Discrimination of smoking status by MRI based on deep learning method
Introduction
Smoking is a major public health problem and the leading cause of preventable deaths worldwide (1,2). More than six million people die from smoking related diseases all over the world each year. The World Health Organization (WHO) has classified nicotine dependence as a disease. Nicotine dependence is a dependence on psychoactive substance, its initiation and development are closely related to the changes in the brain structure and function, and it can cause cerebral vascular disease and Alzheimer’s disease (3,4).
Studies have shown that nicotine dependence leads to the changes in brain structure (5-8), but these changes are typically subtle and hard to macroscopically identify. Currently, it is difficult for clinicians to identify and interpret the changes in the brain by magnetic resonance imaging (MRI), and thus the assessment and prediction of smoke cessation are still a challenge in clinical practice. Despite the rapid development of artificial intelligence in recent years, little is known about the application of deep learning methods in the diagnosis and treatment of clinical diseases, especially in the studies about the discrimination of human brain structure. In the present study, advanced deep learning methods were employed to discriminate the brain structure of smokers, aiming to find a new way to investigate the brain characteristics of smokers.
Methods
Subjects
Both smoking and non-smoking subjects were recruited and included as smoking group (SG) and control group (CG). The SG and CG subjects were enrolled based on the following inclusion criteria: subjects were aged 23–45 years; subjects were healthy; the education level was bachelor degree or higher; they had normal hearing and vision; they had no color blindness and were right-handed, and there were no contraindications to MR examination; smokers started to smoke when they were 18 years old or older and had a history of smoking for more than 5 years. The exclusion criteria were as follows: subjects had central nervous system or mental illness, cardiovascular disease (CVD), respiratory system diseases, the endocrine system diseases, diseases of the digestive system or other important diseases; subjects were susceptible to depression or anxiety.
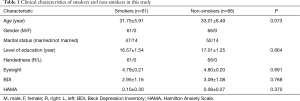
A total of 127 subjects were recruited into this study. In SG group, 61 smokers were enrolled from the Smoking Cessation Clinic of Beijing Chaoyang Hospital between August 2012 and October 2014; in CG group, 66 non-smokers were enrolled from the Clinic Health Center of Beijing Chaoyang Hospital between September 2013 and October 2014. Both groups had comparable demographics and personality characteristics, including age, gender, marital status, dietary habits, education level, eyesight, depression and anxiety [Beck Depression Inventory (BDI); Hamilton Anxiety Scale (HAMA)] (Table 1).
Full table
MRI
MRI data were acquired in the Imaging Center of Beijing Chaoyang Hospital Affiliated to the Capital University of Medical Sciences using a Siemens Magnetom Trio 3T MRI system (gradient field strength: 45 mT/m; gradient field switching rate: 200 mT/m/ms).
MRI images were acquired using 3D T1-weighted MPRAGE T1 sequence with a sagittal view. The dataset contains 176 slices with a slice thickness of 1 mm. The Field of view is 256 mm × 256 mm with 256×256 pixels. The sequence was repeated twice. Figure 1 shows an example volume of 176 slices.
Deep learning decision
The convolutional neural network (CNN) was proposed in LeNet-5 in 1998 (9). The conventional CNN can only deal with 2D input images. Each image has a corresponding tag, which describes the outcome of the image and is entered into the network for training. The experimental data in this study were from the brain MR images. For each sample, there were 176 images corresponding to 176 slices of the MRI volume, and each sample had a corresponding tag. In order to process the volumetric datasets in this study, a deep 3D CNN model (Conv3D) was used.
Approximate 25% of objectives from each group were enrolled randomly as the testing samples, including 15 smokers and 16 non-smokers. The remaining subjects served as training samples. The MRI volumes with their tags as training samples were inputted into the Conv3D network to train the model. In the training phase, the parameters in the network are updated after optimization of the loss function. Then, the trained model was used to test the MRI volume in test dataset. The test results were got finally. This model was trained with the training data and tested on the test datasets. The accuracy, sensitivity and specificity on the smoking status were evaluated.
