BIGDOSE: software for 3D personalized targeted radionuclide therapy dosimetry
Introduction
With the growing number of applications in targeted radionuclide therapy (TRT), patient-specific internal dosimetry is increasingly important to ensure effective treatment (1). Moreover, assessment of 3D absorbed dose distribution is of high clinical value as low dose regions might lead to potential lesion recurrence while high dose regions could cause necrosis in tissues. While for normal organs, different sub-compartments might have different tolerance to radiation and dose distribution could be non-uniform, resulting in different level of toxicity (2). To obtain the 3D absorbed dose information for treatment planning or dose verification, one can measure the 3D cumulative activity distribution of the therapeutic agent in patients non-invasively by quantitative emission computed tomography (ECT), i.e., single photon emission computed tomography (SPECT) and positron emission tomography (PET) and combined with blood sampling if necessary (3). However, the accuracy for individualized organ dose assessment is still limited by the uncertainties in cumulative activity quantification due to calibration, insufficient or inaccurate physic modeling in the imaging process, segmentation of volume-of-interest (VOI) and dose conversion (4). Since serial scans are usually acquired to obtain the time activity curve (TAC), misalignments among images at different time point is also a main source of errors for dose estimation (5). The use of computed tomography (CT) or magnetic resonance imaging (MRI) has been proposed to improve image registration and segmentation (6). For dose conversion, dose-point kernel (DPK) convolution (7), voxel S value (VSV) convolution (8) or Monte Carlo simulation (MCS) based method (9) are developed to convert the voxelized activity/cumulative activity to the 3D dose rate/absorbed dose images.
Generally, 3D internal dosimetric software dosimetry tools involve steps of optional image registration (rigid or non-rigid, manual or automatic registration), VOIs segmentation, curve fitting for generating TAC and dose conversion, given the input of patient imaging data. While early voxelized dosimetry software does not contain all four modules mentioned above and would need to be used together with other image processing or dosimetric software (10-12), more one-stop research and commercial dosimetric software is developed recently, such as STRATOS® (13), PLANET® (14), HERMES® (15), VoxelMed (16), VIDA (17), RAYDOSE (18), VRAK (19), OEDIPE (20) and JADA (21). A review of the recent codes for internal dosimetry can be found from the work from Ramos et al. (22). Most of the developed dosimetric software only provides manual or rigid image registration which might not be adequate especially for small organs or lesions (23).
However, non-rigid registration based on whole body images is relatively computational intensive. Previously, we proposed that organ-based non-rigid registration on sequential quantitative SPECT or CT images can improve the dosimetric estimation (24,25). On the other hand, we also developed a virtual CT (vCT) method to generate CT at different time points when only a single CT is available in the sequential imaging protocol to sustain the dose calculation accuracy (26). In this paper, we aim to develop a 3D voxelized internal dosimetry software, BIGDOSE v1.0, with new features of organ-based image registration and vCT. We evaluated BIGDOSE v1.0 dose estimation result with OLINDA/EXM v1.1 in a simulation study with ground truth, and demonstrated the effectiveness of this software in two patient studies.
Methods
Software flowchart
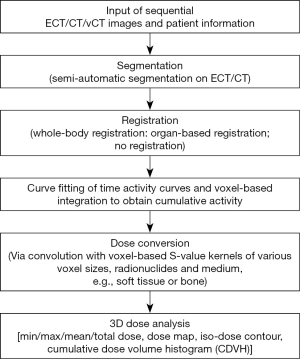
BIGDOSE includes a portable wizard based graphical user interface (GUI) written in Python. It consists of six modules as demonstrated in Figure 1: (I) input of sequential ECT/CT/vCT images; (II) ECT- or CT-based segmentation; (III) whole-body or organ-based, ECT or CT registration; (IV) curve fitting of TACs and voxel-based integration to obtain cumulative activity; (V) dose conversion via convolution with VSV kernels and (VI) 3D dose analysis.
