Virtual museum of congenital heart defects: digitization and establishment of a database for cardiac specimens
Introduction
Over the centuries, specimens removed from the corpse were the ultimate models that anatomy was studied upon. Sir William Withey Gull (from 1816 to1890), an eminent English physician and Governor of Guy’s Hospital, London (also a suspect in the Jack-the-Ripper murders) postulated: “The road to medical knowledge is through the pathological museum and not through an apothecary’s shop.” (1). Generations of physicians and surgeons were educated with the help of morphological archives, especially of cardiac specimens. Dr. Maude Abbott (from 1869 to 1940), founder of pathomorphology for congenital heart disease, began “museum demonstrations” in 1904 that have become part of the medical school curriculum (2,3). In recent years however, the availability of these archives has become limited due to stiffened data protection regulations (4), a reduced number of autopsies, natural attrition of specimens, and most importantly that patients with congenital heart disease survive (5). Due to these factors the source of specimens has dropped dramatically. There are, however, inherent problems with specimens as they are rigid, have friable tissues owing to formaldehyde fixation and storage that also leads to shrinking and distortion; some parts are missing and/or open due to post-autopsy; delicate (valvar) structures are difficult to handle, and are vulnerable to injury. Furthermore, specimens present anatomy in a fixed spatial relationship for the purpose of demonstration rather than the dynamic correlation of the structures in the body.
Transfer of morphological specimens on a digital 3D platform, and the creation of a virtual museum from clinical data could solve the problem (6). High-resolution is mandatory to preserve imaging information of delicate valvar structures (7). First, a specimen is scanned with a high-resolution 3D scanner (that only registers surface information), or preferably with micro-computed tomography (CT) as it can achieve in the 10-micron resolution range and/or magnetic resonance imaging (MRI) (8). Next, digital information is segmented and a 3D virtual model is created. Availability of a virtual museum offers innumerable opportunities for training and education, academic reference, research, and also clinical applications e.g., presurgical planning and virtual surgery, patient-family education, etc. Similar free-to-use, publicly accessible interactive educational sites are available and study material from our project could be linked with similar collections (9,10).
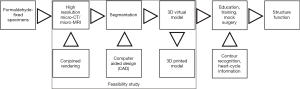
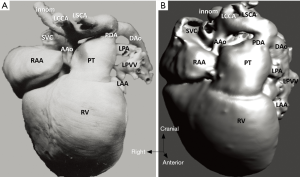
Inclusion of contour recognition and heart-cycle information to the 3D virtual models may conjoin structure and function in the future as shown in Figure 1. This report aims to achieve the following objectives:
- To rescue heart specimens in the collection at Hungarian Institute of Cardiology, Budapest, Hungary (i.e., the Archive) from disintegration, natural attrition, and to bequeath them to future generations. Without this project, these precious objects may be lost for good;
- To establish a comprehensive digital database of malformed cardiac specimens, so that information becomes open for wider communities of researchers, clinicians, trainers and trainees;
- To create a digital platform that can serve as a basis for future research, conduction of meta-analyses (i.e., in comparison with specimens in other international cardiac archives), and machine learning;
- To generate collective awareness about the pilot’s goals and results.
Methods
Archive
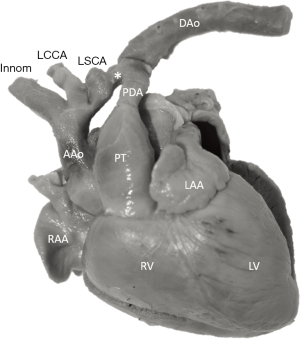
The study material consists of a historic archive of malformed cardiac specimens collected between 1960 and 1999 and maintained at the Hungarian Institute of Cardiology, Budapest, Hungary. Cardiac specimens were routinely retrieved from deceased patients with congenital heart disease at the time of autopsy. Hearts suffered the usual pathological examination by opening cardiac chambers and cutting away certain segments for demonstration purposes. In order to restore their geometry, specimens were filled and supported with soft material, although closure of the post-mortem incision was not attempted at the time of their fixation in 4% buffered formaldehyde (methanal). The slow evaporation of formaldehyde necessitates regular refilling of the containers. The first author acted as curator of the Archive between 1990 and 2006 and catalogued the specimens according to established nomenclature (11). A corresponding photo-library was also set up. There are about 400 cardiac specimens spanning almost 60 years and some of them had been collected in the pre-surgical era (early 1960s). Some specimens feature very unique anatomies, e.g., unoperated tetralogy of Fallot (TOF), aortic coarctation, transverse aortic arch hypoplasia, etc. (Figure 2).
