
Investigating accuracy of 3D printed liver models with computed tomography
Introduction
Visualization techniques in liver surgery have always been a crucial part of preoperative planning as hepatic resections remain to be challenging due to complex anatomy with many variations. Standard slice-by-slice analysis of computed tomography (CT), magnetic resonance imaging and positron emission tomography are important for all surgeons, but these methods have limitations which can be overcome with novel advancements in imaging, specifically computer-assisted three-dimensional (3D) image processing. Most commonly, 3D preoperative analysis and intraoperative guidance is performed with volume rendering techniques, however these do not provide sufficient image clarity of internal structures, especially for soft tissue structures which appear as similar gray-scale intensities (1). To address this limitation, 3D methods based on image segmentation and surface mesh generation, including 3D printing, virtual reality, and augmented reality, are becoming promising for pre-surgical planning as they allow the surgeon to better visualize pertinent surgical anatomy (2).
3D printing allows for easy and comprehensive assessment of 3D anatomical topography. There is a number of evidences on the feasibility of 3D printing for pre-surgical planning (3-8). In addition, the utility of 3D printing continuously grows across various surgical disciplines, making it not only a fad but also a legitimate part of treatment process that can impact perioperative outcomes including operative resection, blood loss, better decision making or intraoperative adverse events (9-11). Currently, 3D printing tends to be expensive, but potential use can become more widespread with emerging cost-effective 3D printing solutions.
Although there has been a recent review article on methods that can be utilized to verify the accuracy of 3D printed models (12), and some small studies have reported on the accuracy of 3D printed anatomical models (13,14), the field still is missing detailed analyses on accuracy of 3D printed models. This is particularly noticeable in regards to liver procedures that require complex, multi-material models, and there are no studies that would evaluate them thoroughly (15,16).
Studies verifying the accuracy of 3D printed anatomical models serve as a necessary bridge to link the feasibility or proof-of-concept of the studies increasingly available in the literature with the trials evaluating major clinical endpoints. The purpose of this study was to evaluate the accuracy of 3D printed liver models developed with a cost-effective approach in order to establish validity of using these models in a clinical setting.
Methods
Fifteen patients undergoing laparoscopic liver resection in a single surgical department were included in this study, which was approved by local bioethics committee at Jagiellonian University Medical College. 3D printed personalized liver models that include the liver, tumor, and associated vasculature were created using a previously described unique cost-effective approach (16). The 3D printed liver models were developed prospectively, based on 15 CT examinations performed in agreement with hospital protocols, prior to the surgical procedure.
Image acquisition
All CT imaging was performed using a GE Optima CT660 scanner (GE Medical Systems, Milwaukee, WI, USA) and protocol parameters of 120 kV, table feed per rotation about 39.375 mm, total collimation width 40 mm.
Patient imaging
All patients underwent contrast-enhanced abdominal CT (PCT) images in the portal venous phases with slice thickness ranging from 1.25 to 2.5 mm.
Imaging of 3D printed models
3D printed models were CT scanned (MCT) with parameters described above and slice thickness of 1.25 mm (Figures 1,2).
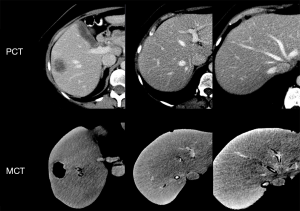
Image segmentation and registration
Both PCT and MCT images were segmented in a 3D Slicer (version 4.8.1) open-source software in order to generate digital mesh-based models of liver anatomy including the liver parenchyma, tumors, portal and hepatic veins (17). While the PCT segmentation is necessary for 3D printing, image segmentation was also performed on MCT for further comparisons of mesh accuracy. Semi-automatic algorithms including thresholding and seed growing were used and regions of interests were created to refine manually by Jan Witowski with Anna Grochowska supervision (radiologist with 15 years of experience in liver imaging) (Figure 3).
