Rapid tissue histology using multichannel confocal fluorescence microscopy with focus tracking
Introduction
In histology, hematoxylin and eosin (H&E) is widely used as a gold standard because it can provide precise morphological information on various tissue types (1). Hematoxylin stains nuclei blue or purple, and eosin stains cytoplasm and extracellular matrix pink (2). Since the tissue being examined is made into a slide to be viewed with a bright-field microscope, H&E histology generally requires physically sectioning tissue to around 5 to 10 µm thick. Paraffin sectioning is routinely used and regarded as the gold standard for H&E histology. Although paraffin sectioning provides clear visualization of tumor tissue, it requires up to days to fix and embed the tissue in paraffin (1). Instead of embedding the tissue, freezing before sectioning can be used to acquire rapid results (3) known as frozen histology. This cryo-sectioning allows for histology results in 30 to 40 minutes compared to days required for paraffin sectioning (4-7).
Although cryo-sectioning provides credible result within a reasonable time, during surgeries such as Mohs micrographic surgery where repeated histological results are needed, faster histology results are needed. In addition, cryo-sectioned tissue sometimes undergo freeze artifacts that make histological interpretation difficult. Although techniques have been developed to overcome such artifacts, freezing the tissue is still time consuming (8).
To overcome the limitations of current histology process, various optical biopsy techniques have been proposed, including optical coherence tomography (9-11), spectroscopy (12-15), terahertz imaging (16,17) and confocal microscopy (18-23). Recently, imaging techniques such as MUSE (Microscopy with ultraviolet surface excitation) (24-26) and light-sheet microscopy (27-29) showed promising results in obtaining pathological results from fresh tissue. MUSE uses an inherent low penetration depth of ultraviolet rays to image only the surface of the tissue. However, use of high NA objective is limited as optical sectioning thickness should not exceed the image depth of field (30), which hinders the high resolution imaging. Light sheet-microscopy uses two objectives placed at a 90 degree angle for excitation and imaging. In particular, an open-top light-sheet microscopy can obtain non-destructive slide-free pathology of clinical specimens with large field-of view. But it requires certain means to correct aberration induced by the coverslip placed at a 45-degree angle from the objective, such as deformable mirrors, spatial light modulators or solid immersion lenses (27,31). Additional image processing is also required to show the cross-sectional images of a tissue, which slows down the image acquisition speed (27). Multimodal nonlinear microscopy also provides various information of fresh tissue (32,33). But there are limitations such as a higher risk of photo-damage or photo-bleaching (34,35), low signal intensity (36,37), and higher cost due to use of pulsed laser (38,39). Among the many imaging techniques, confocal fluorescence microscopy has demonstrated promising results due to its ability to optically section thick specimens with high axial and lateral resolution (40,41), similar to a histology slide (42-44). Previous studies have shown fluorescence confocal microscopy provides strong contrast of nuclear signals (45-47). As with most optical systems, high resolution comes with a limited field of view. As a solution, a line scanning algorithm (48) has been developed. The algorithm uses line scanning to acquire large images in strips, which are then stitched together to produce a much larger image. Line scanning has significantly reduced the scanning time of large biological samples (49). The technique employs a mechanical stage to scan the sample along slow axis and a cylindrical lens (50) or a resonant scanner (49) to scan the sample along the fast axis. Various studies have shown promising results with this technique (51-53). A limitation of this technique is that it requires an additional mechanical tool to adjust the tilt and time to match the sample to the moving focus plane of the objective lens throughout the scan (49).
In this study, we developed a confocal microscope with multiple fluorescence channels to visualize nuclei and cellular structures of a tissue with high spatial resolution over a large field of view. We applied a newly developed fast staining method of freshly excised tissue with Hoechst 33342 and Eosin Y (54). A focus tracking algorithm was developed to keep the sample in focus while acquiring a large image. Using a virtual H&E conversion algorithm, colour codes of grayscale confocal images were converted to mimic H&E histology using a virtual H&E conversion algorithm (55). Also, a simple method was suggested to flatten the tissue and enable uniform imaging over the large field.