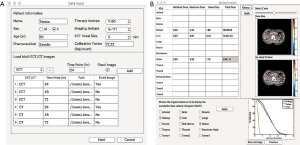
The patient and imaging information, i.e., imaging isotope, therapeutic isotope, therapeutic pharmaceutical and system calibration factor (Bq/count) are first input to the software for a study. The co-registered ECT/CT/vCT data acquired at multiple time points after activity administration are then loaded. The vCT method, which required only a single CT acquisition and vCTs at other time point could be generate by non-rigid image registration, provides comparable registration accuracy of sequential CT scans (27). The vCTs could be used for the attenuation correction, scatter correction and organ segmentation especially for organs with low uptake on SPECT images, while reducing the radiation dose as compared to repeated CT scans. The vCT option was implemented as an add-on plugin for BIGDOSE. The user then selected a reference image as the fixed time point for registration and segmentation (Figure 2A). The 3D activity data are corrected, voxel by voxel, for the physical half live difference between the surrogate imaging isotope and therapeutic isotope according to the following equation (28):
|
| [1] |
In this software, three registration modes are provided, including organ-based, whole-body based registration and no registration. For organ-based registration, all organs-of-interests are segmented out from either ECT or CT data at each time point semi-automatically using an open source program, ITK-SNAP (29). For whole-body based registration, the organs-of-interest are only segmented on the reference image. The non-rigid affine + b-spline image registration was implemented based on an open source program “Elastix” (30), applying mutual information (MI) as the similarity measurement. For the CT registration, the acquired motion vectors would later be applied to register the corresponding ECT images. No registration mode is available for studies whose sequential ECT or CT images are already aligned or for the application of Y-90 microspheres with only one time point ECT/CT acquisition.
For studies with more than three imaging sessions, a voxel-by-voxel numerical integration based on the trapezoidal rule is used to obtain the cumulative activity images, assuming only physical decay after the last time point. While for studies with three imaging sessions, a bi-exponential function was used to fit the TACs for each organ on the registered images before integration to obtain the cumulative activity. For Y-90 microsphere cases with only 1-time point images, cumulative activities were calculated based on a mono-exponential model.
The obtained cumulative activity image is then convoluted with a corresponding VSV kernel for various voxel sizes and tissues to generate 3D absorbed dose distribution images.
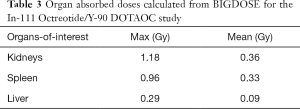
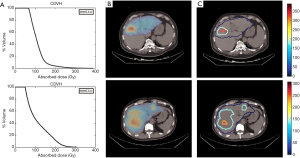
The 3D dose analysis includes organ absorbed dose information (max/mean), dose map, iso-dose contour and cumulative dose volume histogram (CDVH) for the organs-of-interest (Figure 2B).
Dosimetric evaluation
Simulation study
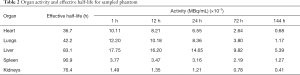
We used a population of nine digital 4D extended cardiac-torso (XCAT) (31) phantoms which model detailed anatomical structures and physiological functions for three male anatomies (Table 1), each with three In-111 Zevalin distributions and biokinetics (32) (Table 2) which served as the imaging surrogate of Y-90 Zevalin. The time-varying activity distributions of each target organ were used to simulate SPECT scans acquired at 1, 12, 24, 72, and 144 hours post-injection. The deformation for organs-of-interest between each scan was modeled by translation and rotation randomly within 5 pixels/degrees while keeping the volume change within 5%. The whole torso rigid transformation was modeled within 5 pixels or degrees of translation or rotation randomly to mimic the whole body movement between each scan (33). The attenuation maps for attenuation modeling and correction in reconstruction were generated along with the corresponding XCAT phantoms at an effective energy of In-111 to serve as the CT images (32).