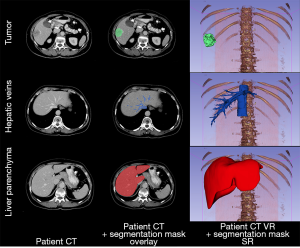
Surface rendering algorithms were used to create 3D triangular meshes from segmentations regions of interests. These meshes could be compared between each other after alignment. Point set registration of liver parenchyma was performed in Meshlab (v.2016.12), an open-source mesh processing tool (18).
Landmark measurements
In order to measure the accuracy of models, we conducted 2D landmark measurements on both axial MCT and PCT images (Figure 4):
- PVD: portal vein diameter measured 1 cm before bifurcation;
- RHD: right hepatic vein diameter measured 1 cm from inferior vena cava (IVC) edge;
- MHD: middle hepatic vein diameter measured 1 cm from IVC edge;
- LHD: left hepatic vein diameter measured 1 cm from IVC edge;
- TD: distance from tumor center (at its widest diameter) to IVC center, on same slice;
- EDGE: shortest distance from tumor center (at its widest diameter) to liver edge. This measurement is not performed if tumor emerges on the surface of liver.
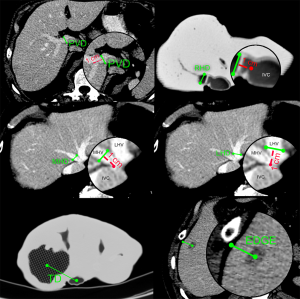
Comparing volumes and surfaces
We performed volumetric analysis of liver parenchyma and tumor segmentation masks to assess their volumes and computed relative errors between PCT and MCT. In addition, to evaluate the quality of the 3D printed models surface-wise, we used the Hausdorff distance to compute errors between meshes of PCT and MCT rendered surfaces. Meshes were aligned through point registration, as described above. Heat maps of distances were generated for visual inspection to help better understand which areas of printed models were most problematic and inaccurate.
Statistical analysis
Bland-Altman plots with 95% confidence intervals and 1.96 standard deviation limits of agreement and Pearson’s correlation coefficient were used to report mean difference between patient CT and model CT in terms of 3D (liver and tumor volume) and 2D measurements. P<0.05 was considered to denote statistical significance. For statistical analyses we used R 3.5.0 software.
Results
Volumes and surfaces
Liver volumes [median (interquartile range)] were 1,281.84 (296.86) cm3 for MCT and 1,448.03 (413.23) cm3 for PCT (Table 1). Although there was a trend towards lower volumes in MCT, after analysis of Bland-Altman plots the bias was not statistically significant (Figure 5). There are no established maximum acceptable differences in terms of these measurements, but we believe that our results (95% CI of mean differences in liver volumes: −273.80 to 7.97 cm3) would not cause problems in clinical use. In 3D printed models where silicone casting was successful, relative error in parenchyma volume does not exceed 10% and mean Hausdorff distance for liver surface is under 5 mm. We investigated two cases where those errors were significant. Case 8 had significant relative error of −40.1% due to problems with silicone leakage during casting, similar issues were encountered with case 1. Those results are confirmed with surface analysis that revealed larger Hausdorff distance values in those models, and heatmaps show missing areas (Figure 6).
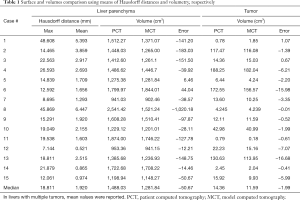
Full table
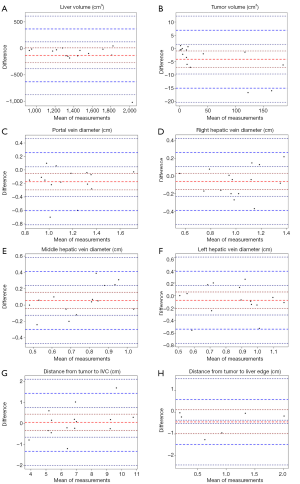

Tumor volume measurements and Bland-Altman plot analysis revealed statistically significant undersizing of lesions in MCT (Figure 5B; 95% CI of mean differences: −7.14 to −0.95 cm3), however its clinical significance can be a subject of discussion.