Nowadays, these malformations are unfortunately impossible to encounter in pathological specimens as patients receive correctional surgery and survive. Therefore, this collection is now closed. Unfortunately, attrition rate due to desiccation and natural decay has been increasing in recent years, prompting preservation by digitization.
Ethical considerations
During the years of collection of the Archive, pertinent national regulation permitted retention and anonymous storage of human tissues and organs of scientific significance for education and research (12). Having been closed down in 1999, no more new specimens are added to the collection. The Institutional Review Board of the Hungarian Institute of Cardiology, Budapest, Hungary had approved maintenance of the Archive provided that anonymity is granted and “the person cannot be recognised simply by examination of the part” (13).
Project and pilot study description
Rapid attrition rate of cardiac specimens in the Archive prompted efforts to save the collection by digitization. A feasibility study team consisting of a pediatric cardiac surgeon with expertise in cardiac morphology, a biophysicist with expertise in multimodal imaging, and a bioengineer with experience in segmentation and virtual modelling was formed. Scanning and segmentation were performed at the Department of Biophysics and Radiation Biology, Semmelweis University, Budapest, Hungary. First, a pilot study was conducted to ascertain whether scanning methods with high-resolution micro-CT/MRI could result in a full digital imaging and communications in medicine (DICOM) dataset allowing representation of all cardiac segments including the cardiac valves. Next, 3D virtual models were created by image segmentation and rendering. Models were further refined by computer aided design (CAD), so that missing and/or dislocated parts could be corrected. Virtual 3D models presented as 3D rotating pdf files can be further investigated and edited; one can circumspect them, open the chambers in any trans-sectional plane, delete and annotate segments, etc. In our case, segmental anatomy, i.e., cardiac chambers, details of the viscero-atrial situs, atrioventricular and ventriculo-arterial connexions, and other structures were identified and labelled for reference.
In the next phase of the project, with a full morphological spectrum of models with congenital cardiac anomalies, a comprehensive database will be established, in which specific entities can be searched by diagnosis and morphological label (according to the segmental approach and internationally adopted classification of congenital cardiac nomenclature) (14). The database also connects descriptions of the segmental anatomy, and existing photo/video documentation to the newly-created virtual 3D models, DICOM data, stereolithography file, etc. for each of the scanned specimens. A website is about to be set up to provide an interactive platform, acting as a virtual museum, for study, education, and research with open-access by registration and/or subscription.
In a further research phase of the project we plan to add wall-motion and flow information rendered to the virtual 3D models. By adding heart-cycle metadata to segmental identification of the models, intracardiac flow could be studied and quantified alongside with “virtual intracardiac tours” in pulsating virtual hearts. This will be additional to augmented visualization and virtual reality in presurgical planning and surgical emulation.
Methods of image scanning and creation of virtual models
Scanning
Before scanning all specimens were partly cleansed of formaldehyde using water, and they were placed on the scanning table in the proper anatomical position.
MRI was performed with a nanoScan PET/MRI system (Mediso, Hungary), which is equipped with a permanent magnetic field of 1T and with a 450 mT/m gradient system using a volume coil for both reception and transmission. The diameter of the coil is 60 mm, therefore only small enough specimens (less than 60 mm in diameter) could be investigated with MRI. T1-weighted 3D gradient echo sequence was acquired using 4 excitations with TR =25 ms repetition and TE =2.6 ms echo times and a 30° flip angle. The in-plane resolution and the slice thickness were both set to 0.3 mm. The size of the field of view (FOV) was set according to the actual size of each specimen, therefore the acquisition time varied around an average of 30 minutes.
CT measurements were performed on a NanoX-CT (Mediso, Hungary) cone-beam micro-CT imaging system. Semi-circular CT scans were acquired with an 8W power X-ray source with 45 kV tube voltage, 3.6× zoom, 1,300 ms exposure time, 1:4 binning and 360 projections in 16–32 minutes (depending on the size of each particular specimen). For reconstruction, we used filtered back projection with a Butterworth filter. In-plane resolution and slice thickness were set to 0.2 mm. In the case of the CT scans, the maximal FOV size was big enough for all investigated specimens.