Full table
Full table
An analytical projector modeling attenuation, scatter, and geometric collimator detector response (GCDR) (34) was used to simulate a standard dual head clinical SPECT/CT scanner (Discovery VH Hawkeye, GE Healthcare) with a crystal thickness of 2.54 cm mounted with MEGP collimators. The scatter modeling and correction were achieved by the effective source scatter estimation (ESSE) method based on the scattering schemes of In-111 (35). We simulated 128 projections with x-y dimension of 128×170 and 30 s/projection over 360°, using an energy window with a 14% width centered at two photopeaks of 171 and 245 keV. A system calibration factor of 1.43×10−4 cts·s−1·Bq−1 was used to scale the noise-free projections to a clinical count level and then modeled with Poisson noise to obtain realistic noisy projections (36). These noisy projections were then reconstructed using OS-EM algorithm with 8 iterations and 16 subsets, incorporating attenuation, scatter and detector-collimator response compensations. No post-filtering was applied to the reconstructed images.
BIGDOSE v1.0 vs. OLINDA/EXM v1.1
For each phantom, five sets of simulated SPECT/CT images were input in BIGDOSE. Our previous study showed that the CT organ-based registration was superior to other registration schemes (25) and was used in this evaluation study for organs-of-interest of liver, spleen, kidneys and lungs. The trapezoidal rule was applied to calculate the cumulative activity followed by the Y-90 VSV kernel convolution.
OLINDA/EXM v1.1 is a conventional MIRD-based dosimetry software with weight adjustment which calculates absorbed dose on organ level and the user would need to input organ activities at different time points (37). We used the same VOIs as defined in BIGDOSE to map out the target organs and obtain their activities on serial SPECT/CT images. The TAC of each organ was fit based on the mono-exponential model to estimate the dose.
GATE dosimetry
In this study, we calculated the absorbed dose of Y-90 for phantoms without any misalignment in a MCS code (GATE v6.1) with physics modelling of photoelectric absorption, Compton interaction, Rayleigh scattering, ionizations, multiple scatter and bremsstrahlung photons, to serve as the gold standard (38). The electron energy spectrum was acquired from the Brookhaven National Laboratory database (39). The density maps of the target organs were input to the program separately and the S-values for Y-90 beta particles were output and stored in the unit of Gy∙MBq−1∙s−1for each voxel. A total of 2×107 photon histories were traced to keep the statistical uncertainty to be <10% in each voxel. The cumulative activity for each target organ was calculated by integration along the TAC with known effective half-lives. We assumed beta particles were locally absorbed inside each organ owing to their short ranges, thus absorbed doses for target organs were originated from their own and were obtained from multiplying their cumulative activity by the corresponding S values output from GATE (40).
We calculated the total absorbed dose error of BIGDOSE vs. OLINDA/EXM for different organs-of-interests as compared to GATE:
|
| [2] |
An independent t-test analysis was performed with MATLAB to compare results of different software. A P value of <0.05 was considered to be statistically significant.
Clinical study
In-111 octreotide
To evaluate the clinical feasibility of BIGDOSE, a female patient (age: 73; weight: 82 kg) with known history of neuroendocrine tumors was enrolled. A written informed consent was obtained following the guidelines of the local ethics board. Three-time point In-111 Octreo SPECT/CT scans were obtained using a clinical SPECT/CT scanner (Bright View XCT, Philips Healthcare) at 24, 48, and 72 hours post-injection of 222 MBq In-111 DTPAOC for Y-90 DOTAOC dosimetry. Sixty-four projections were acquired over 360˚ with 30 s/projection for each scan. Energy windows of 171 keV ±10% and 245 keV ±10% were used for acquisition, while 140 keV ±10% was used for scatter correction. The CT scanning parameters were 120 kV, 20 mA and slice thickness of 1 mm. The reconstructed CT matrix size was 512×512×406, with a voxel size of 1×1×1 mm3. A CT-based attenuation correction and dual energy window scatter correction were used in the SPECT OS-EM reconstruction with a voxel size of 6.4×6.4×6.4 mm3. Target organs, i.e., liver, kidneys and spleen, were segmented out from the CT images at all time points in BIGDOSE with organ-based registration for dose analysis.