Methods
Multichannel confocal microscope system
A multichannel inverted confocal fluorescence microscope, K1-Fluo (Nanoscope Systems Inc., Daejeon, Republic of Korea) system, was modified for line scanning and focus tracking (Figure 1). The system consists of a laser module as the excitation source and a microscope module to detect the fluorescence signal. The laser module contains 3 wavelength lasers (405, 488, 638 nm). In this study, the 405- and 488-nm lasers were used as excitation sources for Hoechst 33342 and Eosin Y, respectively.
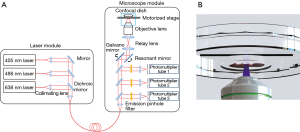
A motorized stage was placed above the objective lens to scan the sample in the axial and lateral directions. A 20× objective lens (UPlanSApo, Olympus, Shinjuku, Japan) with a numerical aperture of 0.75 was used. A galvano and resonant mirror were used for raster scanning the sample on a confocal dish (101350, SPL Life Sciences, Pocheon-si, Republic of Korea). We selected a confocal dish with a large diameter cover glass on the bottom for imaging large samples. Photomultiplier tubes with matching emission filters were selected according to the emission spectra of the fluorescent dyes.
Tissue preparation and staining procedure
Tissue samples were prepared from a nude mouse. After euthanasia, the mouse organs were collected and cut in half. Half the tissue was stained with fluorescent dyes and imaged with the confocal microscope to assess the performance of the microscope. The other half was cryo-sectioned to create an H&E slide and a confocal slide, which was stained with Hoechst 33342 and eosin Y. The confocal slide was used to validate the microscope performance and the focus-tracking algorithm. The study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Cancer Center Research Institute (NCC-15-229B).
The staining method was modified from our previous research (54), and numerous staining trials were performed to determine the appropriate staining time and dye concentrations (Table 1). Eosin-Y solution (Eosin Y alcoholic counterstain, Cat# 3600) was used as it provided sufficient staining time and quality. Hoechst 33342 (Hoechst 33342, Cat# MOP-H-3570, ThermoFisher Scientific, Waltham, MA, USA) was diluted to 1% in PBS (phosphate buffered saline) because, although a higher concentration shortened the staining time, the nuclear signal was saturated.

Full table
The sample was fixed with ethanol in between Eosin-Y and Hoechst 33342 staining because fluorescent dyes tend to leak out of tissue during imaging and degrade the image quality. Since prolonged fixation time cause the fluorescent dyes to wash out, the sample was exposed to ethanol repeatedly for a short time. The total staining time was 80 seconds.
Tissue holding and H&E pseudocolor
When the excised tissue is placed on a flat surface, the edges tend to float due to the soft property of the tissue. In our experience, staining tissue with fluorescent dyes slightly shrinks the tissue, creating space between the tissue edges and cover glass. A gap or air pocket as small as a few tens of micrometers between the tissue and the cover glass will prevent the confocal microscope from obtaining an adequate image in that area as a shift in focus is created. Therefore, a method to keep the tissue flat on the cover glass was necessary.
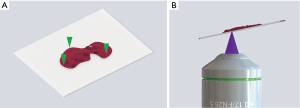
Using PDMS (polydimethylsiloxane), which is a type of silicone, we created a cover to keep the sample in place by lightly pressing the tissue down. The PDMS press was molded into a cup shape and attached to the cover of the confocal dish to gently push down on the tissue (Figure 2). The press was made with a curing agent to elastomer base ratio of 1:20. A ratio of 1:10 is typical, but the flexibility of the material can be controlled by adjusting the ratio. We found that more elastomer base makes the PDMS more flexible, but too much elastomer base makes the PDMS too sticky. After trying different ratios from 1:5 to 1:40, we found that a ratio of 1:20 provided sufficient flexibility to evenly press the tissue onto the cover glass without sticking to the tissue. The thickness of the glass bottom of the confocal dish was 0.13 mm, which was sufficient enough to withstand the pressure from the PDMS press.