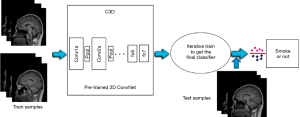
C3D (10) is a neural network which supports 3D convolution operations. The parameters were first pre-trained on the Sports-1M data set, and then this model was fine-tuned with the training data to predict smoking status from MR images. Figure 2 shows the flow chart of the training and testing procedures of the Conv3D model. Each training dataset on the left contains multiple images, and each test dataset on the right contains the same number of images.
Because the Conv3D method is based on C3D model, the input training and test data has to follow the format that is recognized by the C3D model [1, 3, 16, 112, 112]. Each sample should have 3 groups of images, and each group contains 16 images with the resolution of 112×112 pixels. In order to maximize the information from each brain MRI volume, processing was initiated from the middle slice out of the 176 slices, every other image going in both directions was extracted. Twenty-four images were extracted from each direction, and 48 images were inputted into the model.
The CNN models are commonly used in the processing of image data, and another neural network structure (11), the recurrent neural network (RNN), is widely used in the processing of sequential datasets (12). RNN permits data persistence. Figure 3 shows the block diagram of a typical RNN module A, which receives an input xi and outputs hi. The recurrent nature of RNN allows the information to propagate to the next step. RNN can be regarded as implementing the same neural network block multiple times, and each block passes the information to the next. The chain structure in Figure 3 indicates that RNN is a sequence of operations, and is an appropriate method to process sequential data. In the past few years, RNN has achieved success in the field of voice recognition, language modeling and translation, as well as photo description.

The long short-term memory (LSTM) is a special RNN architecture, which enables learning from experience. LSTM was proposed by Hochreiter & Schmidhuber in 1997 (13,14), and has been improved over year (15). Figure 4 shows a block diagram of the ConvLSTM model, which uses the LSTM architecture. The input data is a series of images, which correspond to the multiple slices of an MRI volume. Image features were extracted from each image using CNN models, and the features were inputted into the LSTM model. The entire volume data were trained with its corresponding tag (smoking or non-smoking). In this study, a 3-layer LSTM model was used, and the central 100 slices were applied as the input data. A 1,024-dimensional feature set was extracted from each slice using GoogleNet. The last layer of the ConvLSTM model was a full-connected layer. We also compared with support vector machine (SVM) model using the features extracted by GoogleNet. First, the features for slices are concatenated. Then, PCA was used for dimension reduction. Finally, the linear SVM was trained by the features.

The deep learning model was implemented by using the Lasagne (Lightweight library to build and train neural networks in Theano. http://lasagne.readthedocs.org/) framework based on Theano (Theano Development Team. Theano: A Python framework for fast computation of mathematical expressions. http://arxiv.org/abs/1605.02688), on a workstation with NVIDIA Titan X GPU that has 12GB memory. Based on implementation and hardware configuration, the model took 0.021 s to process every image during testing.
Results
Four-fold cross validation was used in this study, and all the data mentioned were the averages from four examinations.
Training and testing results based on the Conv3D deep learning model
The accuracy curve based on the Conv3D deep learning model is shown in Figure 5. The accuracy generally increased with the increase in the number of epochs. After 55 epochs, the model became stable, and achieved high accuracy on both training and test data. Eventually, the accuracy of the Conv3D model was 93.7% on the training set and 80.6% on the test set.
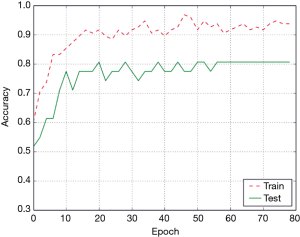
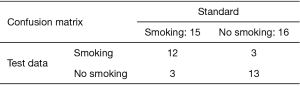
Figure 6 shows the confusion matrix of the Conv3D results on the test data. The sensitivity, specificity and overall accuracy of Conv3D deep learning model was 80%, 81.3% and 80.6%.
Training and test results Based on the ConvLSTM model
The accuracy curve based on ConvLSTM deep learning model is shown in Figure 7. The accuracy on the training data reached 100% beyond 70 epochs. However, the accuracy on the test data continued to increase gradually with the increase in the number of epochs. Eventually, the accuracy of ConvLSTM model was 100% on the training set and 93.5% on the test set.