Y-90 microsphere
We recruited 2 patients with Y-90 microspheres embolization under the local ethics approval. Patient#1 was treated with glass microspheres (TheraSphere®, MDS Nordion, Ottawa, Canada) while Patient#2 was treated with resin microspheres (SIR-Sphere®, Sirtex Medical, Sydney, Australia). They underwent one time point bremsstrahlung/CT scan using a dual head SPECT/CT scanner (Discovery NM/CT 670, GE Healthcare) 4 hrs post embolization. Projection data were acquired over 360˚ with 30 s/projection. Three 10% wide energy windows centered at 70, 135, 167 keV were used to collect the bremsstrahlung photons. The CT scanning parameters were 120 kV, 42 mA, helical mode, 0.938:1 pitch, scan rotation speed of 18.75 (mm/rot) and slice thickness of 3.75 mm. The reconstructed CT matrix size was 512×512×73, with a voxel size of 1×1×3.75 mm3. A CT-based attenuation correction was used in the SPECT OS-EM reconstruction with a voxel size of 4.42×4.42×4.42 mm3. Additionally, contrast CT images were performed for each patient with a voxel size of 0.72×0.72×5 mm3. No registration mode was chosen and we assumed physical decay after drug delivery. After calculating the cumulative activity of SPECT images, a Y-90 dose kernel with a voxel size of 4.42 mm was employed to convolve with them to achieve dose conversion. The VOIs of healthy liver and tumor were segmented on SPECT images by setting an activity threshold such that the volumes of the VOIs were equal to those of the corresponding contrast CT images for each patient (41).
Results
Simulation study
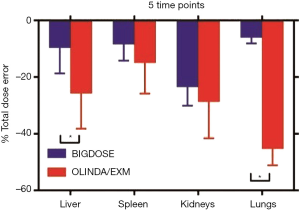
When compared with OLINDA/EXM, large improvement could be observed in absorbed dose estimation in target organs especially for lungs. The average absorbed dose errors of BIGDOSE for the nine phantoms were −9.59%±9.06%, −8.36%±5.82%, −23.41%±6.67%, −6.05%±2.06% for liver, spleen, kidneys and lungs, while they were −25.72%±12.52%, −14.93%±10.91%, −28.63%±12.97% and −45.30%±5.84% for OLINDA/EXM (Figure 3). The differences are statistically significant for the liver and lungs.
Clinical study
In-111 octreotide
Absorbed doses of kidneys, spleen and liver calculated from BIGDOSE are listed in Table 3, while the CDVHs, dose maps and iso-dose contours provide their 3D dose distribution information (Figure 4). The total computational time for 3 organs-of-interest was about 3 hours on a Macintosh Operating System, with a 2.6 GHz Intel Core i5 processor and 8 GB RAM.
Full table
Y-90 microsphere
Healthy liver and tumor absorbed doses for different patients from BIGDOSE are listed in Table 4. Sampled CDVH, dose map and iso-dose contour of liver for Patient #1 and Patient #2 are shown in Figure 5.
Full table
Discussion
While OLINDA/EXM is still the main stream organ-based dosimetric software in the clinic, the increased popularity of Y-90 and Lu-177-based TRT raises the demand of 3D voxel-based personalized dosimetry. A new feature for BIGDOSE is the implementation of organ-based registration, with flexible number of input images. Our previous study showed that organ-based registration generally performed better than whole-body based registration (25), and can reduce the dosimetric error of up to 25% as compared to no registration performed. However, organ-based registration is relatively operational intensive as segmentations need to be performed for multiple time point images and organs-of-interest. Both organ-based and whole-body registrations are available in BIGDOSE.
Monte Carlo based techniques provide the highest accuracy for internal dose estimates yet they are hampered by the long computational time for clinical practice. A previous study showed that the 3D dose distributions produced by MCS and VSV are nearly identical while the processing time for VSV is more clinically feasible (42). In this study, the more pronounced difference between BIGDOSE and OLINDA/EXM could be due to several factors. The use of different curve fitting methods may lead to a discrepancy in cumulative activities. OLINDA/EXM uses organ-level S values based on fixed phantoms, while the average organ S values varies for each phantom based on its own geometry for BIGDOSE. OLANDA/EXM also assumes the cumulative activities and organ absorbed doses are uniformly distributed.
