IMAge/enGINE: a freely available software for rapid computation of high-dimensional quantification
Introduction
Routine modalities of MR imaging such as diffusion weighted imaging (DWI) and dynamic contrast-enhanced MRI (DCE-MRI) generate high dimensional image data which require processing at a further step for interpreting.
DWI enables the identification of the structure and organization of tissue at the microscopic level (1,2). And it has been widely performed on the brain, prostate, liver, or kidney (3,4). DWI enables converting MR imaging into lots of quantified models, such as apparent diffusion coefficient (ADC) (5), intra-voxel incoherent motion (IVIM) (6), diffusional kurtosis imaging (DKI) (7) and neurite orientation dispersion and density imaging (NODDI) (8). The response of tumors (9,10) and brain development can be assessed by these quantified models (8).
Dynamic susceptibility enhanced (DSC) (11) and dynamic contrast enhanced (DCE) (10) MRI are often used to identify the pathological tissues by dynamically monitoring the change in image after using the contrast agent (12). Contrast agents, like gadolinium, with good detective sensitivity can be detected with routine acquisition protocols. Blood flow, permeability and other parameters could be computed by kinetic analysis of these high dimensional data.
Indeed, the application of these techniques holds great potential to improve clinical outcomes and to potentially guide patient managements. However, with several challenges such as technical complexity and methodological challenges, it is challenging for a researcher with clinical background to compute these quantified datasets. In order to overcome this limitation, several integrated software such as commercial workstations, OsiriX (13) and ImageJ (14) are designed to view and process medical image. The problem is a trade-off between being easy to use and extensive for the software.
IMAge/enGINE is free and extensible software for high-dimensional medical image computing. Comparing to other free software, IMAge/enGINE is equipped with a graphic user interface which is designed specifically for users without engineering or programming skills. Most of the operations are simplified as clicking buttons and dragging sliders. In addition, IMAge/enGINE offers an open-source and extensible framework. Developers with proper C++ programming skills can integrate and implement their own algorithms into an application.
IMAge/enGINE was released in 2017 on a freely available basis. Since then it has been downloaded by more than 300 unique users from approximately 100 different hospitals and institutions.
Methods
IMAge/enGINE is integrated out-of-box software with easy-to-use graphic user interface. Its binary package of WINDOW platform can be downloaded freely at http://www.vusion.com.cn/?page_id=14971. The usage for research only will be verified during a simple sign-up process. Afterwards, an account will be created for logging into this software and accessing most of its functions. The framework part of IMAge/enGINE is open-source and archived at https://github.com/VusionMed/IMAge-enGINE.
We chose C++ as the primary language for implementing IMAge/enGINE to integrate with the existing open source libraries and tools. IMAge/enGINE was built upon Qt (15), DICOM toolkit (DCMTK) (16), visualization toolkit (VTK) (17), insight toolkit (ITK) (18), common toolkit (CTK) (19), CMake (20) and TEEM (21). Among them, Qt provides a cross-platform GUI front. DICOM toolkit is used for reading and writing DICOM objects. The DICOM widget of CTK is used for managing the DICOM files. ITK is used for image registration, segmentation and implementing algorithms of computing blocks. VTK is used for image visualization and interaction. TEEM is used for providing support for files with a suffix of nrrd.
Overview
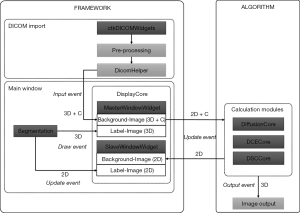
IMAge/enGINE consists of 7 main functional blocks and three of them contain calculation modules. An overview of how these blocks communicate and relate to each other is given in Figure 1. Though the calculation modules are not included in the source code, a developer can implement his own calculation module by inheriting the sample class.
DICOM import
IMAge/enGINE employs DICOM browser widget in CTK to manage the DICOM database. User can select and load DICOM data. After data loading, the DICOM data will be read into memory as a VTKImageData with 4 dimensions, X-Y-Z and components (C). The C dimension is used for indicating the b-value and diffusion direction for Diffusion data and time frame for dynamic data. Because most of the MRI scanner manufactures keep the b-value information into the private tag of DICOM metadata, it is difficult to generally support the DICOM from all manufactures. For GE, Philips and Siemens DICOM data, current DicomHelper class can parse the b-value and gradient direction correctly. But for other manufactures data, users have to provide a .bvec and a .bval text files as defined by Chris Rorden’s DCM2Nii tool (22) to describe the b-value and gradient direction of loading data. After the data are correctly loaded, users can apply rigid motion correction on the C dimension.
