate fasciculus/superior longitudinal fasciculus crossing transcallosal fibers from rostrum, genu and rostral body, were delineated explicitly by DSI (Figure 4A,C,E) but not by DTI.
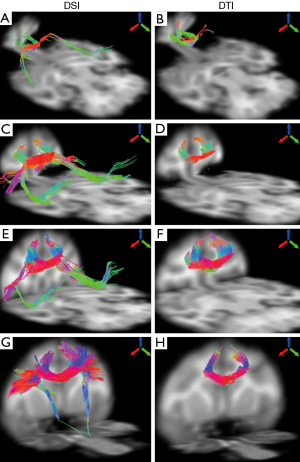
The fiber bundles across posterior midbody, isthmus, and splenium were evaluated with DSI and DTI as well. Overall, the DSI and DTI tractography results look similar in these segments (Figure 5). However, the DSI tractography from posterior midbody showed longer corticofugal fibers to the brainstem (Figure 5A), and as similar as that from anterior midbody (Figure 4G). In addition, DSI tractography showed more extensive lateral cortical fibers than DTI tractography which only revealed the tracking fibers passing through primary motor cortex (Figures 5A,B). Such differences between DSI and DTI tractography results were also observed in other fiber bundles (Figure 5C-F).
Previous results have demonstrated that DTI could partially reveal the transcallosal fiber networks in the macaque brain (3). Although the corpus callosum segmentation in the prior study was slightly different from Witelson’s (24), the transcallosal fibers across the anterior part of corpus callosum mainly comprised the cortices in dorsal orientation (3), consistent with the DTI findings in the present study. In addition, as indicated in the present study, DTI tractography failed to reveal the long association pathways (such as arcuate fasciculus/superior longitudinal fasciculus and descending corticofugal fibers).
Even though NHPs are allowed to be scanned for hours under anesthesia, the current scanning duration may still be a concern in some disease models and hence hinder the DSI’s application in translational researches. As raw image SNR is the major consideration in DSI data acquisition, a low GRAPPA acceleration factor (R =2) and low bandwidth were selected and applied in the present study. To further reduce the scanning duration but with the image SNR maintained, a better phased-array coil (with more channels and/or optimal dimensions) can be used in order to reduce the number of signal averages. Also, DSI data acquisition may be accelerated by using compressed sensing (35) or reduced-encoding DSI (36).
In addition, the DSI image quality may be further improved by using multi-shot EPI acquisition (37), spiral acquisition (38), iterative self-consistent parallel imaging reconstruction strategy (39), or steady-state radial acquisition (40).
In conclusion, a parallel imaging based DSI protocol for in vivo macaque brain imaging was implemented on a clinical setting. The preliminary results demonstrate its capacity to examine the white matter fiber bundles with crossing fibers and visualize long fiber pathways and a comprehensive map of neuronal circuits. The novel approach may allow for evaluating the integrity of fiber pathways and understanding the neural substrates in NHP models of various diseases and neuroscience researches.
Acknowledgements
This project was funded by the National Center for Research Resources P51RR000165 and currently by the Office of Research Infrastructure Programs/OD P51OD011132. The authors thank Sudeep Patel, Ruth Connelly and Doty Kempf (DVM) for animal care and experimental setup.
Disclosure: The authors declare there are no conflicts of interest.
References
- Nakahara K, Adachi Y, Osada T, et al. Exploring the neural basis of cognition: multi-modal links between human fMRI and macaque neurophysiology. Trends Cogn Sci 2007;11:84-92. [PubMed]
- Thiebaut de Schotten M, Dell’Acqua F, Valabregue R, et al. Monkey to human comparative anatomy of the frontal lobe association tracts. Cortex 2012;48:82-96. [PubMed]
- Hofer S, Merboldt KD, Tammer R, et al. Rhesus monkey and human share a similar topography of the corpus callosum as revealed by diffusion tensor MRI in vivo. Cereb Cortex 2008;18:1079-84. [PubMed]
- Jbabdi S, Lehman JF, Haber SN, et al. Human and monkey ventral prefrontal fibers use the same organizational principles to reach their targets: tracing versus tractography. J Neurosci 2013;33:3190-201. [PubMed]
- Bihel E, Pro-Sistiaga P, Letourneur A, et al. Permanent or transient chronic ischemic stroke in the non-human primate: behavioral, neuroimaging, histological, and immunohistochemical investigations. J Cereb Blood Flow Metab 2010;30:273-85. [PubMed]
- Bendlin BB, Canu E, Willette A, et al. Effects of aging and calorie restriction on white matter in rhesus macaques. Neurobiol Aging 2011;32:2319.e1-11.