Modelling
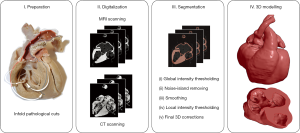
CT and MRI images were loaded into the open-source 3D Slicer 4.10 software (15), by means of which the multistep semi-automatic segmentation procedure was carried out (Figure 3). First, a conservative approximate value was assigned to the segmentation using global thresholding. Second, small noise-origin islands were removed by an automatic algorithm. Third, the built in “hole-closing” smoothing method was applied with a 1-mm kernel. Fourth, the skipped parts caused by local intensity variabilities were corrected by local threshold segmentation, in accordance with the theoretical requirements. As a final step, a 3D model was created from the segmented area, and theoretical and aesthetic corrections were made based on the visual examination of the model (including the mistakes committed by our semi-automatic segmentation procedure and the deterioration of the specimen). The results were double checked by an independent expert who did not take part in the segmentation process.
Results
Six specimens were randomly selected from the Archive and scanned with CT and/or MRI. MRI was performed in all but one case where size constraints did not permit. Presence of formaldehyde contributed to increased contrast in the CT studies, especially in the foreign materials (cotton balls) utilized to fill the cardiac cavities at the time of preservation. The filling material was removed in subsequent scans. Effect of formaldehyde was not observed in the MRI scans. Raw image data were adequate for segmentation and 3D virtual models were created. At the time of initial assessment of the models, no segmental diagnosis or details of anatomy were provided.
Morphological considerations
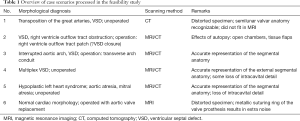
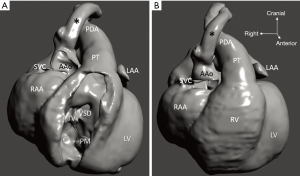
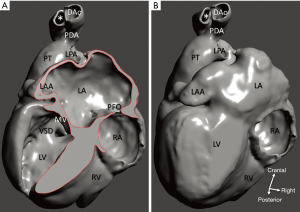
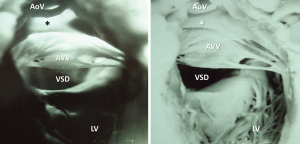
Diagnosis of the segmental cardiac anatomy could be established accurately in 5 out of the 6 models (Table 1). In the remaining case [case 4: multiplex ventricular septal defect (VSD)], malformations primarily affected the interventricular septum and accuracy of intracavital representation of anatomy was suboptimal. In this case scenario, absence of the compact layer of the myocardium and malalignment of the remaining septal components created an impression that both aortic and pulmonary outlets confined to the left ventricle (Figure 4). Technical inadequacy in positioning of the specimen and lack of refolding its structures and tissue flaps resulted in severe distortion and misrepresentation of structures (Figure 5). Viscero-atrial situs was not fully discernible for the lack of abdominal viscera; however, sidedness of the systemic veins was recognizable. Correct segmental diagnosis, i.e., atrio-ventricular (AV) and ventriculo-arterial connexions was uniformly recognised. Representation of finer details of the intracavital anatomy, especially the AV valves, their tensor apparatus and papillary muscles was clearly suboptimal compared to the original specimens (Figure 6). Semilunar (aortic) valves yielded better representation in the models. Aortic sinuses were recognizable by commissural ridges and orifices of the coronaries were indicated (Figure 7). The present resolution range did not allow recognition and follow-up on the course of the epicardial coronary arteries. Arterial segments were represented accurately in cases where the pathological process left the great arteries intact (Figure 8). Morphological information was cross-checked with the actual specimens and existing data from previous classification of the Archive. Although basic segmental diagnosis regarding the external anatomy was correctly established by the models alone, background data confirmed our assessment and elucidated missing details especially the intracavital anatomy.