Line scanning and focus tracking
The field of view of a confocal microscope is limited by the objective lens. In our case, a 20× objective lens was used, and the field of view was 625 µm in the x and y directions. To overcome the limited field of view and quickly scan a large sample, line scanning was used. A resonant mirror was used to create a line in the x axis while the motorized stage scanned the sample in the y direction to create a strip image with the galvano mirror fixed. Although the size of the strip in the x direction is limited by the objective lens (625 µm), the size of the strip in the y direction is only limited by the bottom hole size of the confocal dish and motorized stage. Multiple strip images, depending on the sample size, were acquired with a 5% overlap as a reference for the stitching algorithm. Matlab (The MathWorks, Inc.) and a stitching algorithm provided by the National Institutes of Health (NIH) were used to stitch image strips together into a single large image.
As the microscope scans the entire sample, a difference in focus appears because the objective lens does not keep equal focus throughout the scan. Thus, we developed a focus tracking method to keep the sample within focus. Conventional raster scanning with real-time visualization of a 625 µm × 625 µm field of view was used to find three focal points on the sample that were in focus. The process was performed manually with a z-axis knob, and the three points were kept far apart to keep the scanning plane of the focus-tracking algorithm close to the imaging plane (Figure 3A). The coordinates of the focal points were saved, and an imaging plane was obtained from a plane formula that contains the three points. The objective lens scanned the sample according to the calculated imaging plane thus acquiring an in-focus image of the entire sample (Figure 3B).
We used mouse tissue and conventional raster scanning to obtain a series of axial images and evaluate the depth of the focal plane within the sample. Results have shown that signals can be obtained up to 20 µm into the tissue, but the best depth showing clear results was at 5 µm deep. At this depth, both the Hoechst 33342 and eosin Y signals appeared well. From repeated experiments, we learned that there was a margin of about 3 µm on either side of the best imaging plane for the focus tracking algorithm. Therefore, the focus-tracking algorithm had to keep the focus point of the objective lens from 2 to 8 µm into the sample during the entire scan.
The image contained 1,024 pixels over 625 µm in the x direction, which gives a resolution of 0.61 µm per pixel. To match the x resolution, the speed of the motorized stage was set at 4.88 mm per second. Because the resonant mirror scans the sample at 8 kHz, the y pixel resolution also became 0.61 µm per pixel. The pixel dwell time was 61 ns and the laser power on sample was measured to be 5 mW. The maximum length in the y direction is only limited by the stage travel length, 20 mm. The largest sample measured is 20 mm × 20 mm.
To convert the multichannel grayscale confocal images to resemble H&E histology images, we used a virtual H&E conversion algorithm (55). The algorithm uses Beer-Lambert law to accurately model the absorption of Hematoxylin and Eosin in transillumination microscopy. Using the algorithm, pixel intensity of the grayscale multichannel fluorescence image can be converted to mimic H&E histology.
Results
H&E pseudocolor and tissue holder
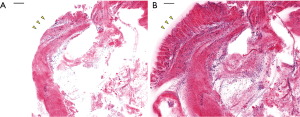
Mouse stomach tissue was stained with Hoechst 33342 and Eosin-Y according to the protocols described in Table 1. The result was first obtained with raster scanning and square mosaic imaging. This method stitches a series of square (625 µm × 625 µm) confocal images together into a single large image. This method was used when imaging small tissues because the focus does not differ significantly between points. When imaging relatively small samples, a focus-tracking algorithm would only lengthen the imaging acquisition time. H&E pseudocolor results with and without the tissue holder are shown in Figure 4. Both the Hoechst and Eosin-Y signals were converted with the colormap to resemble H&E histology. Hoechst 33342 signals appear purple and eosin Y signals appear pink. This step is necessary as H&E is widely used in pathology, so the appearance of a confocal image may play an important role in diagnosing cancer. Imaging was performed with and without the PDMS cover. As shown in Figure 4, the PDMS cover pressing the tissue against the cover glass clearly provides additional morphological information. The glandular mucosa present in Figure 4B (marked with yellow arrows) cannot be seen in Figure 4A due to the loss of focus.