Final absorbed dose errors are attributed to many aspects including activity quantification, organ segmentation, image registration, curve fitting of TAC and dose conversion. In this study, we preliminarily investigated the errors after quantitative image reconstruction, image registration, cumulative activity calculation and the dose conversion on organ absorbed doses. We found that the quantitative reconstruction (quantitative SPECT vs. original phantom) affected most on the absorbed dose calculation, while the residual errors from registration, trapezoidal integration of the TAC and dose conversion were similar and had relatively smaller effects (data not shown). Figure 3 showed that the absorbed dose error is higher for low uptake organs, i.e., kidneys and lungs, possibly due to the “spill-in” effects from the adjacent high uptake organs and background especially in the later time points during the quantitative image reconstruction.
In the In-111 Octreotide/Y-90 DOTAOC study, the calculated dose range from BIGDOSE is similar to the reported dose (43). In the Y-90 microsphere clinical study, the healthy liver absorbed dose of Patient #1 with glass microspheres is 102.37 Gy which is less than maximum dose limit of 120 Gy for clinical studies with intensive dosage (44-46). The tumor absorbed dose of Patient #1 (196.76 Gy) is larger than 120 Gy as recommended by the guideline, considering to be an effective treatment (47). For Patient #2 using resin microspheres, the healthy liver absorbed dose (66.87 Gy) is smaller than the suggested dose limit of 80 Gy while the tumor absorbed dose (165.24/120.43 Gy) is larger than 120 Gy as proposed by SIR-Sphere manual.
There are certain limitations for BIGDOSE. For example, it only considers absorbed dose for beta particle and assuming absorbed dose contributions from organs other than the target organs are negligible. Although TRT mostly relies on short-ranged radiation like beta, alpha particles or Auger electrons to kill the cancer cells, the contribution of long-ranged gamma photons is not negligible for some radionuclide, e.g., Lu-177, whose gamma photons account for about 13% of the total absorbed dose (48). Combined weighted absorbed dose with beta particles and photons would be included to address this problem (19). The S value for gamma photons could be computed as a long-ranged low-resolution volumetric dose kernel. The absorbed dose from gamma photons and beta particles could be obtained by convoluting the cumulative activity images with two different kernels separately, and then normalizing the 2 dose images into the same voxel size before summing them up as the final absorbed dose image. On the other hand, for our current organ-based registration, the marginal absorbed dose from adjacent organs is not considered, e.g., dose attributed from liver to lungs and vice versa. One possible solution is to combine the cumulative activity images of all critical organs as one single cumulative image before the VSV convolution and it requires extra computational processing time, or to select whole-body registration mode which would require more registration time but reduce the number of segmentation required. Besides, ECT and CT images are co-registered in the simulation study which might not be clinically realistic. In the future, we would provide image registration option between ECT and CT images. More patient data with different tracers and applications are warranted to demonstrate the effectiveness of BIGDOSE.
Conclusions
BIGDOSE provides a one-stop platform for voxel-based dose estimation, which can substantially alleviate the activity inhomogeneity problem in dose calculation, with enhanced function of non-rigid registration and vCT options. It is a promising tool to streamline the current clinical TRT dosimetric practice for treatment planning and post-therapy dose verification, with high accuracy and 3D personalized information for potential improved treatment outcome.
Acknowledgments
Funding: This work was supported by research grants from Macau Science and Technology Development Fund (114/2016/A3) and University of Macau (MYRG2017-00060-FST).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: A written informed consent was obtained following the guidelines of the local ethics board.