Accuracy of internal anatomy
All length and diameter measurements are presented in Table 2. Median errors between PCT and MCT were: −0.15, −0.04, 0.05, −0.07, 0.09 and −0.2 cm for PVD, RHD, MHD, LHD, TD and EDGE measurements, respectively. Nine models had lesions protruding onto the liver surface, which made it impossible to calculate EDGE values.
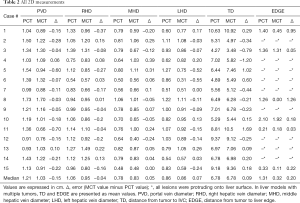
Full table
Bland-Altman plots revealed there was no statistically significant bias in all 2D measurements (Figure 5C,D,E,F,G,H) with the exception of PVD, where the bias was −0.18 (95% CI: −0.30 to −0.05) cm. Taking into consideration slice height of 0.125 to 0.25 cm, this error doesn’t show flaws in methodology, but rather bias due to the source imaging.
Median [median (interquartile range)] errors for 2D measurements between PCT and MCT were: −0.15 (0.17) cm for PVD, −0.04 (0.25) cm for RHD, 0.05 (0.22) cm for MHD, 0.07 (0.225) cm for LHD, 0.09 (0.54) cm for TD and 0.2 (0.685) cm for EDGE.
Discussion
In this study, we evaluated the accuracy of 15 3D printed liver models created with a novel, cost-effective 3D printing technique by performing a set of 2D and 3D measurements on CT images, which confirm the precision of discussed approach.
Currently, there are approximately 20 publications that describe the use of 3D printing in planning or navigating liver surgery, as described in review studies (15,19). However, only few of them attempted to assess quantitative accuracy of the models and done it with very limited number of cases (20,21). In the very first publication in the field, Zein et al. in a simple way assessed accuracy of three 3D printed liver models by comparing liver volumes by means of liquid displacement and compared to patients’ CT volumetry and intraoperatively diameters of portal vein, right and left hepatic veins (20). Their mean error for vascular diameters was less than 1.3 mm. Zein et al. also measured entire models’ diameters, revealing mean error of less than 4 mm. In our study, median error was less than 1 mm for all hepatic veins and −1.5 mm for portal vein diameter.
Additionally, we believe that Zein et al. approach to assessing accuracy is not ideal and thus we decided to utilize modern, highly suitable methods including measuring landmarks and calculating Hausdorff distances. We also overcame the limitation of small number of subjects by analyzing fifteen 3D models, making it one of the largest studies in the field. Bücking et al., printing a single liver model, revealed 1.3% mean percentage error for total height, width and depth, but—as already mentioned—we find this methodology not acceptable (21). Perica et al. measured five anatomical landmarks for a single model and compared them with original CT data (22). Authors of this study also agreed that models are accurate, but—along with other papers—had multiple limitations, including: small number of measurements, few subjects, scaled liver models.
Although our study shows that the analyzed 3D printing technique is by definition accurate, it requires good execution as it is not only time consuming but has many problematic spots, which may be its major limitation to be implemented in different institutions. The whole process can take several days and requires extensive postprocessing, including silicone casting, which is the lengthiest and most problematic stage. In our series, this caused most problems in developing 3D models, which resulted in major differences of some of the models, such as absolute difference of over 1,000 cm3 in liver parenchyma volume (case number 8) and model shape.
There is no doubt that the most important part of the 3D models, the 3D anatomy of internal structures and distances between vessels and tumors, are accurate. It may be considered that even printing those structures themselves, without liver parenchyma, could be clinically sufficient.