Line scanning and focus tracking
A sectioned confocal slide was created from an excised mouse tumor. Nine image strips were acquired with line scanning. The slide was imaged with and without the focus-tracking algorithm. Figure 5 shows the effect of the focus tracking algorithm. As shown in Figure 5A, without the focus-tracking algorithm, the edges of the confocal slide were not imaged as the sample was not in focus. The focus-tracking algorithm allowed the whole sample to be imaged in focus, as shown in Figure 5B. The image acquisition time for a 4 mm × 5 mm slide was around 3 minutes including sample staining and manual focusing. The sample scanning itself took 50 seconds. The manual focusing time varies depending on the operator but does not depend on the size of the sample as only three points are used. In our case, it took one minute to adjust the three focus points and create an imaging plane. The imaging plane is calculated instantly upon entering the three focus point coordinates.
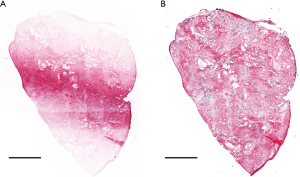
Image acquisition and comparison to histology
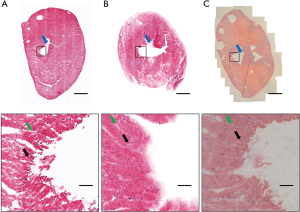
Mouse heart tissue was imaged as the morphological tissue features allowed us to validate the microscope performance. The tissue was prepared and stained as described in Table 1. The tissue was placed on a confocal dish with the PDMS cover. Large morphological features, such as the ventricle (marked with a red arrow in Figure 6), can be seen in the acquired image. The overall shape of the tissue slide (Figure 6A) and thick tissue (Figure 6B) matched the H&E slide (Figure 6C) imaged with the bright-field microscope. The magnified image can be used to visualize small morphological features, such as muscle fibres and nuclei (marked with green and black arrows, respectively). As shown in Figure 6B, entire tissue including the tissue boundaries was in focus due to the use of gently pressing PDMS cover. While too much pressure may cause the curvature of the coverslip and create distortion in the image, Figure 6B and Figure 4B show that the applied pressure by the PDMS cover was adequate to maintain focus throughout the whole tissue without inducing coverslip curved.
Discussion
We developed the multichannel confocal microscopy system to image large tissue areas. To minimize tissue processing steps and time, we applied the newly developed staining method using Hoechst 33342 and Eosin-Y to stain nuclei and cytoplasm of freshly excised tissue (37). Tissue samples did not require any additional processing other than the staining, which only takes about 80 seconds, enabling fast sample preparation. Using the holder made of soft PDMS that keeps the sample in place and gently presses the sample against the cover glass, we were able to obtain a flat image plane. We were able to quickly collect image strips by the line scanning method over a large field of view. Also, the focus-tracking method enabled to keep the sample in focus while scanning the large area of the sample. The obtained two-channel confocal images were pseudocolored to mimic conventional H&E histology. From the obtained images, morphological information of cytoplasm and nucleus could be verified. The entire process took around 3 minutes, which is much faster than cryo-sectioning. The technique will allow intraoperative histology to be possible and will significantly reduce stress on patients undergoing long surgeries requiring repeated histology results.