References
- Li T, Ao ECI, Lambert B, Brans B, Vandenberghe S, Mok GSP. Quantitative imaging for targeted radionuclide therapy dosimetry - Technical review. Theranostics 2017;7:4551-65. [Crossref] [PubMed]
- Loke KS, Padhy AK, Ng DC, Goh AS, Divgi C. Dosimetric considerations in radioimmunotherapy and systemic radionuclide therapies: a review. World J Nucl Med 2011;10:122-38. [Crossref] [PubMed]
- Sgouros G, Roeske JC, McDevitt MR, Palm S, Allen BJ, Fisher DR, Brill AB, Song H, Howell RW, Akabani G, Committee SM, Bolch WE, Brill AB, Fisher DR, Howell RW, Meredith RF, Sgouros G, Wessels BW, Zanzonico PB. MIRD Pamphlet No. 22 (abridged): radiobiology and dosimetry of alpha-particle emitters for targeted radionuclide therapy. J Nucl Med 2010;51:311-28. [Crossref] [PubMed]
- Gustafsson J, Brolin G, Cox M, Ljungberg M, Johansson L, Gleisner KS. Uncertainty propagation for SPECT/CT-based renal dosimetry in (177)Lu peptide receptor radionuclide therapy. Phys Med Biol 2015;60:8329-46. [Crossref] [PubMed]
- Papavasileiou P, Divoli A, Hatziioannou K, Flux GD. The importance of the accuracy of image registration of SPECT images for 3D targeted radionuclide therapy dosimetry. Phys Med Biol 2007;52:N539-48.
- Schillaci O, Simonetti G. Fusion imaging in nuclear medicine—applications of dual-modality systems in oncology. Cancer Biotherapy and Radiopharmaceuticals 2004;19:1-10. [Crossref] [PubMed]
- Giap HB, Macey DJ, Bayouth JE, Boyer AL. Validation of a dose-point kernel convolution technique for internal dosimetry. Phys Med Biol 1995;40:365-81. [Crossref] [PubMed]
- Liu A, Williams LE, Wong JY, Raubitschek AA. Monte Carlo-assisted voxel source kernel method (MAVSK) for internal beta dosimetry. Nucl Med Biol 1998;25:423-33. [Crossref] [PubMed]
- Furhang EE, Chui CS, Sgouros G. A Monte Carlo approach to patient-specific dosimetry. Med Phys 1996;23:1523-9. [Crossref] [PubMed]
- Tagesson M, Ljungberg M, Strand SE. A Monte-Carlo program converting activity distributions to absorbed dose distributions in a radionuclide treatment planning system. Acta Oncol 1996;35:367-72. [Crossref] [PubMed]
- Prideaux AR, Song H, Hobbs RF, He B, Frey EC, Ladenson PW, Wahl RL, Sgouros G. Three-dimensional radiobiologic dosimetry: Application of radiobiologic modeling to patient-specific 3-dimensional imaging–based internal dosimetry. J Nucl Med 2007;48:1008-16. [Crossref] [PubMed]
- Dewaraja YK, Wilderman SJ, Ljungberg M, Koral KF, Zasadny K, Kaminiski MS. Accurate dosimetry in 131I radionuclide therapy using patient-specific, 3-dimensional methods for SPECT reconstruction and absorbed dose calculation. J Nucl Med 2005;46:840-9. [PubMed]
- Berker Y, Goedicke A, Kemerink GJ, Aach T, Schweizer B. Activity quantification combining conjugate-view planar scintigraphies and SPECT/CT data for patient-specific 3-D dosimetry in radionuclide therapy. Eur J Nucl Med Mol Imaging 2011;38:2173-85. [Crossref] [PubMed]
- Dieudonné A, Aouf A, Sabet A, Meyer C, Simon B, Haslerud T, Biersack H, Sanchez-Garcia M, Lebtahi R, Ezziddin S. Correlation between average absorbed dose and metabolic response in the radio-embolization of liver colorectal metastases with resin microspheres. J Nucl Med 2015;56:1237-7.