We do not believe that the above issues should restrain this technique from being utilized clinically. It is important to keep in mind that our approach significantly reduces costs from even $4,000 and extremely expensive hardware—such as $200,000 PolyJet printers—to as little as $100 per model (16,20,23,24). Results from our study are also not only applicable to our method exclusively, since 3D printers themselves have been proven to create models of micron-scale accuracy (25-27). When high-grade printers with proven accuracy (generally higher than our imaging resolution) are available to an institution, in our view, it is safe to assume that 3D printed models will be accurate, granted they are prepared with proper image segmentation and pre-processing.
As a whole, the medical 3D printing field is moving towards stage of long-term studies, we should consider this study conclusive in terms of 3D printed liver models’ accuracy. However, each institution should validate their own 3D printing methods, including acquisition, segmentation, post-processing and printing, not to prove that 3D printing is feasible and accurate in general, but rather to make sure their own workflow is flawless. After years of feasibility, pilot, and proof-of-concept studies it is time for clinical trials. According to IDEAL framework (framework describing the stages of innovation in surgery), we are leaving development and exploration phases and starting the assessment and long-term studies (28,29). It can be expected that by 2020–2022 a number of studies will discuss clinical utility and impact on perioperative outcomes (30).
There are several limitations that need to be mentioned. Firstly, there is no standardized approach to verify accuracy of 3D printed models. We have chosen in our opinion most reasonable tools and statistical methods to do an extensive analysis; however, those are not all available techniques. Secondly, there is no standardized acceptable measure of error, especially taking into consideration the clinical usage. Bland-Altman plots in our study showed that nearly all measurements were without significant bias, but even that doesn’t say much for clinicians. In our opinion, we should always refer to source imaging and clinical situation. We also consider the type of imaging and slice height as two main limitations of models’ accuracy, assuming the workflow and hardware is faultless. In our study we only analyzed models developed based on CT scans acquired on a single CT scanner. Clinical MRI scans usually have inferior spatial resolution which needs to be taken into consideration.
Conclusions
Our cost-effective method of developing patient-specific 3D printed liver models is accurate, allowing to use these models routinely in a clinical setting for planning purposes. This approach has some technical limitations that can be overcome with the development of hardware, especially difficult and time-consuming post-processing, which have led to discrepancies in volume of 3D printed liver models. Institutions utilizing novel, different techniques should validate them before clinical use. However, future studies should not focus on 3D printed models’ accuracy, but rather on the impact that these models can make on clinical care.
Acknowledgements
Funding: This research has been a part of a research project co-funded by the Ministry of Science and Higher Education in Poland and its “Diamentowy Grant” programme (grant number 0054/DIA/2018/47).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by Jagiellonian University Medical College Committee of Bioethics (approval ID: 122.6120.81.2017).
References
- Okamoto T, Onda S, Yanaga K, Suzuki N, Hattori A. Clinical application of navigation surgery using augmented reality in the abdominal field. Surg Today 2015;45:397-406. [Crossref] [PubMed]
- Soler L, Nicolau S, Pessaux P, Mutter D, Marescaux J. Real-time 3D image reconstruction guidance in liver resection surgery. Hepatobiliary Surg Nutr 2014;3:73-81. [PubMed]
- El Sabbagh A, Eleid MF, Al-Hijji M, Anavekar NS, Holmes DR, Nkomo VT, Oderich GS, Cassivi SD, Said SM, Rihal CS, Matsumoto JM, Foley TA. The Various Applications of 3D Printing in Cardiovascular Diseases. Curr Cardiol Rep 2018;20:47. [Crossref] [PubMed]
- Martelli N, Serrano C, van den Brink H, Pineau J, Prognon P, Borget I, El Batti S. Advantages and disadvantages of 3-dimensional printing in surgery: A systematic review. Surgery 2016;159:1485-500. [Crossref] [PubMed]
- Weinstock P, Rehder R, Prabhu SP, Forbes PW, Roussin CJ, Cohen AR. Creation of a novel simulator for minimally invasive neurosurgery: fusion of 3D printing and special effects. J Neurosurg Pediatr 2017;20:1-9. [Crossref] [PubMed]
- Kamali P, Dean D, Skoracki R, Koolen PGL, Paul MA, Ibrahim AM, Lin SJ. The Current Role of Three-Dimensional Printing in Plastic Surgery. Plast Reconstr Surg 2016;137:1045-55. [Crossref] [PubMed]
- Chao I, Young J, Coles-Black J, Chuen J, Weinberg L, Rachbuch C. The application of three-dimensional printing technology in anaesthesia: a systematic review. Anaesthesia 2017;72:641-50. [Crossref] [PubMed]
- Sitkowski M, Witowski JS, Zuzak T, Wdowiak-Filip A, Filip M, Urbańczyk-Zawadzka M, Kapelak B, Litwinowicz R. Low-cost 3D-printed grown-up congenital heart defect models from CT angiography. Eur J Med Tech 2017;4:56-61.