As the interest on the fresh tissue imaging has been increasing, various microscopy technologies have been introduced for non-destructive pathology of tissue, including MUSE (24-26), light sheet-microscopy (27-29), and nonlinear microscopy (32,33). While they can provide excellent pathologic images of fresh tissue, there are several limitations such as UV excitation (24-26,30), limited use of objective lens (30,56), need for complex optics (57,58) or high cost due to pulsed lasers (38,39). In contrast, the current method provides a simple, cost-effective solution for high-speed microscopic assessment of fresh tissue with a large field-of-view. Also, light sources and objective lenses can be easily optimized for specific applications to accommodate various fluorescent agents with high-resolution.
While this preclinical study showed promising results with mouse tissue, the feasibility of the proposed system for diagnosing cancer should be studied. Future studies may also evaluate the H&E pseudocolor method in diagnosing cancer using human tissues. Although not used in this study, the 638-nm laser already installed in the multichannel confocal system allows the use of an additional cyanine dye, which might be able to target cancer cells, and aid in diagnosing cancer. Because the targeting dyes are used in excised tissue, the approval process will be easier than for in vivo applications. The line scanning method with the focus-tracking algorithm can be easily applied to other optical systems requiring large area scanning. The algorithm may also be set up in the axial direction to rapidly acquire z-stacks of large images to provide comprehensive 3-dimensional information for precise diagnosis.
Conclusions
In conclusion, our study demonstrates the use of multichannel confocal microscopy to obtain histological images of fresh tissue. The newly developed tissue staining method using Hoechst 33342 and Eosin-Y enabled rapid tissue preparation. Rapid pathology results were obtained in a wide field of view using the line scanning and focus tracking methods and the PDMS tissue covers. The entire process, including the tissue staining and the scanning, takes only about three minutes. We expect that the presented method will enable rapid intraoperative histology, which can significantly reduce stress on patients during surgeries in which repeated histological examination is performed.
Acknowledgements
Funding: This research was supported by the Chungcheongbuk-do Value Creation Program through Osong Medical Innovation Foundation of Korea funded by Chungcheongbuk-do and by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2017R1E1A1A01074822).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare
Ethical Statement: This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Cancer Center Research Institute (NCC-15-229B). The NCCRI is an Assessment and Accreditation of Laboratory Animal Care International (AAALAC International) accredited facility and abides by the Institute of Laboratory Animal Resources (ILAR) guide.
References
- Bancroft JD, Layton C. The Hematoxylin and eosin. Theory & Practice of histological techniques 7th ed. Philadelphia: Churchill Livingstone of El Sevier, 2013:173-214.
- Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008;2008:pdb. prot4986.
- Shi SR, Liu C, Pootrakul L, Tang L, Young A, Chen R, Cote RJ, Taylor CR. Evaluation of the value of frozen tissue section used as “gold standard” for immunohistochemistry. Am J Clin Pathol 2008;129:358-66. [Crossref] [PubMed]