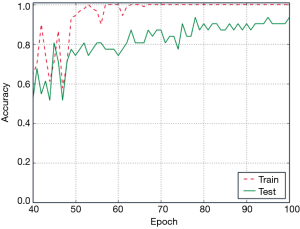
Figure 8 shows the loss value in the process of training of the ConvLSTM model. The loss magnitude decreased with the increase in the number of epochs, indicating the convergence of the model.
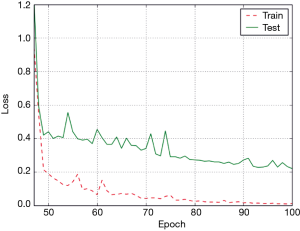
Figure 9 shows the confusion matrix of the ConvLSTM results on the test data. The table above calculated to the deep learning model ConvLSTM the sensitivity, specificity and overall accuracy was 93.33%, 93.75% and 93.5%.
Comparison between Conv3D and SVM and ConvLSTM models
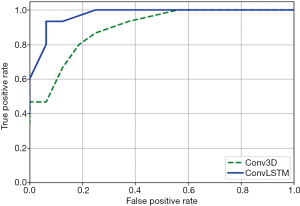
Figure 10 shows the comparison of ROC curves of Conv3D and ConvLSTM models. Table 2 shows the comparison of performances of Conv3D and SVM and ConvLSTM models. As shown in the table above, the image recognition accuracy based on ConvLSTM (93.5%) was better than that on SVM (83.8%) and Conv3D (80.6%).
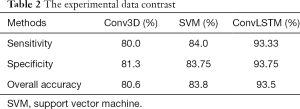
Full table
Discussion
MRI can detect brain anatomical structure characteristics in vivo and reveal the relationship between human behavior and corresponding brain structure. MRI has already been used in the study on brain morphology (such as thickness, volume, brain region, etc.), gray matter development, aging and disease pathology (16,17). High-resolution thin volumetric MRI provides an important method to build the structural characteristics of living human brain, and the modern deep learning models and other artificial intelligence methods can provide us tools to analyze the images. Although RNN are originally designed for temporal features, our results suggest that their application can be expanded for classification of datasets acquired at one snapshot.
Mechelli et al. (18,19) analyzed the structure images of brain, found the correlation of brain gray matter volume among multiple regions, and concluded that the brain morphology and coordination were associated with genetic and acquired plasticity. Zhang et al. (20,21) found that the prefrontal gray thickness of smokers decreased, and the corresponding area was less stimulated by smoking, which was associated with the change in cognitive function of smokers. They analyzed gray matter thickness in 48 smokers and non-smokers with respect to the degree of smoking addiction, smoking amount and duration, and found that the left prefrontal cortex became thinner after long-term smoking, which indicated that long-term heavy smoking could lead to the destruction of gray matter. Many studies have confirmed that long-term smoking may lead to specific brain atrophy, which in turn causes the change in cognitive function (22,23). It has been reported that corpus callosum structural integrity is impaired in smokers, but it can slowly recover in a long time after smoking cessation (4,24,25). Zanchi et al. (26) investigated 18 non-smokers and 14 smokers in 2015, and used MRI diffusion analysis to evaluate white matter and combined VBM method to evaluate the cortex. They found that smoking caused the damage to the connection between right side of the former insula and the anterior cingulate.
In our previous study, DARTEL based VBM method was used to evaluate the gray matter composition in 53 controls and 53 chronic smokers in 2015. Results showed that the smokers had significantly lower brain gray matter volume after the superior temporal gyrus, insula, cingulate, precuneus as compared to controls, and the difference correlated with the amount of cigarettes per year and the age of smokers (17). However, VBM analysis method relies on group analysis, and it is difficult to examine individual brain structure change. Given the blindness of smoking cessation, high relapse rate, and other clinical difficulties, it is necessary to further understand the changes in the brain structure and function of smokers which may provide theoretical basis for individualized cessation treatment and improve the success rate of smoking cessation.