Display
There are two types of window widgets to render the image. Both window widgets can render two layers of images. One is the background image layer which is used for displaying the medical imaging and the other one is the label image layer which is used for drawing and displaying the labelling region of interest. The label image layer is also stored in memory using VTKImageData class and has the same matrix size with its corresponding background image. The background image layer of MasterWindowWidget holds the loaded images whose dimension is 3D+C. The background image layer of SlaveWindowWidget is only used for holding the computed image such as ADC map, IVIM D-star Map which is 2D image of the current slice displaying in MasterWindowWidget. Similarly, the label layer of MasterWindowWidget holds 3D image and SlaveWindowWidget only holds 2D image. There is only one MasterWindowWidget, while SlaveWindowWidget can be as many as the available calculation types of the loaded images.
Calculation
When the users trigger an update event such as adding a new type of computed image, scrolling a new slice or applying a new mask, a QT signal is sent to calculation module with the slice number of currently displaying in the MasterWindowWidget. Then the calculation module responds by calculating the required model to generate the computed images (2D) including an R2 map which indicates the fitting error of each pixel from the informed slice. After the computed images are generated, they will be sent to the Display Core to either update the current SlaveWindowWidget or add a new SlaveWindowWidget. In our design, the calculation will not be performed for slices that are not displayed, and hence largely reduce the computation time. As the only exception, all slices will be calculated automatically when users save the results to local disk. This design is one of the major highlights of IMAge/enGINE.
Fitting algorithms
Linear
One of the most used functions in calculation mode is fitting data to linear models, such as ADC, DTI/DKI. Linear formulation for ADC and DTI mode is simply obtained by taking logarithms on diffusion signals, and the fitting results are ADC values and tensor values respectively. Widely used method least-square regression is used for these two calculation modes. DKI Mode is somewhat complicated; it’s converted to the constrained Linear fitting formulation (23), and tensor values and kurtosis values are combined to the fitting target. A standard quadratic programming algorithm active set is used for this constrained linear fitting. DSC calculation is a deconvolution problem, which is often performed using Singular Value Decomposition (24). The result of DSC deconvolution is the residue function which is used to calculate perfusion parameters.
Non-linear
For models including IVIM, body DKI which compute Kapp (sDKI in our software), and NODDI, the signal intensity curve follows a non-linear decay. Mathematical fitting was decomposed into two steps: (I) exhaustive grid search which was first applied to find the initial guesses of the parameters to be fitted. (II) Nonlinear optimization via the publicly available ALGLIB library (http://www.alglib.net).
Segmentation
The segmentation module allows users drawing the region of interest (ROI) on displaying image. In our current build, nine ROIs can be created at the same time. The ROI drawing is actually filling the image data with different scalar values. Zero indicates background and one to nine indicate the nine ROI labels. The drawing operation can only happen in the MasterWindowWidget which actually modifies the 3D image data in its label image layer. The draw event triggers the update of statistical data in the statistic view. The statistical data include mean, standard deviation, minimum and maximum values of all the ROIs. The label image layer of SlaveWindowWidget only stores the 2D image which is a copy of label image in MasterWindowWidget with current slice. When an update event is triggered, the new slice of label image is copied and sent to the SlaveWindowWidget. The 3D label image can be saved to hard disk as an .nrrd file for later loading.
Data exporting
The data exporting is designed in two ways. One is for writing computing images to local hard disk in the format of DICOM which is handled by calculation module. A QT signal will be emitted by the GUI when users click the saving button, the calculation module will answer the signal accordingly. The other is to upload the computed image within selected ROI to server for further investigate, e.g., radiomics feature extraction as www.vusionmed.com providing. A developer can easily implement the client-server communication part by using the http tools providing by Qt.
Results
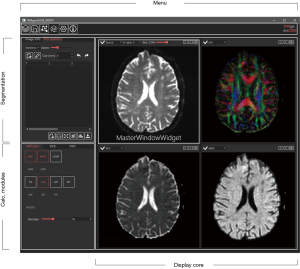
The graphic user interface (GUI) of IMAge/enGINE
The GUI of IMAge/enGINE is shown in Figure 2. The MasterWindowWidget shows the source 3D+C image data. And if a button in calculation modules is toggled on, a related SlaveWindowWidget is added into Display Core layout loaded with related 2D computed Image.
Support of DICOM
DICOM images can be loaded into the software for displaying the diffusion and dynamic MRI data of GE, Siemens and Philips can be loaded into the software for computing. However, the mosaic image of DTI data from Siemens scanner will be treated as one single slice as Figure 3.
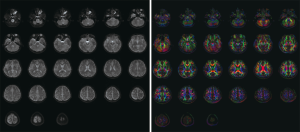
Image display
The basic interactions of image display are implemented:
- Dragging left-button: window/level adjust;
- Dragging middle-button: pan image;
- Dragging right-button: zoom image;
- Wheel: scroll up/down image.
And the buttons on MasterWindowWidget and SlaveWindowWidget provide functions such as image screenshot, applying a full-rainbow sugar color encoding and mouse probing.