- King BJ, Park EP, Christensen BJ, Danrad R. On-Site 3-Dimensional Printing and Preoperative Adaptation Decrease Operative Time for Mandibular Fracture Repair. J Oral Maxillofac Surg 2018;76:1950.e1-8. [Crossref] [PubMed]
- Zhang YD, Wu RY, Xie DD, Zhang L, He Y, Zhang H. Effect of 3D printing technology on pelvic fractures: a Meta-analysis. Zhongguo Gu Shang 2018;31:465-71. [PubMed]
- Liu P, Liu R, Zhang Y, Liu Y, Tang X, Cheng Y. The Value of 3D Printing Models of Left Atrial Appendage Using Real-Time 3D Transesophageal Echocardiographic Data in Left Atrial Appendage Occlusion: Applications toward an Era of Truly Personalized Medicine. Cardiology 2016;135:255-61. [Crossref] [PubMed]
- George E, Liacouras P, Rybicki FJ, Mitsouras D. Measuring and Establishing the Accuracy and Reproducibility of 3D Printed Medical Models. Radiographics 2017;37:1424-50. [Crossref] [PubMed]
- Wake N, Rude T, Kang SK, Stifelman MD, Borin JF, Sodickson DK, Huang WC, Chandarana H. 3D printed renal cancer models derived from MRI data: application in pre-surgical planning. Abdom Radiol (NY) 2017.1501-9. [Crossref] [PubMed]
- Ripley B, Kelil T, Cheezum MK, Goncalves A, Di Carli MF, Rybicki FJ, Steigner M, Mitsouras D, Blankstein R. 3D printing based on cardiac CT assists anatomic visualization prior to transcatheter aortic valve replacement. J Cardiovasc Comput Tomogr 2016;10:28-36. [Crossref] [PubMed]
- Witowski JS, Coles-Black J, Zuzak TZ, Pędziwiatr M, Chuen J, Major P, Budzyński A. 3D Printing in Liver Surgery: A Systematic Review. Telemed J E Health 2017;23:943-7. [Crossref] [PubMed]
- Witowski JS, Pędziwiatr M, Major P, Budzyński A. Cost-effective, personalized, 3D-printed liver model for preoperative planning before laparoscopic liver hemihepatectomy for colorectal cancer metastases. Int J Comput Assist Radiol Surg 2017;12:2047-54. [Crossref] [PubMed]
- Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support BT - Intraoperative Imaging and Image-Guided Therapy. Jolesz FA. editor. New York: Springer New York; 2014:277-89.
- Cignoni P, Callieri M, Corsini M, Dellepiane M, Ganovelli F, Ranzuglia G. Meshlab: an open-source mesh processing tool. Scarano V, De Chiara R, Erra U. editors. London: The Eurographics Association; 2008:129-36.