- Hippeläinen ET, Tenhunen MJ, Maenpaa HO, Heikkonen JJ, Sohlberg AO. Dosimetry software Hermes Internal Radiation Dosimetry: from quantitative image reconstruction to voxel-level absorbed dose distribution. Nucl Med Commun 2017;38:357-65. [Crossref] [PubMed]
- Grassi E, Fioroni F, Ferri V, Mezzenga E, Sarti MA, Paulus T, Lanconelli N, Filice A, Versari A, Iori M. Quantitative comparison between the commercial software STRATOS((R)) by Philips and a homemade software for voxel-dosimetry in radiopeptide therapy. Phys Med 2015;31:72-9. [Crossref] [PubMed]
- Kost SD, Dewaraja YK, Abramson RG, Stabin MG. VIDA: a voxel-based dosimetry method for targeted radionuclide therapy using Geant4. Cancer Biother Radiopharm 2015;30:16-26. [Crossref] [PubMed]
- Marcatili S, Pettinato C, Daniels S, Lewis G, Edwards P, Fanti S, Spezi E. Development and validation of RAYDOSE: a Geant4-based application for molecular radiotherapy. Phys Med Biol 2013;58:2491-508. [Crossref] [PubMed]
- Jackson PA, Beauregard JM, Hofman MS, Kron T, Hogg A, Hicks RJ. An automated voxelized dosimetry tool for radionuclide therapy based on serial quantitative SPECT/CT imaging. Med Phys 2013;40:112503. [Crossref] [PubMed]
- Petitguillaume A, Bernardini M, Broggio D, de Labriolle Vaylet C, Franck D, Desbrée A. OEDIPE, a software for personalized Monte Carlo dosimetry and treatment planning optimization in nuclear medicine: absorbed dose and biologically effective dose considerations. Radioprotection 2014;49:275-81. [Crossref]
- Grimes J, Uribe C, Celler A. JADA: a graphical user interface for comprehensive internal dose assessment in nuclear medicine. Med Phys 2013;40:072501. [Crossref] [PubMed]
- Ramos SMO, Thomas S, Pinheiro MA, Coelho A, Albernaz M. Internal radiation dose and modeling codes in nuclear medicine: A fresh look at old problems. Int J Radiol Radiat Ther 2017;4:439-43.
- Kadoya N. Use of deformable image registration for radiotherapy applications. J Radiol Radiat Ther 2014;2:1042.
- Ao EC, Wu NY, Wang SJ, Song N, Mok GS. Improved dosimetry for targeted radionuclide therapy using nonrigid registration on sequential SPECT images. Med Phys 2015;42:1060-70. [Crossref] [PubMed]
- Li T, Wu NY, Song N, Mok GSP. Evaluation of sequential SPECT and CT for targeted radionuclide therapy dosimetry. Ann Nucl Med 2018;32:34-43. [Crossref] [PubMed]
- Li T, Mok GSP. Technical Note: Virtual CT for reducing CT dose in targeted radionuclide therapy dosimetry. Med Phys 2018;45:5138-44. [Crossref] [PubMed]
- Mok G, Li T. High performance virtual CT for enhanced targeted radionuclide therapy dosimetry. J Nucl Med 2017;58:1303-3.
- Plyku D, Hobbs RF, Huang K, Atkins F, Garcia C, Sgouros G, Van Nostrand D. Recombinant human thyroid-stimulating hormone versus thyroid hormone withdrawal in 124I PET/CT–based dosimetry for 131I therapy of metastatic differentiated thyroid cancer. J Nucl Med 2017;58:1146-54. [Crossref] [PubMed]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31:1116-28. [Crossref] [PubMed]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BM. 4D XCAT phantom for multimodality imaging research. Med Phys 2010;37:4902-15. [Crossref] [PubMed]
- He B, Du Y, Segars WP, Wahl RL, Sgouros G, Jacene H, Frey EC. Evaluation of quantitative imaging methods for organ activity and residence time estimation using a population of phantoms having realistic variations in anatomy and uptake. Med Phys 2009;36:612-9. [Crossref] [PubMed]
- Song N, He B, Frey EC. The effect of volume-of-interest misregistration on quantitative planar activity and dose estimation. Phys Med Biol 2010;55:5483-97. [Crossref] [PubMed]
- Frey E, Ju ZW, Tsui B. A fast projector-backprojector pair modeling the asymmetric, spatially varying scatter response function for scatter compensation in SPECT imaging. IEEE Trans Nucl Sci 1993;40:1192-7. [Crossref]
- Frey EC, Tsui BMW. A new method for modeling the spatially-variant, object-dependent scatter response function in SPECT. In1996 IEEE Nuclear Science Symposium Conference Record 1996;2:1082-6.