Calculation
So far three modules of quantitative image computing were listed in Table 1. We tested the diffusion module on two computers A and B with different configurations. One is equipped with a 4th gen i5 CPU which has a relatively lower performance than most of the using CPUs, while the other equipped with a high-performance Xeon E5 CPU. The calculation speed is listed in Table 2. All the algorithms are implemented using C++ as ITK filter pipeline. For time consuming algorithms: DKI and NODDI, multi-thread computing is employed. In comparison, we used computer A to run a DKI algorithm of DIPY on the same dataset and it costs 30 seconds which was roughly 10 times longer.

Full table

Full table
In the GitHub repository of IMAge/enGINE, the source code of calculating ADC is given as an example. Developers can implement their own algorithm as a new calculation module to process the high-dimensional data. For modules developed in C++, the interface of the example ADC module can be directly copied. If other languages such as MATLAB, Python and Java are used, the calculation module should be built into an executable file (or packed into a Docker image) and be called by IMAge/enGINE framework. Due to the slice-by-slice updating scheme of IMAge/enGINE, the computation will only be applied on the relevant slices. The computing efficiency of calculation modules largely depends on the algorithm and the language used.
Segmentation
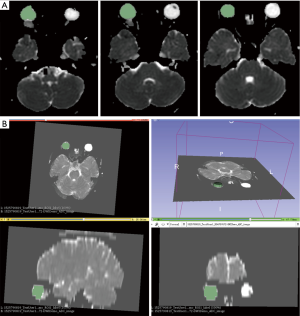
Two classic ways of drawing ROI are provided. One is to click on the image to add control points to form a closed contour which labels the area inside the contour with ROI index. The other is to directly paint on the image with input brush size and ROI index. Since the ROIs are stored in MasterWindowWidget as different values in a 3D label map, drawing on different slice of source image with the same ROI index forms a 3D ROI. The 3D label map is written as a .nrrd file onto local hard disk by clicking the save button. The coordinates of the 3D label map are handled to keep its geometry consistent with the source image, shown in Figure 4.
Discussion
Prospect of IMAge/enGINE will continue to be used by radiology research groups with or without professional engineers. For researchers with pure clinical background, the current software will provide them an efficient tool to complete diffusion, DCE model calculation and the ROI statistics measuring. For research groups with engineering backgrounds, they can write home-made plugins in python or C++ and integrate them into IMAge/enGINE, then deliver to clinical researchers to practice.
Comparision with other free softwares
There is a trade-off between being easy to use and extensive for medical imaging processing softwares. The phrase “easy-to-use” in this article indicates that even a researcher with clinical but without programming or engineering background still can have a friendly and convenient using experience. Figure 5 lists some medical image softwares. Most of the well-integrated softwares such as commercial workstations optimize with usability and usually provide a dedicated workflow for anatomy and modality. Though some of these workstations provide research interface for adopting home-made algorithm, e.g., FUNC2 of GE and PRIDE of Philips, their flexibility of extension is limited. OsiriX is a well-integrated DICOM viewer focusing on image fusion, rendering and annotation while being partial open-source and providing a plug-in mechanic for other developers to add their own tool. 3D-Slicer and Image-J are open-source and broaden their extensibility to provide multiple extension interfaces for plugins developed by other research groups. However, clinicians and researchers may feel confused about the amount of parameters exposed in the GUI. It usually requires user to be able to attend workshop, find proper plugins and/or carefully read documentations to be able to complete their research tasks. Toolkits with GUI such as MITK provide greater flexibility to allow users to combine the various command line tools and libraries they provide to generate their own task-oriented tool for handling images. However, without proper programming knowledge, it is unlikely to use this type of software for the research group without engineering background members. There are also toolkits without GUI such as DIPY 19 which is popular among research groups with developing abilities.
In terms of balancing easy-to-use and easy-to-extend, IMAge/enGINE is similar to OsiriX by providing documented plug-in mechanism for extensibility. But IMAge/enGINE is freely available, ad-free, more open-source and focused on image computation instead of image fusion as well as render. Moreover, MRI is vastly used in brain research and there are many software focused on processing neuro imaging such as FSL (26), MRTrix (27), SPM (28), PANDA (29) and DSI studio (30). Though IMAge/enGINE provides similar functions such as DTI, DKI, NODDI calculation, IMAge/enGINE is not designed for any specific anatomy parts but for high dimensional data. So far the majority of the users of IMAge/enGINE have been using it to process body MRI data, such as breast, prostate and liver.
Terms of license
IMAge/enGINE is freely available for academic research use (studies paid by government-derived funds or donations) provided that the original research publications relevant to the software are cited. The software is also free for educational purposes. Note that the license terms do not generally include trials paid by pharmaceutical companies. For commercial use, IMAge/enGINE is sold and supported by the company Vusion Tech, Hefei, China. Individuals or organizations are not allowed to compile software products derived from IMAge/enGINE that are to be sold commercially or shipped together with other commercial products.