- Li C, Zhang X, Komery A, et al. Longitudinal diffusion tensor imaging and perfusion MRI investigation in a macaque model of neuro-AIDS: a preliminary study. Neuroimage 2011;58:286-92. [PubMed]
- Howell BR, Godfrey J, Gutman DA, et al. Social Subordination Stress and Serotonin Transporter Polymorphisms: Associations With Brain White Matter Tract Integrity and Behavior in Juvenile Female Macaques. Cereb Cortex 2013. [Epub ahead of print]. [PubMed]
- Liu Y, D’Arceuil HE, Westmoreland S, et al. Serial diffusion tensor MRI after transient and permanent cerebral ischemia in nonhuman primates. Stroke 2007;38:138-45. [PubMed]
- Yan Y, Li L, Preuss TM, et al. In vivo evaluation of optic nerve aging in adult rhesus monkey by diffusion tensor imaging. Quant Imaging Med Surg 2014;4:43-9. [PubMed]
- Dauguet J, Peled S, Berezovskii V, et al. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage 2007;37:530-8. [PubMed]
- Wiegell MR, Larsson HB, Wedeen VJ. Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology 2000;217:897-903. [PubMed]
- Wedeen VJ, Hagmann P, Tseng WY, et al. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med 2005;54:1377-86. [PubMed]
- Schmahmann JD, Pandya DN, Wang R, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain 2007;130:630-53. [PubMed]
- Granziera C, Schmahmann JD, Hadjikhani N, et al. Diffusion spectrum imaging shows the structural basis of functional cerebellar circuits in the human cerebellum in vivo. PLoS One 2009;4:e5101. [PubMed]
- Bilgic B, Setsompop K, Cohen-Adad J, et al. Accelerated diffusion spectrum imaging with compressed sensing using adaptive dictionaries. Magn Reson Med 2012;68:1747-54. [PubMed]
- Chiu CH, Lo YC, Tang HS, et al. White matter abnormalities of fronto-striato-thalamic circuitry in obsessive-compulsive disorder: A study using diffusion spectrum imaging tractography. Psychiatry Res 2011;192:176-82. [PubMed]
- Nezamzadeh M, Wedeen VJ, Wang R, et al. In-vivo investigation of the human cingulum bundle using the optimization of MR diffusion spectrum imaging. Eur J Radiol 2010;75:e29-36. [PubMed]
- Kuo LW, Chen JH, Wedeen VJ, et al. Optimization of diffusion spectrum imaging and q-ball imaging on clinical MRI system. Neuroimage 2008;41:7-18. [PubMed]
- Wedeen VJ, Wang RP, Schmahmann JD, et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage 2008;41:1267-77. [PubMed]
- Bloom JS, Hynd GW. The role of the corpus callosum in interhemispheric transfer of information: excitation or inhibition? Neuropsychol Rev 2005;15:59-71. [PubMed]
- Paul LK. Developmental malformation of the corpus callosum: a review of typical callosal development and examples of developmental disorders with callosal involvement. J Neurodev Disord 2011;3:3-27. [PubMed]
- Lee SK, Mori S, Kim DJ, et al. Diffusion tensor MR imaging visualizes the altered hemispheric fiber connection in callosal dysgenesis. AJNR Am J Neuroradiol 2004;25:25-8. [PubMed]
- Witelson SF. Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain 1989;112:799-835. [PubMed]
- Bhagat YA, Emery DJ, Naik S, et al. Comparison of generalized autocalibrating partially parallel acquisitions and modified sensitivity encoding for diffusion tensor imaging. AJNR Am J Neuroradiol 2007;28:293-8. [PubMed]
- Ardekani S, Selva L, Sayre J, et al. Quantitative metrics for evaluating parallel acquisition techniques in diffusion tensor imaging at 3 Tesla. Invest Radiol 2006;41:806-14. [PubMed]
- Zhang X, Tong F, Li CX, et al. A fast multiparameter MRI approach for acute stroke assessment on a 3T clinical scanner: preliminary results in a non-human primate model with transient ischemic occlusion. Quant Imaging Med Surg 2014;4:112-22.
- Yan Y, Nair G, Li L, et al. In vivo evaluation of optic nerve development in non-human primates by using diffusion tensor imaging. Int J Dev Neurosci 2014;32:64-8. [PubMed]
- Li CX, Patel S, Auerbach EJ, et al. Dose-dependent effect of isoflurane on regional cerebral blood flow in anesthetized macaque monkeys. Neurosci Lett 2013;541:58-62. [PubMed]
- Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202-10. [PubMed]
- Jenkinson M. Improving the registration of B0-disorted EPI images using calculated cost function weights. 10th International Conference on Functional Mapping of the Human Brain, 2004.
- Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage 2012;62:782-90. [PubMed]
- Parrish TB, Gitelman DR, LaBar KS, et al. Impact of signal-to-noise on functional MRI. Magn Reson Med 2000;44:925-32. [PubMed]
- Yeh FC, Verstynen TD, Wang Y, et al. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One 2013;8:e80713. [PubMed]
- Menzel MI, Tan ET, Khare K, et al. Accelerated diffusion spectrum imaging in the human brain using compressed sensing. Magn Reson Med 2011;66:1226-33. [PubMed]
- Yeh CH, Cho KH, Lin HC, et al. Reduced encoding diffusion spectrum imaging implemented with a bi-Gaussian model. IEEE Trans Med Imaging 2008;27:1415-24. [PubMed]
- Heidemann RM, Porter DA, Anwander A, et al. Diffusion imaging in humans at 7T using readout-segmented EPI and GRAPPA. Magn Reson Med 2010;64:9-14. [PubMed]
- Heidemann RM, Griswold MA, Seiberlich N, et al. Direct parallel image reconstructions for spiral trajectories using GRAPPA. Magn Reson Med 2006;56:317-26. [PubMed]
- Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med 2010;64:457-71. [PubMed]
- Jung Y, Samsonov AA, Block WF, et al. 3D diffusion tensor MRI with isotropic resolution using a steady-state radial acquisition. J Magn Reson Imaging 2009;29:1175-84. [PubMed]