With the rapid development of artificial intelligence in recent years, machine-learning methods have been widely used for image recognition, speech recognition and other procedures. In the present study, we for the first time used Conv3D and ConvLSTM models to predict nicotine dependence based on brain MR images. The ConvLSTM model achieved an overall accuracy of 93.5%, which was better than that of Conv3D model (80.6%). The present study showed the accuracy was significantly higher than that reported in a previous study (64.04%) on SVM (27). In addition, our methods do not require manual extraction of the features from the images. The models rely on CNN to extract features from the volumetric datasets, which may guide radiologists to study the brain composition of smokers and predict the effectiveness of cessation plans in the future.
In conclusion, state-of-the-art deep learning models are used to predict smoking status based on MRI images of individual human subject and provide a tool for the intelligent medical image interpretation. Though there is no direct clinical issues, we can see that the potential of deep learning methods to assist the doctor in the aspect of image feature recognition. It provides a tool to analyze nicotine dependence and develop cessation plans. However, more studies with larger sample size are needed to refine our models to differentiate nicotine dependence of different degrees and to confirm our findings.
Acknowledgements
Funding: This study was supported by Capital’s Funds for Health Improvement and Research (No. 2018-2-4066).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethnical Statement: The study was conducted in accordance with the Committee for Human Research in our institution and followed all regulations (No. 12-KE-21). Informed consent was obtained before scans.
References
- Gu D, Kelly TN, Wu X, Chen J, Samet JM, Huang JF, Zhu M, Chen JC, Chen CS, Duan X, Klag MJ, He J. Mortality attributable to smoking in China. N Engl J Med 2009;360:150-9. [Crossref] [PubMed]
- Yang GH, Ma JM, Liu N, Zhou LN. Smoking and passive smoking in Chinese, 2002. Zhonghua Liu Xing Bing Xue Za Zhi 2005;26:77-83. [PubMed]
- Yu R, Deochand C, Krotow A, Leao R, Tong M, Agarwal AR, Cadenas E, de la Monte SM. Tobacco Smoke-Induced Brain White Matter Myelin Dysfunction: Potential Co-Factor Role of Smoking in Neurodegeneration. J Alzheimers Dis 2016;50:133-48. [Crossref] [PubMed]
- Viswanath H, Velasquez KM, Thompson-Lake DG, Savjani R, Carter AQ, Eagleman D, Baldwin PR, De La Garza R 2nd, Salas R. Alterations in interhemispheric functional and anatomical connectivity are associated with tobacco smoking in humans. Front Hum Neurosci 2015;9:116. [Crossref] [PubMed]
- Crunelle CL, Kaag AM, van Wingen G, van den Munkhof HE, Homberg JR, Reneman L, van den Brink W. Reduced frontal brain volume in non-treatment-seeking cocaine-dependent individuals: exploring the role of impulsivity, depression, and smoking. Front Hum Neurosci 2014;8:7. [Crossref] [PubMed]
- Wang C, Xu X, Qian W, Shen Z, Zhang M. Altered human brain anatomy in chronic smokers: a review of magnetic resonance imaging studies. Neurol Sci 2015;36:497-504. [Crossref] [PubMed]
- Csabai D, Cseko K, Szaiff L, Varga Z, Miseta A, Helyes Z, Czeh B. Low intensity, long term exposure to tobacco smoke inhibits hippocampal neurogenesis in adult mice. Behav Brain Res 2016;302:44-52. [Crossref] [PubMed]
- Li C, Sun H, Arrick DM, Mayhan WG. Chronic nicotine exposure exacerbates transient focal cerebral ischemia-induced brain injury. J Appl Physiol (1985) 2016;120:328-33. [Crossref] [PubMed]
- Nebauer C. Evaluation of convolutional neural networks for visual recognition. IEEE Trans Neural Netw 1998;9:685-96. [Crossref] [PubMed]
- Tran D, Bourdev L, Fergus R, Torresani L, Paluri M. Learning Spatiotemporal Features with 3D Convolutional Networks. Proceedings - 2015 IEEE International Conference on Computer Vision, ICCV 2015.