- Kim SH, Hubbard GB, McManus WF, Mason AD Jr, Pruitt BA Jr. Frozen section technique to evaluate early burn wound biopsy: a comparison with the rapid section technique. J Trauma 1985;25:1134-7. [Crossref] [PubMed]
- Chao C, Wong SL, Ackermann D, Simpson D, Carter MB, Brown CM, Edwards MJ, McMasters KM. Utility of intraoperative frozen section analysis of sentinel lymph nodes in breast cancer. Am J Surg 2001;182:609-15. [Crossref] [PubMed]
- Sato Y, Mukai K, Watanabe S, Goto M, Shimosato Y. The AMeX method. A simplified technique of tissue processing and paraffin embedding with improved preservation of antigens for immunostaining. Am J Pathol 1986;125:431-5. [PubMed]
- Sainte-Marie G. A paraffin embedding technique for studies employing immunofluorescence. Journal of Histochemistry & Cytochemistry 1962;10:250-6. [Crossref]
- Erickson QL, Clark T, Larson K, Minsue Chen T. Flash Freezing of Mohs Micrographic Surgery Tissue Can Minimize Freeze Artifact and Speed Slide Preparation. Dermatol Surg 2011;37:503-9. [Crossref] [PubMed]
- Fujimoto JG, Brezinski ME, Tearney GJ, Boppart SA, Bouma B, Hee MR, Southern JF, Swanson EA. Optical biopsy and imaging using optical coherence tomography. Nat Med 1995;1:970-2. [Crossref] [PubMed]
- Fujimoto JG, Pitris C, Boppart SA, Brezinski ME. Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia 2000;2:9-25. [Crossref] [PubMed]
- Tearney GJ, Brezinski ME, Southern JF, Bouma BE, Boppart SA, Fujimoto JG. Optical biopsy in human gastrointestinal tissue using optical coherence tomography. Am J Gastroenterol 1997;92:1800-4. [PubMed]
- van Veen RL, Amelink A, Menke-Pluymers M, van der Pol C, Sterenborg HJ. Optical biopsy of breast tissue using differential path-length spectroscopy. Phys Med Biol 2005;50:2573. [Crossref] [PubMed]
- Masters BR, So PT, Gratton E. Multiphoton Excitation Microscopy of In Vivo Human Skin: Functional and Morphological Optical Biopsy Based on Three‐Dimensional Imaging, Lifetime Measurements and Fluorescence Spectroscopy. Ann N Y Acad Sci 1998;838:58-67. [Crossref] [PubMed]
- Bigio IJ, Bown SG, Briggs G, Kelley C, Lakhani S, Pickard D, Ripley PM, Rose IG, Saunders C. Diagnosis of breast cancer using elastic-scattering spectroscopy: preliminary clinical results. J Biomed Opt 2000;5:221-8. [Crossref] [PubMed]
- Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Lyons J, Hicks D, Fitzmaurice M, Dasari RR, Crowe JP, Feld MS. In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res 2006;66:3317-22. [Crossref] [PubMed]
- Sun Q, He Y, Liu K, Fan S, Parrott EP, Pickwell-MacPherson E. Recent advances in terahertz technology for biomedical applications. Quant Imaging Med Surg 2017;7:345. [Crossref] [PubMed]
- Grootendorst MR, Fitzgerald AJ, de Koning SGB, Santaolalla A, Portieri A, Van Hemelrijck M, Young MR, Owen J, Cariati M, Pepper M. Use of a handheld terahertz pulsed imaging device to differentiate benign and malignant breast tissue. Biomed Opt Express 2017;8:2932-45. [Crossref] [PubMed]
- González S, Tannous Z. Real-time, in vivo confocal reflectance microscopy of basal cell carcinoma. J Am Acad Dermatol 2002;47:869-74. [Crossref] [PubMed]
- Yoshida S, Tanaka S, Hirata M, Mouri R, Kaneko I, Oka S, Yoshihara M, Chayama K. Optical biopsy of GI lesions by reflectance-type laser-scanning confocal microscopy. Gastrointest Endosc 2007;66:144-9. [Crossref] [PubMed]
- Masters B, So P. Confocal microscopy and multi-photon excitation microscopy of human skin in vivo. Opt Express 2001;8:2-10. [Crossref] [PubMed]
- Le Goualher G, Perchant A, Genet M, Cavé C, Viellerobe B, Berier F, Abrat B, Ayache N. Towards optical biopsies with an integrated fibered confocal fluorescence microscope. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2004 2004:761-8.