Full table
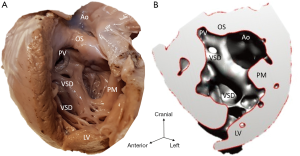
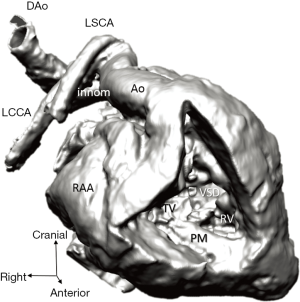
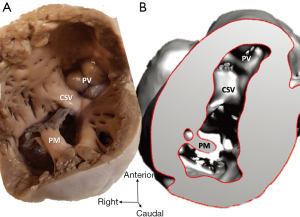
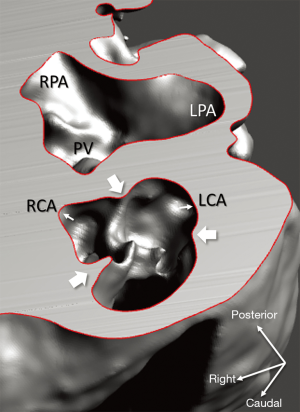
Technical considerations
Scanning and evaluation of the cardiac specimens has been a learning process. Proper preparation before scanning by adequate rinsing, removal of foreign material from the cardiac cavities, adequate positioning of distorted structures, e.g., aortic arch its branches, and refolding of flaps created during the post-mortem examination resulted in improved scanning datasets. Conjoined rendering of CT/MRI data and applying higher resolution could translate into better presentation of the intracavital structures, especially the cardiac valves. We utilized CAD to close openings of the cardiac chambers, and complete missing segments (Figures 9,10). These methods may improve demonstration and education value of the models; however, one potential query is about their overuse as they may introduce artificial relationships and shape of the structures that never existed in the original study material.
Observations
- Currently, cardiac specimens are scanned in a certain position, lying unsupported on a platform resulting in limitations for a perfect anatomical reconstruction, in contrast to clinical scenarios with living hearts that are surrounded by anatomic structures and filled with blood. There are limitations in size: our current MRI setup can handle specimens up to 60 mm in diameter.
- Apart from the size limitations of the MRI, generally speaking there was no significance difference between the information content of the images acquired by the means of the two modalities. However, in special cases (depending on the preparation, the deterioration and the partial thinning of the specimen) the double imaging approach proved to be quite useful in the segmentation process.
- The image resolutions described above seem to be satisfactory at the moment, but if later on higher resolutions are to be required, the resolution of the CT scanning could be upgraded even to as small as 10 microns.
- Segmentation and model design take longer for the specimens than for a clinical case, as there are additional iterations and refinements to be done. According to our experience, segmentation of a complex clinical scenario usually takes less than two hours, whereas specimens in the feasibility study needed more than three hours and repeated sessions to create a virtual model. It is expected that with participation of an anatomist with intimate knowledge of raw specimens and additional experience, segmentation time may be reduced to the range of the clinical cases.
- The flexibility of the reconstruction is limited; additional CAD software is needed to rebuild missing or dislocated segments (i.e., consequences of the pathological examination).
- Accuracy of the 3D models: representation of the external and segmental anatomy seems accurate; loss of intracavital detail with current processes is still significant.
Discussion
In 16th century Europe anatomy was the most popular subject in universities and was taught by demonstrations on human specimens (16). Modern application of 3D imaging and modelling techniques and 3D-printed prototypes may bring back the same intellectual excitement into present-day medical education and clinical practice that pioneers of anatomy could have experienced during preparation and studying of early anatomical specimens (17,18). In the clinical imaging scene, 3D echocardiography/ultrasound, 3D rotational angiography, CT-angiography and MRI greatly contribute to the development of individualized medicine and surgery (19). Image data integration of these modalities will enhance 3D modelling with grossly improved reliability, accuracy and resolution (20). Based on the general, commonly shared features of anatomy, clinical practice focuses on the individual phenotype of structures. 3D virtual modelling is especially useful as it often reveals patient-specific new information not obvious from simple planar (2D) projection.
Cardiac specimens offer the opposite approach: although they feature anatomy of an individual, their characteristics of the patho-morphology stimulate abstraction into general rules. This makes them especially useful for education and training (21). They are reviewed binocularly with interactivity and hands-on training (22). Touching and feeling an object activates the same area in the brain as vision, thus neural responses are modulated by sensory input from other modalities (23). Multisensory convergence enhances visual processing, and haptic input significantly contributes to the fine-tuning of visual information (24). Given the essentially 3D nature of the heart, photographic or even video archiving and documentation have a limited value (Figure 11).