- Hou Z, Yang Y, Li S, Yan J, Ren W, Liu J, Wang K, Liu B, Wan S. Radiomic analysis using contrast-enhanced CT: predict treatment response to pulsed low dose rate radiotherapy in gastric carcinoma with abdominal cavity metastasis. Quant Imaging Med Surg 2018;8:410-20. [Crossref] [PubMed]
- Wang S, Wang R, Zhang S, Li R, Fu Y, Sun X, Li Y, Sun X, Jiang X, Guo X, Zhou X, Chang J, Peng W. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters</=3 cm using HRCT. Quant Imaging Med Surg 2018;8:491-9. [Crossref] [PubMed]
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. [Crossref] [PubMed]
- Hochreiter S, Schmidhuber J. Flat minima. Neural Comput 1997;9:1-42. [Crossref] [PubMed]
- Cao R, Nosofsky RM, Shiffrin RM. The development of automaticity in short-term memory search: Item-response learning and category learning. J Exp Psychol Learn Mem Cogn 2017;43:669-79. [Crossref] [PubMed]
- Gallinat J, Meisenzahl E, Jacobsen LK, Kalus P, Bierbrauer J, Kienast T, Witthaus H, Leopold K, Seifert F, Schubert F, Staedtgen M. Smoking and structural brain deficits: a volumetric MR investigation. Eur J Neurosci 2006;24:1744-50. [Crossref] [PubMed]
- Peng P, Wang Z, Jiang T, Chu S, Wang S, Xiao D. Brain-volume changes in young and middle-aged smokers: a DARTEL-based voxel-based morphometry study. Clin Respir J. 2017;11:621-31. [Crossref] [PubMed]
- Mechelli A, Crinion JT, Noppeney U, O'Doherty J, Ashburner J, Frackowiak RS, Price CJ. Neurolinguistics: structural plasticity in the bilingual brain. Nature 2004;431:757. [Crossref] [PubMed]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci 2005;25:8303-10. [Crossref] [PubMed]
- Zhang X, Salmeron BJ, Ross TJ, Gu H, Geng X, Yang Y, Stein EA. Anatomical differences and network characteristics underlying smoking cue reactivity. Neuroimage 2011;54:131-41. [Crossref] [PubMed]
- Zhang X, Salmeron BJ, Ross TJ, Geng X, Yang Y, Stein EA. Factors underlying prefrontal and insula structural alterations in smokers. Neuroimage 2011;54:42-8. [Crossref] [PubMed]
- Cho H, Kim C, Kim HJ, Ye BS, Kim YJ, Jung NY, Son TO, Cho EB, Jang H, Lee J, Kang M, Shin HY, Jeon S, Lee JM, Kim ST, Choi YC, Na DL, Seo SW. Impact of smoking on neurodegeneration and cerebrovascular disease markers in cognitively normal men. Eur J Neurol 2016;23:110-9. [Crossref] [PubMed]
- Sutherland MT, Riedel MC, Flannery JS, Yanes JA, Fox PT, Stein EA, Laird AR. Chronic cigarette smoking is linked with structural alterations in brain regions showing acute nicotinic drug-induced functional modulations. Behav Brain Funct 2016;12:16. [Crossref] [PubMed]
- Umene-Nakano W, Yoshimura R, Kakeda S, Watanabe K, Hayashi K, Nishimura J, Takahashi H, Moriya J, Ide S, Ueda I, Hori H, Ikenouchi-Sugita A, Katsuki A, Atake K, Abe O, Korogi Y, Nakamura J. Abnormal white matter integrity in the corpus callosum among smokers: tract-based spatial statistics. PLoS One 2014;9:e87890. [Crossref] [PubMed]
- Lin F, Wu G, Zhu L, Lei H. Heavy smokers show abnormal microstructural integrity in the anterior corpus callosum: a diffusion tensor imaging study with tract-based spatial statistics. Drug Alcohol Depend 2013;129:82-7. [Crossref] [PubMed]
- Zanchi D, Brody AL, Montandon ML, Kopel R, Emmert K, Preti MG, Van De Ville D, Haller S. Cigarette smoking leads to persistent and dose-dependent alterations of brain activity and connectivity in anterior insula and anterior cingulate. Addict Biol 2015;20:1033-41. [Crossref] [PubMed]
- Ding X, Yang Y, Stein EA, Ross TJ. Multivariate classification of smokers and nonsmokers using SVM-RFE on structural MRI images. Human Brain Mapping 2015;36:4869-79. [Crossref] [PubMed]