- Yoo H, Kang D, Katz AJ, Lauwers GY, Nishioka NS, Yagi Y, Tanpowpong P, Namati J, Bouma BE, Tearney GJ. Reflectance confocal microscopy for the diagnosis of eosinophilic esophagitis: a pilot study conducted on biopsy specimens. Gastrointest Endosc 2011;74:992-1000. [Crossref] [PubMed]
- Kang D, Suter MJ, Boudoux C, Yoo H, Yachimski PS, Puricelli WP, Nishioka NS, Mino-Kenudson M, Lauwers GY, Bouma BE. Comprehensive imaging of gastroesophageal biopsy samples by spectrally encoded confocal microscopy. Gastrointest Endosc 2010;71:35-43. [Crossref] [PubMed]
- Fereidouni F, Harmany ZT, Tian M, Todd A, Kintner JA, McPherson JD, Borowsky AD, Bishop J, Lechpammer M, Demos SG. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Eng 2017;1:957. [Crossref]
- Fereidouni F, Mitra AD, Demos S, Levenson R. editors. Microscopy with UV Surface Excitation (MUSE) for slide-free histology and pathology imaging. Optical Biopsy XIII: Toward Real-Time Spectroscopic Imaging and Diagnosis, 2015: International Society for Optics and Photonics.
- Levenson RM, Fereidouni F. MUSE: a new, fast, simple microscopy method for slide-free histology and surface topography. FASEB J 2016;30:51.3-.3.
- Glaser AK, Reder NP, Chen Y, McCarty EF, Yin C, Wei L, Wang Y, True LD, Liu JT. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng 2017;1:0084.
- Glaser AK, Wang Y, Liu JT. Assessing the imaging performance of light sheet microscopies in highly scattering tissues. Biomed Opt Express 2016;7:454-66. [Crossref] [PubMed]
- Migliori B, Datta MS, Dupre C, Apak MC, Asano S, Gao R, Boyden ES, Hermanson O, Yuste R, Tomer R. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol 2018;16:57. [Crossref] [PubMed]
- Yoshitake T, Giacomelli MG, Quintana LM, Vardeh H, Cahill LC, Faulkner-Jones BE, Connolly JL, Do D, Fujimoto JG. Rapid histopathological imaging of skin and breast cancer surgical specimens using immersion microscopy with ultraviolet surface excitation. Scientific reports 2018;8:4476. [Crossref] [PubMed]
- Mcgorty R, Xie D, Huang B. High-NA open-top selective-plane illumination microscopy for biological imaging. Opt Express 2017;25:17798-810. [Crossref] [PubMed]
- Bower AJ, Chidester B, Li J, Zhao Y, Marjanovic M, Chaney EJ, Do MN, Boppart SA. A quantitative framework for the analysis of multimodal optical microscopy images. Quant Imaging Med Surg 2017;7:24. [Crossref] [PubMed]
- Tao YK, Shen DJ, Sheikine Y, Ahsen OO, Wang HH, Schmolze DB, Johnson NB, Brooker JS, Cable AE, Connolly JL, Fujimoto JG. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A 2014;111:15304-9. [Crossref] [PubMed]
- Tauer U. Advantages and risks of multiphoton microscopy in physiology. Experimental physiology 2002;87:709-14. [Crossref] [PubMed]
- Ustione A, Piston D. A simple introduction to multiphoton microscopy. J Microsc 2011;243:221-6. [Crossref] [PubMed]
- Arora R, Petrov GI, Yakovlev VV. Analytical capabilities of coherent anti-Stokes Raman scattering microspectroscopy. J Mod Opt 2008;55:3237-54. [Crossref] [PubMed]
- Wurpel GW, Schins JM, Müller M. Chemical specificity in three-dimensional imaging with multiplex coherent anti-Stokes Raman scattering microscopy. Opt Lett 2002;27:1093-5. [Crossref] [PubMed]
- Carter M, Shieh J. Chapter 5 - Microscopy. In: Carter M, Shieh J. editors. Guide to Research Techniques in Neuroscience (Second Edition). San Diego: Academic Press, 2015:117-44.