Comments on the feasibility of the study
Technical aspects of scanning and data processing introduced an aspect of learning into our study. Theoretically, stand-alone specimens fixed in position offered benefits of maximum data acquisition in the micrometer range. Representation of the intracavital structures needs improvements. Direct involvement of cardiac morphologist and conscientious utilization of CAD methods in the modelling process can significantly ameliorate accuracy and detail of the 3D virtual models (Table 2). Static reconstructions of the 3D virtual models are still projected on the visual screen in two dimensions. Understanding of complex patho-anatomy rests on mental reconstruction only aided by a two-dimensional, unisensory visual input (25).

Full table
Although our pilot project was impelled by an imperative to rescue the Archive, in its future phases it will also open avenues for applications of visual analytics and new knowledge derived from dynamic 3D models. As an example, in the segmentation process, predictive modelling may be applied, i.e., machine-learning will learn the cardiac segments and complete them. Our future database can operate as a mixed data reservoir where raw data (e.g., DICOM datasets) are alongside structured and processed data, and they can be accessed on a highly agile and secure platform designed for low-cost storage for a wide range of professionals (26). Such new knowledge can improve decision support algorithms, exploit deep learning, and underlie novel future applications.
As cardiac specimens with congenital heart disease become increasingly difficult to collect and maintain, it may be very helpful to create a collection of 3D printed physical models based on the scanned data. Among the many applications of these physical models, they can be utilized for education, postgraduate training and presurgical planning and simulation (27,28).
Another direction of 3D technology is image-guided surgery in augmented reality. With this modality, patient-specific 3D models or holograms can be projected to a fixed point in space or directly superimposed on the operative area (29). In combination with robotics, optical displays developed into intelligent interfaces could revolutionize surgery: they could allow for procedures in the heart with preserved perfusion/organ function while being operated (30). Such prospects are anticipated to revive an intellectual excitement comparable to the one that established anatomy as medical science five hundred years ago.
Acknowledgements
The authors express their gratitude to Prof. Peter Andreka MD PhD and Dr. Zsolt Prodan MD at Gottsegen Hungarian Institute of Cardiology, Budapest, Hungary, current guardians of the Archive for allowing access to the collection; Mr. Carlos Perez, clinical engineer, Materialise, Leuven, Belgium for performing the initial segmentation for 3D virtual model in case 1. The authors are grateful for consultation on information technology opportunities with Dr. Konstantinos Avgerinakis and Dr. Efstratios Kontopoulos, CERTH, Thessaloniki, Greece.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Famous medical quotes. Sir William Withey Gull. Available online: http://izquotes.com/quote/297534
- Maude Abbott. Available online: http://www.mcgill.ca/medicalmuseum/introduction/history/physicians/abbott
- Abbott ME. Atlas of Congenital Cardiac disease. New York: The American Heart Association, 1936.
- Smith R. All changed, changed utterly. British medicine will be transformed by the Bristol case. BMJ 1998;316:1917-8. [Crossref] [PubMed]
- Wren C, O’Sullivan JJ. Survival with congenital heart disease and need for follow up in adult life. Heart 2001;85:438-43. [Crossref] [PubMed]
- Mostefa-Kara M. 3D imaging of heart specimens: a new teaching tool for understanding the anatomy of double outlet right ventricle. Available online: http://www.uni-kiel.de/aepc/aepcAbstractsFinalPrint/O4_6fin.pdf
- Hutchinson JC, Shelmerdine SC, Simcock IC, Sebire NJ, Arthurs OJ. Early clinical applications for imaging at microscopic detail: microfocus computed tomography (micro-CT). Br J Radiol 2017;90:20170113. [Crossref] [PubMed]