Acknowledgements
Funding: This work was supported by Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) and Shanghai Natural Science Foundation (17ZR1401600).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Dietrich O, Biffar A, Baur-Melnyk A, Reiser MF. Technical aspects of MR diffusion imaging of the body. Eur J Radiol 2010;76:314-22. [Crossref] [PubMed]
- Le Bihan D, Mangin JF, Poupon C, Clark C, Pappata S, Molko N, Chabriat H. Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging 2001;13:534-46. [Crossref] [PubMed]
- Seyedmehdi Payabvash. Quantitative diffusion magnetic resonance imaging in head and neck tumors. Quant Imaging Med Surg 2018;8:1052-65. [Crossref] [PubMed]
- Ma W, Zhang G, Ren J, Pan Q, Wen D, Zhong J, Zhang Z, Huan Y. Quantitative parameters of intravoxel incoherent motion diffusion weighted imaging (IVIM-DWI): potential application in predicting pathological grades of pancreatic ductal adenocarcinoma. Quant Imaging Med Surg 2018;8:301-10. [Crossref] [PubMed]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 1986;161:401-7. [Crossref] [PubMed]
- Luciani A, Vignaud A, Cavet M, et al. Liver cirrhosis: intravoxel incoherent motion MR imaging--pilot study. Radiology 2008;249:891-9. [Crossref] [PubMed]
- Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: the quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med 2005;53:1432-40. [Crossref] [PubMed]
- Jelescu IO, Veraart J, Adisetiyo V, Milla SS, Novikov DS, Fieremans E. One diffusion acquisition and different white matter models: how does microstructure change in human early development based on WMTI and NODDI? Neuroimage 2015;107:242-56. [Crossref] [PubMed]
- Moffat BA, Chenevert TL, Meyer CR, McKeever PE, Hall DE, Hoff BA, Johnson TD, Rehemtulla A, Ross BD. The functional diffusion map: an imaging biomarker for the early prediction of cancer treatment outcome. Neoplasia 2006;8:259-67. [Crossref] [PubMed]
- Bonekamp S, Jolepalem P, Lazo M, Gulsun MA, Kiraly AP, Kamel IR. Hepatocellular carcinoma: response to TACE assessed with semiautomated volumetric and functional analysis of diffusion-weighted and contrast-enhanced MR imaging data. Radiology 2011;260:752-61. [Crossref] [PubMed]
- Ostergaard L. Principles of cerebral perfusion imaging by bolus tracking. J Magn Reson Imaging 2005;22:710-7. [Crossref] [PubMed]
- Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Curr Med Imaging Rev 2009;3:91-107. [Crossref] [PubMed]
- Osiri X. an open-source software for navigating in multidimensional DICOM images J Digit Imaging 2004;17:205-16. [Crossref] [PubMed]
- Barboriak DP, Padua AO, York GE, Macfall JR. Creation of DICOM--aware applications using ImageJ. J Digit Imaging 2005;18:91-9. [Crossref] [PubMed]
- Qt cross-platform application and UI framework. Available online: http://qt.io
- DCMTK — DICOM Toolkit. Available online: http://dicom.offis.de/dcmtk
- VTK: Visualization Toolkit. Available online: http://vtk.org
- ITK: Insight Toolkit. Available online: http://itk.org
- CTK: The Common Toolkit. Available online: http://commontk.org
- CMake: cross-platform build system. Available online: http://cmake.org
- NRRD: Nearly Raw Raster Data. Available online: http://teem.sourceforge.net/nrrd
- DCM2NII: DICOM to NIFTI Conversion. Available online: https://github.com/rordenlab/dcm2niix
- Tabesh A, Jensen JH, Ardekani BA, Helpern JA. Estimation of Tensors and Tensor-Derived Measures in Diffusion Kurtosis Imaging. Magn Reson Med 2011;65:823-36. [Crossref] [PubMed]
- Wu O, Østergaard L, Weisskoff RM, Benner T, Rosen BR, Sorensen AG. Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med 2003;50:164-74. [Crossref] [PubMed]
- Rosenkrantz AB, Padhani AR, Chenevert TL, Koh DM, De Keyzer F, Taouli B, Le Bihan D. Body diffusion kurtosis imaging: Basic principles, applications, and considerations for clinical practice. J Magn Reson Imaging 2015;42:1190-202. [Crossref] [PubMed]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23 Suppl 1:S208-19. [Crossref] [PubMed]
- MRTrix. Available online: http://www.mrtrix.org
- SPM. Available online: https://www.fil.ion.ucl.ac.uk/spm
- PANDA. Available online: https://www.nitrc.org/projects/panda
- Studio DSI. Available online: http://dsi-studio.labsolver.org