- Bousso P, Robey EA. Dynamic Behavior of T Cells and Thymocytes in Lymphoid Organs as Revealed by Two-Photon Microscopy. Immunity 2004;21:349-55. [Crossref] [PubMed]
- Webb RH. Confocal optical microscopy. Rep Prog Phys 1996;59:427. [Crossref]
- Nie S, Chiu DT, Zare RN. Real-time detection of single molecules in solution by confocal fluorescence microscopy. Analytical Chemistry 1995;67:2849-57. [Crossref]
- Wilson T. Optical sectioning in confocal fluorescent microscopes. J Microsc 1989;154:143-56. [Crossref]
- Conchello JA, Lichtman JW. Optical sectioning microscopy. Nat Methods 2005;2:920. [Crossref] [PubMed]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004;305:1007-9. [Crossref] [PubMed]
- Bennàssar A, Vilata A, Puig S, Malvehy J. Ex vivo fluorescence confocal microscopy for fast evaluation of tumour margins during Mohs surgery. Br J Dermatol 2014;170:360-5. [Crossref] [PubMed]
- Gareau DS, Li Y, Huang B, Eastman Z, Nehal KS, Rajadhyaksha M. Confocal mosaicing microscopy in Mohs skin excisions: feasibility of rapid surgical pathology. J Biomed Opt 2008;13. [Crossref] [PubMed]
- Patel YG, Nehal KS, Aranda I, Li Y, Halpern AC, Rajadhyaksha M. Confocal reflectance mosaicing of basal cell carcinomas in Mohs surgical skin excisions. J Biomed Opt 2007;12. [Crossref] [PubMed]
- Abeytunge S, Li Y, Larson B, Toledo-Crow R, Rajadhyaksha M. Rapid confocal imaging of large areas of excised tissue with strip mosaicing. J Biomed Opt 2011;16. [Crossref] [PubMed]
- Abeytunge S, Li Y, Larson BA, Peterson G, Seltzer E, Toledo-Crow R, Rajadhyaksha M. Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue. J Biomed Opt 2013;18:61227. [Crossref] [PubMed]
- Im KB, Han S, Park H, Kim D, Kim BM. Simple high-speed confocal line-scanning microscope. Opt Express 2005;13:5151-6. [Crossref] [PubMed]
- Larson B, Abeytunge S, Seltzer E, Rajadhyaksha M, Nehal K. Detection of skin cancer margins in Mohs excisions with high‐speed strip mosaicing confocal microscopy: a feasibility study. Br J Dermatol 2013;169:922-6. [Crossref] [PubMed]
- Gareau DS, Jeon H, Nehal KS, Rajadhyaksha M. Rapid screening of cancer margins in tissue with multimodal confocal microscopy. J Surg Res 2012;178:533-8. [Crossref] [PubMed]
- Abeytunge S, Larson BA, Peterson G, Morrow M, Rajadhyaksha M, Murray MP. Evaluation of breast tissue with confocal strip-mosaicking microscopy: a test approach emulating pathology-like examination. J Biomed Opt 2017;22:34002. [Crossref] [PubMed]
- Yoon WB, Kim H, Kim KG, Choi Y, Chang HJ, Sohn DK. Methods of Hematoxylin and Erosin Image Information Acquisition and Optimization in Confocal Microscopy. Healthc Inform Res 2016;22:238-42. [Crossref] [PubMed]
- Giacomelli MG, Husvogt L, Vardeh H, Faulkner-Jones BE, Hornegger J, Connolly JL, Fujimoto JG. Virtual hematoxylin and eosin transillumination microscopy using epi-fluorescence imaging. PLoS One 2016;11. [Crossref] [PubMed]
- Qorbani A, Fereidouni F, Levenson R, Lahoubi SY, Harmany ZT, Todd A, Fung MA. Microscopy with ultraviolet surface excitation (MUSE): A novel approach to real-time inexpensive slide-free dermatopathology. J Cutan Pathol 2018;45:498-503. [Crossref] [PubMed]
- Turaga D, Holy TE. Image-based calibration of a deformable mirror in wide-field microscopy. Appl Opt 2010;49:2030-40. [Crossref] [PubMed]
- McGorty R, Liu H, Kamiyama D, Dong Z, Guo S, Huang B. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt Express 2015;23:16142-53. [Crossref] [PubMed]