- Happel CM, Klose C, Witton G, Angrisani GL, Wienecke S, Groos S, Bach FW, Bormann D, Männer J, Yelbuz TM. Non-destructive, high-resolution 3-dimensional visualization of a cardiac defect in the chick embryo resembling complex heart defect in humans using micro-computed tomography: double outlet right ventricle with left juxtaposition of atrial appendages. Circulation 2010;122:e561-4. [Crossref] [PubMed]
- The Atlas of Human Cardiac Anatomy, maintained by the Visible Heart® Laboratory at the University of Minnesota in collaboration with Medtronic, Inc. The Visible Heart Project. Available online: http://www.vhlab.umn.edu/atlas/histories/dicom-request.shtml
- Cardiac Morphology. Available online: http://www.cardiacmorphology.com
- Anderson RH, Becker AE, Freedom RM, Macartney FJ, Quero-Jimenez M, Shinebourne EA, Wilkinson JL, Tynan M. Sequential segmental analysis of congenital heart disease. Pediatr Cardiol 1984;5:281-7. [Crossref] [PubMed]
- Magyar Közlöny (Official Bulletin of National Regulations). Regulations of the removal and retention of human tissues and organs during autopsy. 18/1998.XII.27. National Decree of the Hungarian Ministry of Health, with its modification No. 12/2012/VIII.6/EMMI Art(§) 6. Available online: http://www.kozlonyok.hu/nkonline/MKPDF/hiteles/MK12105.pdf
- Human Tissue Act 2004, Section 10. Regulations for existing anatomical specimens. Available online: http://www.legislation.gov.uk/ukpga/2004/30/section/10
- Anderson RH, Shirali G. Sequential segmental analysis. Ann Pediatr Cardiol 2009;2:24-35. [Crossref] [PubMed]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Rippa Bonati M. Some traditions regarding the old anatomy theatre of Padua University. Archivio Antico dell'Università di Padova, Atti dell'Università Artista, Raccolta Minato. Available online: http://www.unipd.it/esterni/visiteweb/english/pagine/teatro.htm
- O’Malley CD, Saunders CM. Leonardo da Vinci on the Human Body. New York: Winks, 1982.
- Ghosh SK. Evolution of illustrations in anatomy: a study from the classical period in Europe to modern times. Anat Sci Educ 2015;8:175-88. [Crossref] [PubMed]
- Fagan TE, Truong UT, Jone PN, Bracken J, Quaife R, Hazeem AA, Salcedo EE, Fonseca BM. Multimodality 3-Dimensional Image Integration for Congenital Cardiac Catheterization. Methodist Debakey Cardiovasc J 2014;10:68-76. [Crossref] [PubMed]
- Kurup HK, Samuel BP, Vettukattil JJ. Hybrid 3D printing: a game-changer in personalized cardiac medicine? Expert Rev Cardiovasc Ther 2015;13:1281-4. [Crossref] [PubMed]
- Kiraly L. 3D-modelling and printing of the heart and great vessels in medical education and clinical practice – a historical review. Kaleidoscope 2016;7:284-93.
- Preece D, Williams SB, Lam R, Weller R. “Let’s get physical”: advantages of a physical model over 3D computer models and textbooks in learning imaging anatomy. Anat Sci Educ 2013;6:216-24. [Crossref] [PubMed]
- James TW, Humphrey GK, Gati JS, Servos P, Menon RS, Goodale MA. Haptic study of three-dimensional objects activates extrastriate visual areas. Neuropsychologia 2002;40:1706-14. [Crossref] [PubMed]
- Lunghi C, Alais D. Touch Interacts with Vision during Binocular Rivalry with a Tight Orientation Tuning. PLoS One 2013;8:e58754. [Crossref] [PubMed]
- Biaggi P, Fernandez-Golfín C, Hahn R, Corti R. Hybrid Imaging During Transcatheter Structural Heart Interventions. Curr Cardiovasc Imaging Rep 2015;8:33. [Crossref] [PubMed]
- Chang AC. Medical Intelligence: Principles and Applications of Artificial Intelligence in Medicine and Healthcare. Available online: http://ai-med.io/wp-content/uploads/2018/08/CHANG_MITextbook_0718.pdf
- Hibino N. Three-Dimensional Printing: Applications in Surgery for Congenital Heart Disease. World J Pediatr Congenit Heart Surg 2016;7:351-2. [Crossref] [PubMed]
- Baker CJ, Sinha R, Sullivan ME. Development of a cardiac surgery simulation curriculum: from needs assessment results to practical implementation. J Thorac Cardiovasc Surg 2012;144:7-16. [Crossref] [PubMed]
- RealViewHearth. Surgery using 3D hologram. Available online: http://www.realviewimaging.com/?page_id=158
- Hurley J. Building robots for the cutting edge of medicine. A Cambridge-based company is intent on leading a ‘revolution in surgery’. The Times, 16 August 2017. Available online: https://www.thetimes.co.uk/past-six-days/2017-08-16/business/building-robots-for-the-cutting-edge-of-medicine-nhnk5c65j#