- Perica ER, Sun Z. A Systematic Review of Three-Dimensional Printing in Liver Disease. J Digit Imaging 2018;31:692-701. [Crossref] [PubMed]
- Zein NN, Hanouneh IA, Bishop PD, Samaan M, Eghtesad B, Quintini C, Miller C, Yerian L, Klatte R. Three-dimensional print of a liver for preoperative planning in living donor liver transplantation. Liver Transpl 2013;19:1304-10. [Crossref] [PubMed]
- Bücking TM, Hill ER, Robertson JL, Maneas E, Plumb AA, Nikitichev DI. From medical imaging data to 3D printed anatomical models. PLoS One 2017;12:e0178540. [Crossref] [PubMed]
- Perica E, Sun Z. Patient-specific three-dimensional printing for pre-surgical planning in hepatocellular carcinoma treatment. Quant Imaging Med Surg 2017;7:668-77. [Crossref] [PubMed]
- Quintini C, Aucejo F, Hashimoto K, Zein N, Miller C. State of the Art and Future Developments for Surgical Planning in LDLT. Curr Transpl Rep 2014;1:35-42. [Crossref]
- Shallan AI, Smejkal P, Corban M, Guijt RM, Breadmore MC. Cost-effective three-dimensional printing of visibly transparent microchips within minutes. Anal Chem 2014;86:3124-30. [Crossref] [PubMed]
- Brouwers L, Teutelink A, van Tilborg FA, de Jongh MA, Lansink KW, Bemelman M. Validation study of 3D-printed anatomical models using 2 PLA printers for preoperative planning in trauma surgery, a human cadaver study. Eur J Trauma Emerg Surg 2018. Epub ahead of print. [Crossref] [PubMed]
- Ionita CN, Mokin M, Varble N, Bednarek DR, Xiang J, Snyder K V, Siddiqui AH, Levy EI, Meng H, Rudin S. Challenges and limitations of patient-specific vascular phantom fabrication using 3D Polyjet printing. Proc SPIE Int Soc Opt Eng 2014;9038:90380M.
- Ibrahim D, Broilo TL, Heitz C, de Oliveira MG, de Oliveira HW, Nobre SM, dos Santos Filho JH, Silva DN. Dimensional error of selective laser sintering, three-dimensional printing and PolyJetTM models in the reproduction of mandibular anatomy. J Craniomaxillofac Surg 2009;37:167-73. [Crossref] [PubMed]
- McCulloch P, Altman DG, Campbell WB, Flum DR, Glasziou P, Marshall JC, Nicholl J, Aronson JK, Barkun JS, Blazeby JM, Boutron IC, Campbell WB, Clavien P-A, Cook JA, Ergina PL, Feldman LS, Flum DR, Maddern GJ, Nicholl J, Reeves BC, Seiler CM, Strasberg SM, Meakins JL, Ashby D, Black N, Bunker J, Burton M, Campbell M, Chalkidou K, Chalmers I, de Leval M, Deeks J, Ergina PL, Grant A, Gray M, Greenhalgh R, Jenicek M, Kehoe S, Lilford R, Littlejohns P, Loke Y, Madhock R, McPherson K, Meakins J, Rothwell P, Summerskill B, Taggart D, Tekkis P, Thompson M, Treasure T, Trohler U, Vandenbroucke J. No surgical innovation without evaluation: the IDEAL recommendations. Lancet 2009;374:1105-12. [Crossref] [PubMed]
- Lemke HU. Editorial for innovative clinical investigations: example of a framework for determining the stages of innovation in surgery or other interventional procedures. Int J Comput Assist Radiol Surg 2017;12:2033-7. [Crossref] [PubMed]
- Witowski J, Sitkowski M, Zuzak T, Coles-Black J, Chuen J, Major P, Pędziwiatr M. From ideas to long-term studies: 3D printing clinical trials review. Int J Comput Assist Radiol Surg 2018;13:1473-8. [Crossref] [PubMed]