- He B, Frey EC. Comparison of conventional, model-based quantitative planar, and quantitative SPECT image processing methods for organ activity estimation using In-111 agents. Phys Med Biol 2006;51:3967-81. [Crossref] [PubMed]
- Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med 2005;46:1023-7. [PubMed]
- Villoing D, Marcatili S, Garcia MP, Bardies M. Internal dosimetry with the Monte Carlo code GATE: validation using the ICRP/ICRU female reference computational model. Phys Med Biol 2017;62:1885-904. [Crossref] [PubMed]
- Stabin MG, da Luz LC. Decay data for internal and external dose assessment. Health Phys 2002;83:471-5. [Crossref] [PubMed]
- Bolch WE, Eckerman KF, Sgouros G, Thomas SR. MIRD pamphlet No. 21: a generalized schema for radiopharmaceutical dosimetry--standardization of nomenclature. J Nucl Med 2009;50:477-84. [Crossref] [PubMed]
- Bastiaannet R, Viergever MA, de Jong H. Impact of respiratory motion and acquisition settings on SPECT liver dosimetry for radioembolization. Med Phys 2017;44:5270-79. [Crossref] [PubMed]
- Grimes J, Celler A. Comparison of internal dose estimates obtained using organ‐level, voxel S value, and Monte Carlo techniques. Med Phys 2014;41:092501. [Crossref] [PubMed]
- Förster GJ, Engelbach MJ, Brockmann JJ, Reber HJ, Buchholz HG, Macke HR, Rosch FR, Herzog HR, Bartenstein PR. Preliminary data on biodistribution and dosimetry for therapy planning of somatostatin receptor positive tumours: comparison of (86)Y-DOTATOC and (111)In-DTPA-octreotide. Eur J Nucl Med 2001;28:1743-50. [Crossref] [PubMed]
- Garin E, Lenoir L, Rolland Y, Edeline J, Mesbah H, Laffont S, Poree P, Clement B, Raoul JL, Boucher E. Dosimetry based on 99mTc-macroaggregated albumin SPECT/CT accurately predicts tumor response and survival in hepatocellular carcinoma patients treated with 90Y-loaded glass microspheres: preliminary results. J Nucl Med 2012;53:255-63. [Crossref] [PubMed]
- Garin E, Lenoir L, Edeline J, Laffont S, Mesbah H, Poree P, Sulpice L, Boudjema K, Mesbah M, Guillygomarc'h A, Quehen E, Pracht M, Raoul JL, Clement B, Rolland Y, Boucher E. Boosted selective internal radiation therapy with 90Y-loaded glass microspheres (B-SIRT) for hepatocellular carcinoma patients: a new personalized promising concept. Eur J Nucl Med Mol Imaging 2013;40:1057-68. [Crossref] [PubMed]
- Srinivas SM, Natarajan N, Kuroiwa J, Gallagher S, Nasr E, Shah SN, DiFilippo FP, Obuchowski N, Bazerbashi B, Yu N. Determination of radiation absorbed dose to primary liver tumors and normal liver tissue using post-radioembolization 90Y PET. Front Oncol 2014;4:255. [Crossref] [PubMed]
- Salem R, Thurston KG. Radioembolization with 90Yttrium microspheres: a state-of-the-art brachytherapy treatment for primary and secondary liver malignancies: part 1: technical and methodologic considerations. J Vasc Interv Radiol 2006;17:1251-78. [Crossref] [PubMed]
- Bé M, Chisté V, Dulieu C, Browne E, Baglin C. Monographie BIPM-5—Table of Radionuclides (Comments on Evaluation). Bureau International Des Poids et Mesures, Sèvres 2010;7.